Significance
The substrate binding and specificity are essential features for the amino acid–polyamine–organocation superfamily members. Here, we report two crystal structures of an alanine transporter, alanine or glycine:cation symporter, capable of transporting both L- and D-alanine and discovered that both the size and polarity of the substrate binding pocket are important structural determinants for specificity and stereo-selectivity. The size of the binding pocket determines what amino acids could fit in the transporter, while electrostatics control whether L-type or D-type amino acids could bind. Mutations in the binding site can broaden the specificity of this transporter to allow uptake of larger amino acids, such as valine and leucine.
Keywords: secondary transporter, substrate binding, specificity, membrane protein, crystal structure
Abstract
The amino acid, polyamine, and organocation (APC) superfamily is the second largest superfamily of membrane proteins forming secondary transporters that move a range of organic molecules across the cell membrane. Each transporter in the APC superfamily is specific for a unique subset of substrates, even if they possess a similar structural fold. The mechanism of substrate selectivity remains, by and large, elusive. Here, we report two crystal structures of an APC member from Methanococcus maripaludis, the alanine or glycine:cation symporter (AgcS), with l- or d-alanine bound. Structural analysis combined with site-directed mutagenesis and functional studies inform on substrate binding, specificity, and modulation of the AgcS family and reveal key structural features that allow this transporter to accommodate glycine and alanine while excluding all other amino acids. Mutation of key residues in the substrate binding site expand the selectivity to include valine and leucine. These studies provide initial insights into substrate selectivity in AgcS symporters.
Controlling nutrient balance in cells and by extension entire organisms is critical. Bacteria and archaea evolved sophisticated mechanisms to adapt to life in the most extreme environments. One such example is Methanococcus maripaludis that exists in salt-marsh plains (1) and has the ability to use both l- and d-alanine as an important nitrogen source to support life in high salt and anaerobic conditions (2–4). As a transporter that uptakes both enantiomers of alanine, the alanine or glycine:cation symporter (AgcS) from M. maripaludis plays a crucial role in this process (3). Furthermore, though l-amino acids are the dominant substrate in all kingdoms of life, it is now clear that the uptake system of d-amino acids also fulfills essential functions in many organisms (5, 6). Several recent studies show that the uptake system of d-amino acids in bacteria and archaea are crucial for stationary-phase cell wall remodeling (5), host metabolism, and virulence (6) although the mechanism by which such amino acids are transported into cells is poorly understood.
The amino acid–polyamine–organocation (APC) superfamily [Transporter Classification DataBase (TCDB)] (7) represents the second largest family of secondary carriers currently known (8–11) and plays essential roles in a wide spectrum of physiological processes, transporting a large range of substrates across the membrane. Members of this superfamily have since been identified and expanded to 18 different families in diverse organisms (12). Recent structures have shown that APC superfamily members contain 10–14 transmembrane helices and exhibit sufficiently similar folds characterized by a five or seven transmembrane helix inverted-topology repeat motif (13). However, some of these proteins have exceptionally broad selectivity, others are restricted to just one or a few amino acids or related derivatives. It is of much interest to investigate the structural basis for substrate binding and specificity of the APC superfamily.
The AgcS family (TC# 2.A.25) belongs to the APC superfamily and shows limited sequence similarities with other members. In contrast to ApcT and LeuT of which substrate specificity are broader (14, 15), members of the AgcS family have been reported to act as symporters, transporting l- or d-alanine or glycine with Na+ and/or H+ but no other amino acids (3, 16–20). Here, we present the crystal structures of AgcS from M. maripaludis in complex with l- or d-alanine as a model protein for members in the AgcS family. The two structures of AgcS were captured in a fully occluded conformation with the transmembrane architecture like other APC superfamily members. Functional assays demonstrated that purified AgcS binds only glycine and both enantiomers of alanine, while strictly excluding other amino acids that were assayed. Further structural analyses combined with mutagenesis and biochemical studies suggest that the residues at the intracellular face of the binding pocket play a key role in substrate binding and specificity. Moreover, structural comparisons of AgcS with LeuT, which is selective for l-amino acids, also pave the way for a better understanding of stereo-selectivity adopted by the APC superfamily transporters.
Results
Overall Architecture of AgcS.
A M. maripaludis AgcS was expressed in Escherichia coli and purified in various detergents for crystallization. Although successfully crystallized, AgcS alone produced only anisotropically diffracting crystals. The diffraction quality was poor and could not be improved despite major effort. Fab fragments were then generated and cocrystallized with AgcS to improve crystal contacts (SI Appendix, Figs. S1 and S2A). Crystals of AgcS in a complex with Fab fragment diffracted to ∼3.2 Å. The phases obtained by a combination of molecular replacement, using a generic Fab structure, and experimental phasing from Se-Met labeled AgcS were of high quality, allowing us to unambiguously discern two AgcS molecules and two Fab fragments in one asymmetric unit (SI Appendix, Fig. S2B). In total, two structures of AgcS, one with l-alanine bound and the other with d-alanine bound, were determined (SI Appendix, Table S1).
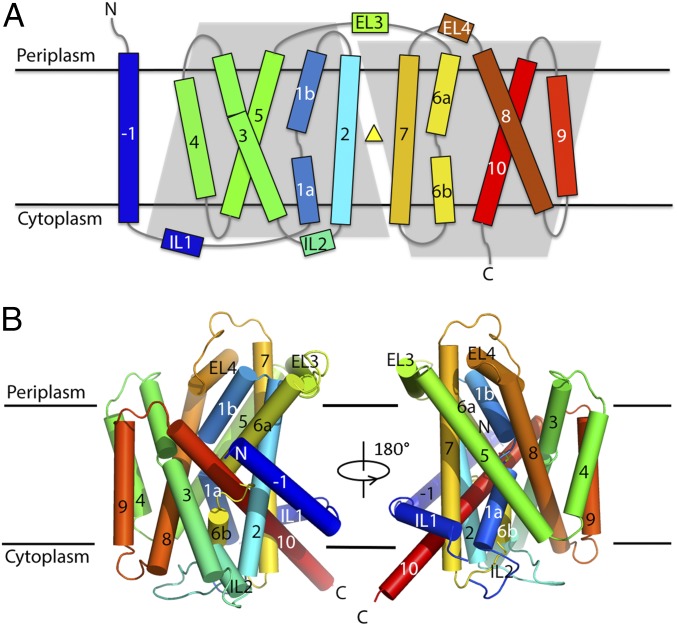
AgcS forms a roughly cylindrical shape, which is ∼40 Å tall and ∼35 Å in diameter comprising 11 transmembrane (TM) helices connected by short cytoplasmic or periplasmic loops (Fig. 1 and SI Appendix, Figs. S3A and S4). Its N terminus is at the periplasm while its C terminus is cytoplasmic. While the N-terminal helix -1 resides outside of the transmembrane bundle and does not seem to participate in translocation of substrates, AgcS appears to maintain pseudo twofold symmetry even though it contains an uneven number of TMs (Fig. 1A and SI Appendix, Fig. S3B). One half contains TMs 1, 2, 5, 6, and 7 while the second half contains TMs 3, 4, 8, 9, and 10. The TMs vary greatly in their length and angle with respect to the plane of the membrane: some helices such as TM2 and TM8 appear almost perpendicular to the membrane plane while TM10 is heavily tilted. TMs 2, 4, 5, 7, 9, and 10 surround the functional core of AgcS, which is made of TMs 1, 3, 6, and 8. Despite the lack of sequence identity (15.1% to LeuT, 14.3% to ApcT, and 14.8% to AdiC), the overall fold of AgcS is similar to that of LeuT (15, 21–23) from the NSS (neurotransmitter:sodium symporter) family. Superposing the crystal structures of AgcS and LeuT gives an overall root mean square deviation (r.m.s.d.) of 4.5 Å. The present structure of AgcS mirrors a common model of LeuT fold Na+ and/or H+-coupled symporters (24, 25), including ApcT (14) and AdiC (26–28) of the APC family, BetP (29) of BCCT (betaine/carnitine/choline transporter) family, vSGLT (30, 31) of the SSS (solute:sodium symporter) family, and Mhp1 (32) of NCS1 (nucleobase:cation symporter-1) family.
Fig. 1.
Overall structure of AgcS. (A) The topology of AgcS. The position of the bound alanine is depicted as a yellow triangle. (B) View in the plane of the membrane. Coloring and numbering of helices (cylinders) are the same as in A.
Notably, as with other LeuT-like symporters, two of the functional core helices in AgcS are kinked (Fig. 2, Inset). TMs 1 and 6 line the substrate translocation pathway in AgcS and are broken into two segments. In TM1, Ile-76 and Gly-77 adopt an extended, nonhelical conformation, linking segments 1a and 1b. TM6 contains a longer nonhelical region from Glu-276 to Ser-281 connecting segments 6a and 6b. The residues at these nonhelical loops expose carbonyl and amide groups at the center of the membrane bilayer for hydrogen bonding and ion coordination, which are involved in substrate binding and transport.
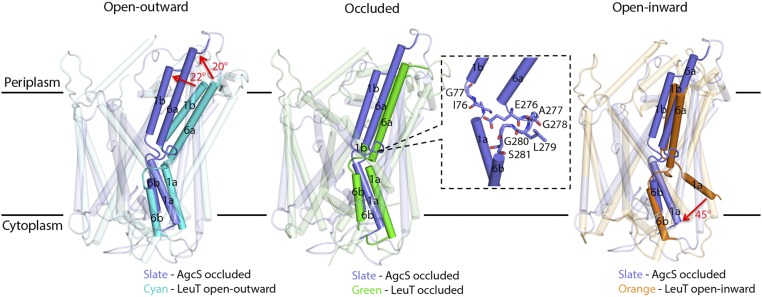
Fig. 2.
AgcS in the occluded state. AgcS structure was compared with LeuT open-outward state (Left, Protein Data Bank [PDB]: 3tt1, cyan), LeuT occluded state (Center, PDB: 2qei, green), and LeuT open-inward state (Right, PDB: 3tt3, orange). Key structural changes are marked with red arrows. A close-up view of the unwound regions splitting TMs 1 and 6 highlighting residues that are involved in substrate binding is shown in the Inset.
AgcS Structure in an Occluded State.
It is generally accepted that secondary transporters function through an alternate access model (33). The LeuT-fold transporters naturally alternate among three major conformations during their functional cycle: outward-open, occluded, and inward-open state. In the outward-open state, a hydrophilic tunnel emerges and connects the substrate binding site located in the center of transmembrane helical bundle with the periplasm. When in the inward-open state, a hydrophilic tunnel leads from the substrate at the central binding site to the cytosol. During the occluded state, the substrate binding site is isolated from either the periplasm or cytoplasm. Within a complete switch cycle, substrates can be transported across the lipid membrane as the transporter alternates from one conformation to the others.
Two structures of AgcS bound with l-alanine or d-alanine were solved in this study and appeared to be both at the occluded state (Fig. 2). No solvent accessible tunnels or vestibules were apparent leading from the substrate binding pocket across the membrane portion of the transporter. Previous studies reported the structures of LeuT in the open-outward, occluded, and open-inward states (22, 23). APC superfamily transporters such as AgcS and LeuT appear to gate through movements of the functional core helices. As discussed above, these helices are broken (Fig. 2, Inset) to allow free movement of the top half of the helices with respect to the bottom half and this in turn opens the binding pocket either to the outside or the inside of the cell. The pivot point appears to be at the substrate binding pocket. In the overlay of AgcS with the open-outward LeuT (Fig. 2, Left), or the overlay of AgcS with the open-inward LeuT (Fig. 2, Right), core helices deviate by more than 20° or 45°, respectively. No such deviation was observed when the occluded LeuT was overlaid with AgcS (Fig. 2, Center). This analysis clearly suggests that AgcS was captured in the occluded state.
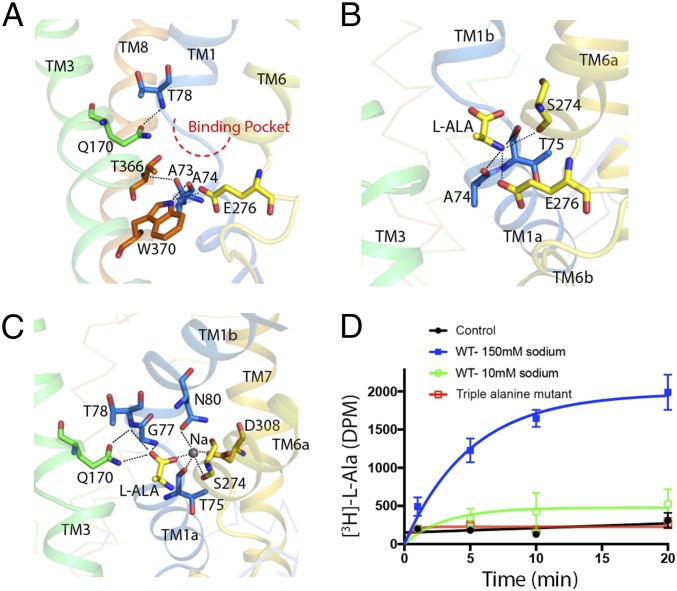
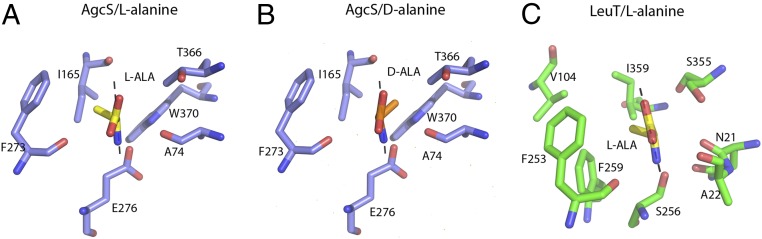
A system of intricate amino acid interactions forms the extracellular and cytoplasmic gates of AgcS. The extracellular gate of AgcS consists of residues from TM1, TM3, and TM8 (Fig. 3A). Specifically, Thr-78 and Ala-73, on the two ends of the unwound region in TM1, expose their Cα nitrogen and carbonyl oxygen atoms for hydrogen bonding with Gln-170 (TM3) and Thr-366 (TM8). On the cytoplasmic side of the binding pocket, the main chain carbonyl oxygen atoms of Ala-74 (TM1) and one of the carboxylate oxygen atoms of Glu-276 (TM6) accept an additional hydrogen bond from the nitrogen atom of indole ring of Trp-370 (TM8), forming a zipper-like motif and adopting the occluded state.
Fig. 3.
Gating interactions and substrate binding site in AgcS. (A) A close-up view of gating interaction network around the binding pocket in AgcS with l-alanine bound. (B) Hydrogen bonds between amino group of the substrate and the residues of AgcS. (C) Hydrogen bonds and ionic interactions in the hydrophilic part of binding pocket are depicted as dashed lines. Bound Na+ ion shown as a gray sphere. (D) The uptake ability for missense mutant (triple alanine mutation at N80A, S274A, and D308A) compared with that of the WT protein.
AgcS Substrate Binding and Ion Modulation.
AgcS binds glycine and both enantiomers of alanine, while strictly excluding other amino acids that were assayed and its transport activity is sodium coupled (SI Appendix, Figs. S5 and S6). In the current study, both structures of AgcS with alanine bound reveal a small amphipathic substrate binding cavity lined by residues from TMs 2, 4, 7, and 8. These residues define two distinct regions in the binding pocket: a hydrophilic charged region close to the extracellular gate and a hydrophobic region embedded deeply in the pocket toward the cytoplasmic gate. A spherical density that could fit the substrate alanine was observed in the composite omit maps at the center of this putative binding pocket (SI Appendix, Figs. S7–S9). Because the resolution of the structures was relatively modest, we relied on a superposition of AgcS with LeuT for placement of the substrate into the binding pocket of AgcS and for confirming the position of the bound Na+ ion. A similar approach was used for d-alanine.
The hydrophilic region of the binding pocket interacts with both the amino and carboxyl groups of the bound alanine as well as a bound sodium ion. The amino group of the bound alanine is coordinated by the main-chain carbonyl oxygens of Ala-74, Thr-75 in TM1, and Ser-274 in TM6, and a side-chain carboxyl group from Glu-276 in TM6 (Fig. 3B). Notably, Glu-276 is an ionizable residue whose protonation state may affect substrate binding or transport although this was not investigated in this study. On the opposite side, the exposed main-chain of Gly-77 and Thr-78, which are in the unwound region of TM1, make contacts with the carboxyl group of alanine by direct hydrogen bonds, along with side-chain amide of Gln-170 (TM3), which is also fully conserved (SI Appendix, Fig. S4). This glutamine may also stabilize the irregular structure around the unwound region in TM1 by interaction between its side-chain carbonyl oxygen to the main-chain amino group of Thr-78 (Fig. 3C).
The carboxylic group of the bound alanine is further coordinated to Asn-80 (TM1), Ser-274 (TM6), and Asp-308 (TM7) through the salt-bridge interactions mediated by a single sodium ion (Fig. 3C). These residues are highly conserved in the AgcS family (SI Appendix, Fig. S4). The coordination of the sodium ion is provided by the side-chain carbonyl oxygens of Asn-80 and Asp-308 and the hydroxyl oxygen of Ser-274, further stabilized by main-chain carbonyl oxygen of Ser-274 and Thr-75 (Fig. 3C). The proposed Na+ binding site is a conserved ion-binding site of the LeuT fold (21). It provides the correct geometry and chemistry for binding a Na+ ion. Uptake studies confirmed that the transport of alanine is Na+ coupled (SI Appendix, Fig. S6). To characterize the role of the conserved residues in sodium ion coordination, we generated a triple mutant [triple alanine mutations (3A) at N80A, S274A, and D308A] and evaluated the impact on substrate uptake using proteoliposomes. The mutant markedly reduced the uptake ability of l-alanine, suggesting that the sodium ion plays a key role in facilitating substrate binding and uptake (Fig. 3D).
Substrate Selectivity in AgcS.
The presence of the substrates in AgcS is supported by electron density in the 2mFo-DFc (SI Appendix, Figs. S7–S9) maps as well as the simulated annealing omit maps for both l- and d-alanine (SI Appendix, Fig. S9), but their absolute location and orientation cannot be established due to the limited resolution of the data. The relative positions of the l- and d-substrate also turn out to be difficult to determine: even though the unit cells for the l- and d-alanine structures and the coordinates of the atoms in the corresponding protein molecules are almost identical (SI Appendix, Table S1; all-atom r.m.s.d. 0.18 Å), the AgcS-Fab complexes are at slightly different positions in their respective unit cells (relative translation 0.49 Å). This precludes using an Fo-Fo map to elucidate the differences between the location and orientation of the l- and the d-alanine molecules. There are no significant peaks in the mFo-DFc map for l-alanine, but a pair of opposing positive and negative peaks in the Fo-Fc difference density maps (SI Appendix, Fig. S9F) is revealed after rigid-body refinement of the d-alanine model against the l-alanine data to account for the translation. This indicates a slight shift of the l-alanine toward S274 compared with d-alanine.
AgcS has a high selectivity for small amino acids like glycine and alanine with a dependency on sodium ion. We first evaluated AgcS binding affinities to several amino acids using isothermal titration calorimetry (ITC). While heat exchange was identified for the binding of glycine, and both l- and d-alanine, other amino acids that were tested did not elicit a response (SI Appendix, Fig. S5). Uptake studies using reconstituted proteoliposomes likewise indicated impeccable specificity in AgcS for glycine, l-alanine, and d-alanine in the presence of sodium (Fig. 3D and SI Appendix, Fig. S6).
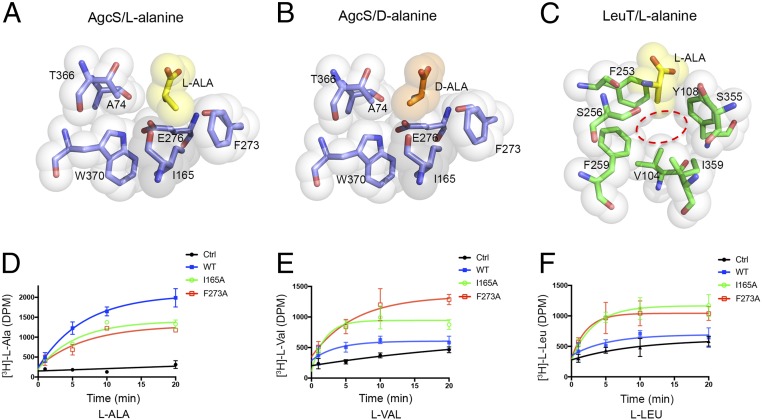
The crystal structures of AgcS with l-alanine or d-alanine bound indicate that the substrate binding site is small and able to accommodate only small amino acids while maintaining an environment favorable to both alanine enantiomers. In contrast to the polar region of the bound alanine (discussed above), the methyl side chain is surrounded within a hydrophobic pocket mainly formed by the side chain of Ile-165 in TM3, Phe-273 in TM6, Thr-366 and Trp-370 in TM8 (Fig. 4 A and B). That hydrophobic pocket could accommodate both alanine enantiomers equally well. The small size of this hydrophobic pocket, shaped by the van der Waals surface of these binding pocket residues, ensures that only glycine or alanine are accommodated while other amino acids are not. In sharp contrast, the substrate binding pocket in LeuT is much larger (Fig. 4C). Correspondingly, LeuT can accommodate a large variety of amino acids, and indeed the substrate selectivity in LeuT is broader. In AgcS, the aliphatic side chains of any amino acid larger than alanine would clash with Ile-165 and Phe-273. Consistent with this postulate, the mutations I165A and F273A show poor substrate selectivity allowing additional amino acids such as l-valine and l-leucine to be efficiently transported by the mutated AgcS (Fig. 4 D–F). These results indicate that the small size of the substrate binding pocket in AgcS is a key determinant in substrate selectivity, a structural feature that may apply to other APC superfamily members.
Fig. 4.
Substrate selectivity in AgcS is dictated by the size of the binding pocket. Comparison of the hydrophobic pocket of alanine-bound AgcS and LeuT (Protein Data Bank [PDB]: 2qei). (A–C) Van der Waals surfaces for alanine substrate and interacting residues are shown as colored spheres. The binding pocket of AgcS (dashed ring in C) is very small and only glycine or alanine can fit while in LeuT, where substrate selectivity is broader, the site is much larger to accommodate larger amino acids. (D–F) AgcS mutants I165A and F273A lose substrate selectivity allowing valine and leucine to be transported. Nonspecific uptake was assessed by using protein-free liposomes under identical conditions as described in Methods Summary. Ctrl, control.
Most amino acid transporters of the APC superfamily have effective stereo-selectivity and tend to transport l-type amino acids exclusively, mirroring the fact that l-amino acids are most commonly used in life while d-amino acids are rarely used. However, AgcS has an ability to transport both enantiomers of alanine as an important nitrogen source (5, 6). This alanine utilization plays a crucial role to support M. maripaludis in harsh environments. As discussed above, the substrate binding site in AgcS has a cytoplasmic-facing hydrophobic surface used to accommodate the aliphatic part of either l- or d-alanine equally well (Fig. 5 A and B). To test this hypothesis, we generated a mutant, W370Q, and assayed the selectivity of the transporter for l- and d-alanine (SI Appendix, Fig. S6). While the ability of W370Q AgcS to uptake l-alanine did not change much, its ability to transport d-alanine was reduced (SI Appendix, Fig. S6E versus SI Appendix, Fig. S6F). This analysis demonstrated that the polarity of the residues at the substrate binding site affects the ability to differentiate between l- and d-amino acids. In sharp contrast, transporters that only allow l-type amino acids to be transported, like LeuT, do not have an even distribution of hydrophobic surface at their substrate binding site. Instead, the cytoplasmic face of their substrate binding site is amphipathic containing both a hydrophobic patch and a charged polarized patch which is also space-limited (Fig. 5C). This feature ensures that only the l-enantiomers of amino acids can bind while d-enantiomers are repelled electrostatically.
Fig. 5.
Stereo-selectivity is afforded by electrostatic repulsion. The amide plane of substrate is shown as gray dash. (A and B) In AgcS both alanine enantiomers fit equally well as the two faces of the binding pocket are hydrophobic. (C) In LeuT, which only binds l-amino acids, one face is hydrophobic while the other is polarized ensuring selection for l-amino acids over d-amino acids.
Concluding Remarks.
The mechanism of substrate binding and selectivity is an essential feature for the APC superfamily members. Here we described the crystal structure of an alanine transporter AgcS capable of transporting both l- and d-alanine and discovered that both the size and polarity of the substrate binding pocket are important structural determinants for specificity and stereo-selectivity. While the resolution of these structures was modest, superposition with LeuT allowed us to identify the position of the bound alanine and that of the Na+ ion. The size of the binding pocket in AgcS determines what amino acids could fit in the transporter while electrostatics control whether l-type or d-type amino acids could bind. We showed that a single point mutation in the binding site of AgcS can broaden the specificity of this transporter to allow uptake of larger amino acids such as valine and leucine. Further, introduction of a charged residue instead of the tryptophan at the binding pocket interfered with the transporters ability to transport d-alanine.
Methods Summary
AgcS (Gene ID: 2761075) from M. maripaludis and its mutants were overexpressed in E. coli C43 (DE3). Fab antibody fragments were generated as described in Methods. The AgcS-Fab complex was purified in the presence of 0.2% (wt/vol) n-decyl-β-d-maltoside and crystallized in the 0.1 M sodium citrate pH 5.6 and 2.5 M ammonium sulfate. Crystals were obtained in the presence of 50 mM l-alanine and d-alanine. Diffraction data sets of all crystals were collected at the Advanced Light Source (Beamline 8.2.1 and 5.0.2) and Advanced Photon Source (NE-CAT 24-ID-C). Data processing and structure determination were performed using the autoPROC, RAPD, Phenix and Coot. Detailed methods for functional studies can be found in the SI Appendix that accompanies this manuscript. Coordinates and structure factors were deposited in the Protein Data Bank (34, 35).
Supplementary Material
Acknowledgments
We thank D. Cawley for development and production of monoclonal antibodies, and staff at the Berkeley Center for Structural Biology, Advanced Light Source (ALS), and the Northeastern Collaborative Access Team (NE-CAT), located at the Advanced Photon Source (APS), for assistance with X-ray data collection. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract DE-AC02-05CH11231. This work is also based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from National Institutes of Health Grant P41 GM103403. The Pilatus 6M detector on 24-ID-C beam line is funded by NIH-ORIP HEI Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Research in the T.G. laboratory is funded by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (Protein Data Bank ID codes 6cse and 6csf).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806206116/-/DCSupplemental.
References
- 1.Jones WJ, Paynter MJB, Gupta R. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch Microbiol. 1983;135:91–97. [Google Scholar]
- 2.Lie TJ, Leigh JA. Regulatory response of Methanococcus maripaludis to alanine, an intermediate nitrogen source. J Bacteriol. 2002;184:5301–5306. doi: 10.1128/JB.184.19.5301-5306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol. 2005;187:972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goyal N, Zhou Z, Karimi IA. Metabolic processes of Methanococcus maripaludis and potential applications. Microb Cell Fact. 2016;15:107. doi: 10.1186/s12934-016-0500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam H, et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly JP, et al. A highly conserved bacterial D-serine uptake system links host metabolism and virulence. PLoS Pathog. 2016;12:e1005359. doi: 10.1371/journal.ppat.1005359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saier MH, Jr, et al. The transporter classification database (TCDB): Recent advances. Nucleic Acids Res. 2016;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 9.Wong FH, et al. The amino acid-polyamine-organocation superfamily. J Mol Microbiol Biotechnol. 2012;22:105–113. doi: 10.1159/000338542. [DOI] [PubMed] [Google Scholar]
- 10.Höglund PJ, Nordström KJV, Schiöth HB, Fredriksson R. The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of Bilaterian species. Mol Biol Evol. 2011;28:1531–1541. doi: 10.1093/molbev/msq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perland E, Fredriksson R. Classification systems of secondary active transporters. Trends Pharmacol Sci. 2017;38:305–315. doi: 10.1016/j.tips.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Vastermark A, Wollwage S, Houle ME, Rio R, Saier MH., Jr Expansion of the APC superfamily of secondary carriers. Proteins. 2014;82:2797–2811. doi: 10.1002/prot.24643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Västermark Å, Saier MH., Jr Evolutionary relationship between 5+5 and 7+7 inverted repeat folds within the amino acid-polyamine-organocation superfamily. Proteins. 2014;82:336–346. doi: 10.1002/prot.24401. [DOI] [PubMed] [Google Scholar]
- 14.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325:1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamata H, et al. Primary structure of the alanine carrier protein of thermophilic bacterium PS3. J Biol Chem. 1992;267:21650–21655. [PubMed] [Google Scholar]
- 17.Rodionov DA, et al. Comparative genomic reconstruction of transcriptional networks controlling central metabolism in the Shewanella genus. BMC Genomics. 2011;12(Suppl 1):S3. doi: 10.1186/1471-2164-12-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida K, et al. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol Microbiol. 2003;49:157–165. doi: 10.1046/j.1365-2958.2003.03567.x. [DOI] [PubMed] [Google Scholar]
- 19.Bualuang A, Kageyama H, Tanaka Y, Incharoensakdi A, Takabe T. Functional characterization of a member of alanine or glycine: Cation symporter family in halotolerant cyanobacterium Aphanothece halophytica. Biosci Biotechnol Biochem. 2015;79:230–235. doi: 10.1080/09168451.2014.968091. [DOI] [PubMed] [Google Scholar]
- 20.Kanamori M, Kamata H, Yagisawa H, Hirata H. Overexpression of the alanine carrier protein gene from thermophilic bacterium PS3 in Escherichia coli. J Biochem. 1999;125:454–459. doi: 10.1093/oxfordjournals.jbchem.a022308. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl–Dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 22.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y. Common folds and transport mechanisms of secondary active transporters. Annu Rev Biophys. 2013;42:51–72. doi: 10.1146/annurev-biophys-083012-130429. [DOI] [PubMed] [Google Scholar]
- 26.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460:1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X, et al. Structure and mechanism of an amino acid antiporter. Science. 2009;324:1565–1568. doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]
- 28.Gao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010;463:828–832. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 29.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature. 2009;458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 30.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe A, et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature. 2010;468:988–991. doi: 10.1038/nature09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drew D, Boudker O. Shared molecular mechanisms of membrane transporters. Annu Rev Biochem. 2016;85:543–572. doi: 10.1146/annurev-biochem-060815-014520. [DOI] [PubMed] [Google Scholar]
- 34.Ma J, et al. 2019 Crystal structure of the sodium-alanine symporter AgcS with L-alanine bound. Protein Data Bank. Available at https://www.rcsb.org/structure/6CSE. Deposited on March 20, 2018.
- 35.Ma J, et al. 2019 Crystal structure of the sodium-alanine symporter AgcS with L-alanine bound. Protein Data Bank. Available at https://www.rcsb.org/structure/6CSF. Deposited on March 20, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.