Significance
Inflammasomes are multiprotein-signaling scaffolds in the innate immune system. Inflammasome-induced inflammation and cell death play critical roles in sensing and responding to pathogen and damage signals. NLRP6 is one of the major inflammasomes in the intestine, whose function has been reported to protect mice from colitis. Here we determined the cryo-EM and crystal structures of NLRP6 pyrin domain (PYD) to reveal a unified model of NLR inflammasome assembly. Additionally, our results show the conformational changes in NLRP6 PYD from its monomeric form in solution to its oligomeric form in filaments. Full-length NLRP6 filaments indicated that the N-terminal PYD domain is the core element for the inflammasome assembly. Collectively, these studies provide mechanistic and structural insight into this biologically important inflammasome.
Keywords: NLRP6, inflammasome, innate immunity, cryo-EM, X-ray crystallography
Abstract
Inflammasomes are large protein complexes that trigger host defense in cells by activating inflammatory caspases for cytokine maturation and pyroptosis. NLRP6 is a sensor protein in the nucleotide-binding domain (NBD) and leucine-rich repeat (LRR)-containing (NLR) inflammasome family that has been shown to play multiple roles in regulating inflammation and host defenses. Despite the significance of the NLRP6 inflammasome, little is known about the molecular mechanism behind its assembly and activation. Here we present cryo-EM and crystal structures of NLRP6 pyrin domain (PYD). We show that NLRP6 PYD alone is able to self-assemble into filamentous structures accompanied by large conformational changes and can recruit the ASC adaptor using PYD–PYD interactions. Using molecular dynamics simulations, we identify the surface that the NLRP6 PYD filament uses to recruit ASC PYD. We further find that full-length NLRP6 assembles in a concentration-dependent manner into wider filaments with a PYD core surrounded by the NBD and the LRR domain. These findings provide a structural understanding of inflammasome assembly by NLRP6 and other members of the NLR family.
Inflammasomes are cytosolic supramolecular complexes containing sensors, adaptors, and effectors in response to pathogen-associated molecular patterns and danger-associated molecular patterns (1–3). Inflammasomes can be classified into canonical and noncanonical inflammasomes. Canonical inflammasomes serve as platforms for caspase-1 activation, while noncanonical inflammasomes activate caspase-11 in mouse and capase-4/5 in humans (4–6). Activated caspases can further process pro-IL-1β or pro-IL-18 to generate the mature cytokines and cleave gasdermin D to form pores to mediate pyroptosis and cytokine release (2, 3, 7–9).
Canonical inflammasome sensors can be classified into two classes: absent in melanoma 2 (AIM2)-like receptors (ALRs) and nucleotide-binding domain (NBD) and leucine-rich repeat (LRR)-containing proteins (NLRs) (2, 3) (Fig. 1A). AIM2 and IFI16 are two known ALRs that are able to assemble into inflammasomes. Both of these ALRs have an N-terminal domain for recruitment of the adaptor protein ASC and a C-terminal HIN domain for recognition of double-stranded DNA (10, 11). NLRs can be further classified into NLRP, NLRC, and NLRB (also known as NAIP) classes according to their N-terminal pyrin domain (PYD), caspase activation and recruitment domain (CARD), and BIR domain, respectively. NLRPs compose the largest subfamily in the NLR family. On activation, NLRP sensors are able to recruit ASC through homotypic PYD–PYD interactions (12). ASC also contains a CARD, and on recruitment by NLRPs, ASC recruits caspase-1 via CARD–CARD interactions, leading to activation of caspase-1 (13).
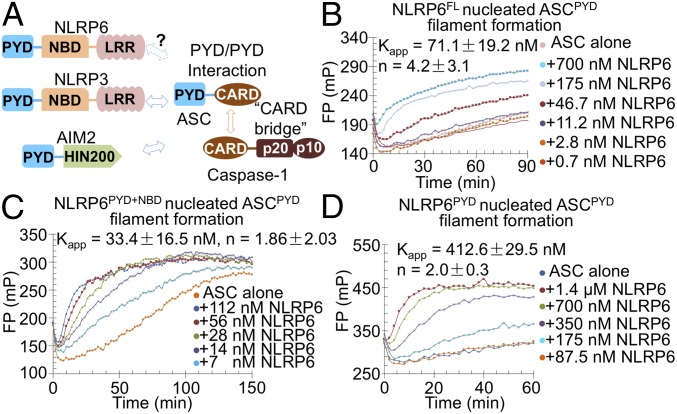
Fig. 1.
NLRP6 promotes assembly of the ASC PYD filament. (A) A cartoon framework of PYD-containing inflammasome assemblies. (B–D) FP assays of ASCPYD filament formation nucleated by NLRP6FL (B), NLRP6PYD+NBD (C), and NLRP6PYD (D). Fitted apparent dissociation constants (Kapp) and Hill coefficients (n) are shown.
NLRP6 is highly expressed in intestine and liver (14, 15) and has a wide range of functions in innate immune signaling. For instance, it has been shown to suppress inflammatory signaling in bone marrow-derived macrophages (16). In the intestine, NLRP6 is associated with the IL-18 production, and NLRP6−/− mice exhibit impaired production of IL-18, suggesting a role of NLRP6 as an inflammasome (17, 18). Moreover, NLRP6 was found to protect mice against dextran sulfate sodium-induced epithelial cell injury (17–19). Further studies showed that NLRP6 activation in epithelial cells is regulated by microbiota-associated metabolites (20). In goblet cells, NLRP6 has been shown to control mucus secretion in response to Toll-like receptor ligands, thereby preventing bacterial invasiveness in the intestine (21). In addition, NLRP6 was found to regulate the expression of a number of IFN-stimulated genes through the mitochondrial adaptor protein MAVS in response to viral RNA (15).
Despite the multifaceted role of NLRP6 in regulating host defense in intestinal epithelial cells and innate immune signaling in myeloid cells, the molecular mechanisms of NLRP6 inflammasome assembly and activation remain unexplored. Here we used biochemical and biophysical methods to show that NLRP6 forms an ASC-dependent inflammasome by helical assembly, with features distinct from various other NLRP inflammasomes.
Results
Different NLRP6 Constructs Nucleate the Polymerization of ASCPYD.
Previous studies have shown that the ASC-containing AIM2 and NLRP3 inflammasomes share a unified assembly mechanism that involves two successive steps of nucleation-induced polymerization through PYD–PYD and CARD–CARD interactions (12, 13) (Fig. 1A). Because NLRP6 has a PYD, we hypothesized that it might also use a similar assembly mechanism as AIM2 and NLRP3 inflammasomes through the adaptor ASC. To test this hypothesis, we first expressed maltose-binding protein (MBP)-tagged NLRP6PYD (1–106), NLPR6PYD+NBD (1–564), and full-length NLRP6 (NLRP6FL) (1–892) in Escherichia coli and insect cells and purified them using affinity and size exclusion chromatography (SI Appendix, Fig. S1A). We used these proteins to induce ASCPYD polymerization in fluorescence polarization (FP) assays on removal of the MBP tags by the tobacco etch virus (TEV) protease. All three constructs of NLRP6 were able to promote the polymerization of ASCPYD with differing potencies (Fig. 1 B–D and SI Appendix, Fig. S1 B–F). One notable feature is that the PYD of NLRP6 alone is a capable nucleator for ASCPYD polymerization, although a higher concentration is required (Fig. 1D and SI Appendix, Fig. S1 D–F). In contrast, the PYD of NLRP3 did not efficiently nucleate ASCPYD polymerization (12).
In comparison, NLPR6PYD+NBD is a stronger nucleator of ASCPYD polymerization than PYD alone, suggesting that the PYD and NBD of NLRP6 synergistically promote the polymerization of ASCPYD (Fig. 1C and SI Appendix, Fig. S1C). Among the three constructs, NLRP6FL showed the greatest dependence on concentration and a large Hill coefficient of 4.2 in promoting ASC polymerization (Fig. 1B and SI Appendix, Fig. S1B). For NLPR6PYD+NBD and NLRP6PYD, the fitted Hill coefficients are smaller (Fig. 1 C and D), and the data also could be fitted well with a Hill coefficient of 1 (SI Appendix, Fig. S1 C–F). This observation might be related to the presumed autoinhibited conformation of the full-length protein (22), so that a cooperative override of autoinhibition is required to generate the platform for ASC polymerization. The apparent dissociation constants for NLPR6PYD+NBD (33 nM) and NLPR6FL (71 nM) are similar, while that for NLPR6PYD is approximately 10-fold higher (412 nM). Nonetheless, the NLRP6 PYD differs from the NLRP3 PYD in its ability to promote ASCPYD polymerization, which prompted us to further explore its assembly.
Cryo-EM Structure Determination of the NLRP6PYD Filament.
To gain an understanding of NLRP6 inflammasome assembly, we first tried to obtain the structure of the NLRP6PYD filament, which is responsible for ASC recruitment. We purified MBP-fused NLRP6PYD overexpressed in E. coli and found that it formed heavily bundled filaments after removal of the MBP tag by the TEV protease. However, the MBP-tagged NLRP6PYD construct with a 3C protease site expressed in insect cells produced NLRP6PYD filaments suitable for further structural studies. Cryo-EM images of NLRP6PYD filaments were collected using a Titan Krios transmission electron microscope (Thermo Fisher Scientific) equipped with a K2 Summit direct electron detector (Gatan) (Fig. 2A).
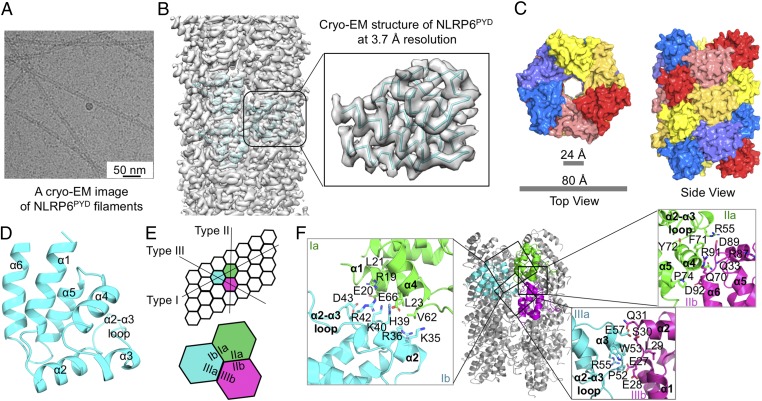
Fig. 2.
Cryo-EM structure of NLRP6PYD filament. (A) A cryo-EM image of NLRP6PYD filaments. (B) Side view of the reconstruction superimposed with three NLRP6PYD subunit models. One subunit is enlarged for a closer view. (C) Surface representation of NLRP6PYD filament structure, top view (Left) and side view (Right). The three main colors of red, yellow, and blue denote the threefold related helical chains. The two shades of each color are used to distinguish neighboring subunits. (D) Cartoon representation of a single NLRP6 PYD domain with six α-helices. (E) Schematic diagram of NLRP6PYD filament, with three adjacent subunits labeled in cyan, green, and magenta. (F) Detailed interactions of type I, II, and III interfaces in the NLRP6PYD filament.
The NLRP6PYD filaments were boxed and segmented in EMAN2 (23), followed by calculation of a power spectrum in Spider (24) (SI Appendix, Fig. S2A). Like the ASCPYD filament (12), the NLRP6PYD filament has C3 point group symmetry. For structure determination, we applied an initial helical parameter of 53° of right-handed rotation and a 13.9-Å axial rise per subunit using helical reconstruction in RELION (25, 26), and these parameters were later refined to 56.8° and 13.8 Å, respectively. The final volume after 3D refinement displayed clear α-helices and achieved a resolution of 3.7 Å as estimated by Fourier shell correlation (Fig. 2 B–D and SI Appendix, Figs. S2 B–D and S3A, and Table S1). Local resolution estimation showed a range of 2.5–5.0 Å (SI Appendix, Fig. S2D).
NLRP6PYD Filament Structure.
The NLRP6PYD filament exhibits a hollow cylinder architecture with an inner diameter of 2.4 nm and an outer diameter of 8 nm (Fig. 2C). The filament assembles through a right-handed helix triplicated by the C3 point group symmetry, with each layer composed of approximately six subunits of NLRP6PYD. Each NLRP6PYD subunit displayed a typical death domain fold with six antiparallel α-helices as shared by members of the death domain superfamily (27) (Fig. 2D and SI Appendix, Fig. S3B). These six α-helices are arranged in a helical bundle with a Greek key motif (SI Appendix, Fig. S3B). Among known PYD structures, NLRP6PYD belongs to the group with a long α2-α3 loop, as described below.
Like other death domain-assembled filaments (27), NLRP6PYD also forms three major asymmetric interfaces in the filament, of which the type I interface mediates intrastrand interactions and type II and type III interfaces mediate interstrand interactions (Fig. 2E). Type I and type II interactions are slightly more extensive, burying approximately 721 Å2 and 749 Å2 of surface area, respectively, while type III interaction buries approximately 668 Å2 of surface area. The type I interface is mediated by the interaction of α1 and α4 helices of one subunit with the α2 helix and the α2-α3 loop of an adjacent subunit (Fig. 2F). The type II interface mainly involves the interaction of the α4-α5 loop region of one subunit with the α5-α6 loop region of a neighboring subunit. The interaction of the α3 helix and the α2-α3 loop of one subunit with the α1-α2 loop region of another subunit defines the type III interaction (Fig. 2F). All three types of interfaces contain charge–charge interactions, hydrophilic interactions, and hydrophobic interactions (Fig. 2F). Notably, the long α2-α3 loop in NLRP6PYD (Fig. 2D) participates in all three types of interactions in the NLRP6 PYD filament (Fig. 2F).
To validate our filament structure model, we picked residues on the different types of filament interfaces and generated mutations on an mCherry-fused NLRP6PYD construct. We expressed and purified wild-type (WT) and mutant mCherry-NLRP6PYD proteins and compared their elution positions from a gel filtration column, because filaments should elute from void fractions. While the WT eluted mainly from void fractions, almost all mutants showed delayed elution and had more proteins at the monomeric fractions (Fig. 3A). R42E on the type I interface and W53E and E27R/E28R on the type III interface were almost totally defective in filament formation. L21R, L23R, and H39R on the type I interface were partially defective in filament formation (Fig. 3A). W53F failed to disrupt filament formation, consistent with our structural analysis indicating that W53 mediates hydrophobic interactions.
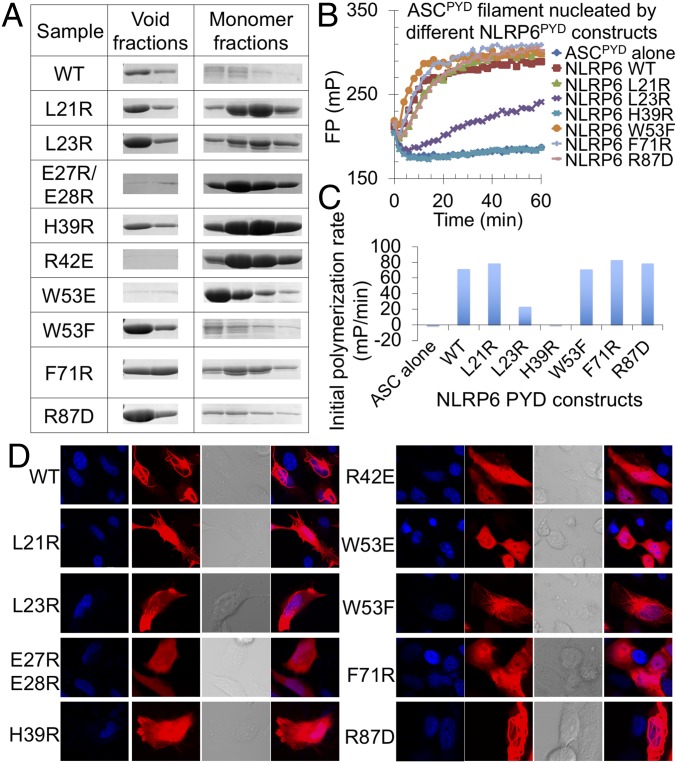
Fig. 3.
Structure validation by mutagenesis. (A) Void and monomer fractions in the gel filtration profile of WT and mutant NLRP6PYD-mCherry proteins. (B) FP assay of ASCPYD filament formation nucleated by void fractions of WT and mutant NLRP6PYD. (C) Plotted initial polymerization rates of ASCPYD from data in B. (D) WT and mutant NLRP6PYD-mCherry overexpressed in HeLa cells examined by confocal microscopy.
To further test whether partially defective mutants identified through gel filtration were still able to nucleate the polymerization of ASC, we performed ASCPYD polymerization assays using the aggregated fractions of these mutants. Among the partially defective mutants, H39R completely lost the ability to promote ASC polymerization, and L23R was less effective in promoting ASC polymerization compared with the WT (Fig. 3 B and C). We further expressed WT and mutant mCherry-fused NLRP6PYD in HeLa cells (Fig. 3D). When overexpressed, WT NLRP6PYD formed nice filaments in cells. R42E, W53E, and E27R/E28R mutants totally disrupted filament formation in cells, whereas L21R, L23R, H39R, F71R, and R87D still formed some filamentous structures, suggesting partial defectiveness of these mutants. In keeping with the gel filtration profile, the W53F mutant was not defective in filament formation.
Crystal Structure of NLRP6PYD.
To gain insight into the conformational changes during NLRP6PYD filament assembly, we further determined the crystal structure of the NLRP6PYD monomer before filament assembly. We were able to obtain monomeric WT NLRP6PYD for crystallization by fusing it rigidly to an engineered MBP tag. MBP-NLRP6PYD was expressed, purified, and crystallized, and the final crystals appeared as clustered plates in 0.2 M lithium sulfate, 0.1 M Bis-Tris pH 6.5, and 25% PEG3350 (SI Appendix, Fig. S4A). The crystals diffracted to 2.3-Å resolution using the 24-ID-C beamline at the Advanced Photon Source, and the crystal structure was solved by molecular replacement (SI Appendix, Table S2). In the crystal structure, the first helix of NLRP6PYD was fused with the last helix of MBP to form a long helix, serving as a rigid linker between NLRP6 PYD and MBP (Fig. 4A). Surprisingly, while the first five helices of NLRP6PYD could be built into the electron density map, little density of the sixth helix was observed (Fig. 4B), suggesting that NLRP6PYD is not fully folded before filament formation.
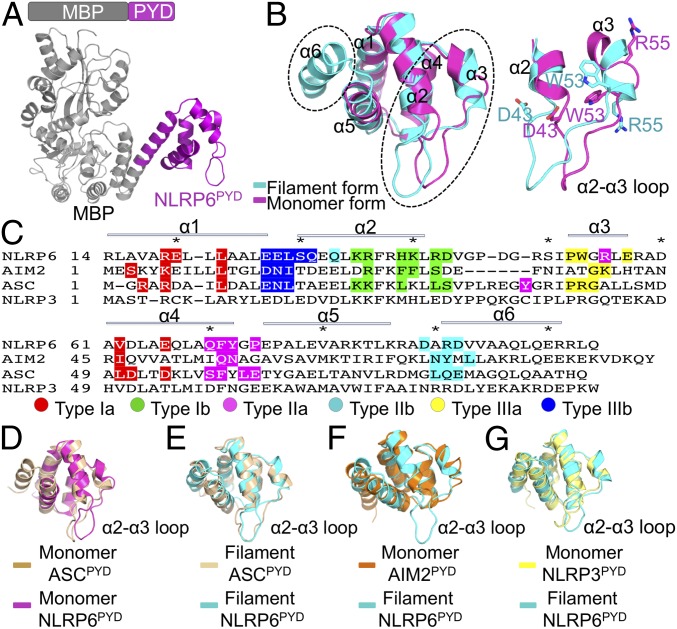
Fig. 4.
Crystal structure of NLRP6 PYD and structural comparison. (A) Overall crystal structure of NLRP6PYD with rigidly linked MBP. (B) Structural comparison of the filament form of an NLRP6PYD subunit (cyan) and the monomeric crystal structure of NLRP6PYD (magenta). Shown is an enlarged view of conformational changes induced by filament formation at the α2-α3 loop. (C) Sequence alignment of the PYDs of NLRP6, AIM2, ASC, and NLRP3. The PYD filament forms of NLRP6, AIM2, and ASC are known, and the different interfacial residues in these filaments are labeled with the indicated colors. (D) Structural comparison of monomeric ASCPYD (PDB ID code 1UCP) and monomeric NLRP6PYD in the crystal structure. (E) Structural comparison of a single subunit of ASCPYD filament (PDB ID code 3J63) and the NLRP6PYD filament form. (F) Structural comparison of the AIM2PYD monomer form (PDB ID code 3VD8) and the NLRP6PYD filament form. (G) Structural comparison of the NLRP3PYD crystal structure (PDB ID code 3QF2) and the NLRP6PYD filament form.
Comparison of the crystal and cryo-EM structures of NLRP6PYD showed that the most notable conformational change lies in the α2-α3 loop (Fig. 4B), which is involved in all three types of interactions (Fig. 2F). D43 on the type I interface, R55 on the type II interface, and W53 on the type III interface all undergo large conformational changes (Fig. 4 B and C). Interestingly, the topologically equivalent α2-α3 loop of ASCPYD also undergoes prominent changes on filament formation (12). These data may suggest a ubiquitous “induced-fit” mode of filament assembly among death domain superfamily proteins, which further dictate a unidirectional filament formation by conformational changes (28).
To investigate whether the lack of a folded α6 and the different α2-α3 loop conformation in the crystal structure are due to crystal packing, we superimposed a NLRP6PYD subunit structure from the filament into the crystal lattice (SI Appendix, Fig. S4B). No steric hindrance was observed, suggesting that the differences in the structure are not caused by crystal packing restraints.
We further performed molecular dynamics (MD) simulations on the crystal and cryo-EM NLRP6PYD structures. We found that the conformation of the α2-α3 loop in the filament structure is more stable than that in the crystal structure, as demonstrated by the smaller root mean square deviation (RMSD) of this loop during the simulation when starting from the filament subunit structure than when starting from the crystal structure (SI Appendix, Fig. S5A). Moreover, during MD simulation, the α3 helix in the filament structure maintained more backbone hydrogen bonds than that in the crystal structure, supporting the greater stability of the filament form (SI Appendix, Fig. S5B). These data indicate that NLRP6PYD filament formation uses not only the favorable energetics from the protein–protein interactions, but also the more stable subunit structure in the filament form.
Given the symmetry similarity of NLRP6PYD and ASCPYD in filament assembly, we superimposed NLRP6PYD with ASCPYD in monomer and filament forms, respectively (Fig. 4 D and E). The monomeric NLRP6PYD crystal structure differs greatly from that of the monomeric ASCPYD structure, particularly in the region of the α2-α3 loop (Fig. 4D), indicating the dynamic properties of this loop in PYD domain evolution. In contrast, the NLRP6PYD subunit structure in the filament is similar to the ASCPYD filament form (Fig. 4E), suggesting a unified assembly mechanism for NLRP6PYD and ASCPYD. On the other hand, the compatible folding of NLRP6PYD and ASCPYD may indicate that NLRP6 is an efficient seed for heterofilament formation and inflammasome activation.
Features of NLRP6 PYD Filament Assembly Compared with Other ALR and NLR Inflammasomes.
A previous study showed that AIM2PYD also assembles into filaments for nucleation of ASC but exhibits less obvious conformational changes during filament formation (29) than NLRP6, which undergoes dramatic conformational changes during filament assembly (Fig. 4B). This observation may be due to the much shorter, and thus likely less dynamic, α2-α3 loop of AIM2PYD (30) compared with NLRP6PYD (Fig. 4F). In contrast, structural comparisons revealed that the PYDs of NLRP3, NLRP7, and NLRP12 possess a long α2-α3 loop (31–33) (Fig. 4G and SI Appendix, Fig. S6 A and B), suggesting that conformational changes may also be required for these NLRPs to form oligomeric platforms to recruit the downstream ASC.
In the NLRP3 inflammasome, the PYD+NBD, but not the PYD alone, is sufficient to provide an effective platform for ASC assembly (12). In contrast, NLRP6PYD alone is able to nucleate ASC polymerization, while the NBD of NLRP6 may play a synergetic role to enhance the NLRP6 platform assembly, as indicated by the lower concentration of the NLRP6PYD+NBD construct required to promote ASCPYD filament formation compared with the NLRP6PYD-alone construct (Fig. 1 C and D and SI Appendix, Fig. S1 C and D). We envision that for both NLRP3 and NLRP6, the NBD aids in the oligomerization and filament formation of the PYD in the context of the full-length proteins, which in turn provides the platform for ASC recruitment and inflammasome assembly. We speculate that the differing polymerization tendencies of NLRP3 and NLRP6 PYDs may lead to differential activation thresholds for these inflammasomes.
To further understand the promotion of ASCPYD polymerization by NLRP6, we overlaid the NLRP6PYD and ASCPYD filament (12) structures using segments of 15 subunits each and found that they superimposed to an RMSD of 3.2 Å, compared with an RMSD of 1.3 Å when single subunits were aligned. When one of the subunits among the 15-subunit segments was aligned, the degree of misalignment increased at subunits away from the aligned subunits (SI Appendix, Fig. S6 C and D). In this regard, homo-oligomerization may be more favorable than hetero-oligomerization. However, we observed variable helical twists in our previous studies within the same PYD filament samples, suggesting plasticity in filament assembly (12, 29) and supporting the validity of hetero-oligomeric filament assembly. In cells, concentrations of NLRP6 or other NLRs may be low, and filament elongation will stop as soon as the concentration of an NLR drops below the critical concentration of polymerization. The assembled NLR platforms may then be able to recruit the downstream signaling protein ASC through PYD–PYD hetero-oligomerization.
NLRP6PYD Filaments Nucleate ASCPYD Assembly Directionally.
The availability of the ASCPYD filament and NLRP6PYD filament cryo-EM structures provides a structural basis for understanding the nucleation and assembly of the NLRP6 inflammasome. Previous studies of the AIM2, NLRC4, and NLRP3 inflammasomes have shown that inflammasome assembly through ASC interaction is unidirectional (12, 13, 34). To address the situation for NLRP6, we analyzed the electrostatic surface of a cross-section of the NLRP6PYD and ASCPYD filaments. For both NLRP6PYD and ASCPYD, the surface (top surface) corresponding to the type Ia, IIa, and IIIa interfaces is largely positively charged, whereas the opposite surface (bottom surface) corresponding to the type Ib, IIb, and IIIb interfaces is largely negatively charged, suggesting the role of charge complementarity in filament assembly and nucleation (Fig. 5A). However, the charge complementarity does not distinguish a potential directional preference in the nucleation-polymerization process.
Fig. 5.
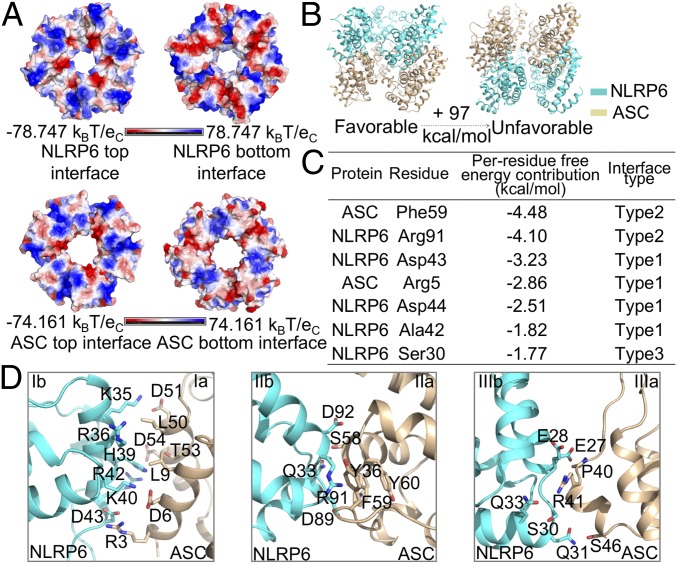
NLRP6 PYD recruits ASC PYD with directional preference. (A) Top and bottom views of the electrostatic surface for one cross-sectional layer from NLRP6PYD and ASCPYD filaments. (B) Computational analysis of the binding energy between the NLRP6PYD bottom surface and the ASCPYD top surface and between the ASCPYD bottom surface and the NLRP6PYD top surface, showing that the former is much preferred. (C) Per-residue free-energy contribution of the favorable model listed with a cutoff of −1.5 kcal/mol. (D) Detailed interactions of type I, II, and III interfaces in the favorable NLRP6PYD-ASCPYD hetero-oligomer model.
To dissect whether NLRP6 recruits ASC with directional preference, we generated two models accounting for ASC recruitment by NLRP6 and calculated their free energy difference based on MD simulations (Fig. 5B). The model in which NLRP6 uses its bottom surface to recruit the top surface of ASC has 97 kcal/mol lower binding free energy than the reverse model, in which NLRP6 uses its top surface to recruit the bottom surface of ASC (Fig. 5B). Residues R5 and F59 in ASC and residues R91, D43, D44, A42, and S30 in NLRP6 contribute to the energetic difference between the two models (Fig. 5C). All these residues are located on the proposed interfaces between NLRP6 and ASC (Fig. 5D).
Detailed analysis showed that the type I interface of NLRP6 and ASC is composed of charge–charge interactions and hydrophobic interactions, that the type II interface contains hydrophobic and hydrophilic interactions, and that the type III interface is dominated by charge–charge and hydrophilic interactions (Fig. 5D). Compared with the unfavorable interfaces (SI Appendix, Fig. S6), more residues are involved in mediating interactions in the favorable model, further supporting the idea that NLRP6 may recruit ASC with a directional preference.
Full-Length NLRP6 Inflammasome Assembly.
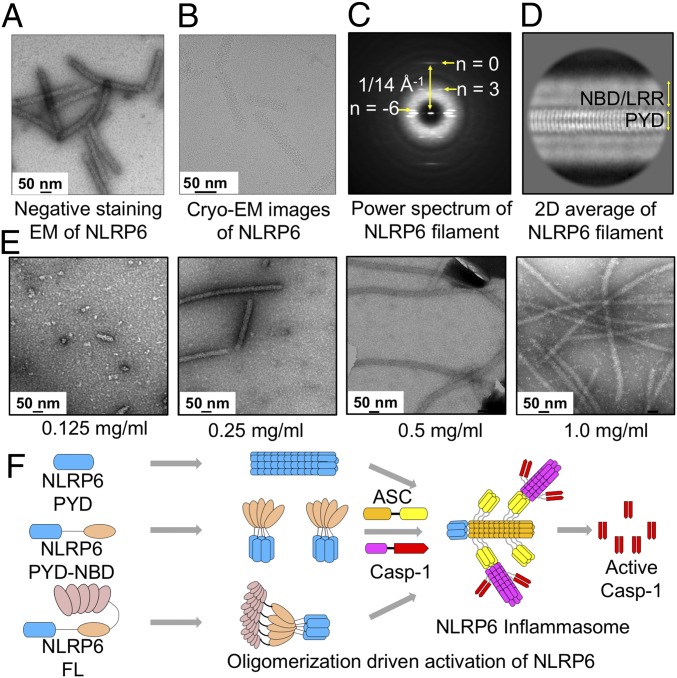
To reveal the structural architecture of full-length NLRP6, we expressed an N-terminal MBP-tagged construct from insect cells and purified it to homogeneity. On removal of the MBP tag, negative-staining EM showed that full-length NLRP6 formed filamentous structures with a diameter of approximately 30 nm (Fig. 6A), similar to the NAIP/NLRC4 inflammasome architecture from cryo-electron tomography (35).
Fig. 6.
Full-length NLRP6 assembles into filaments in a concentration-dependent manner. (A) Negative-staining EM image of NLRP6FL filaments. (B) Cryo-EM image of NLRP6FL filaments. (C) Power spectrum of aligned NLRP6FL filament segments from the cryo-EM data. (D) A 2D class of NLRP6FL filaments. The presumed locations of PYD, NBD, and LRR are shown. (E) Negative-staining EM images of NLRP6FL at different concentrations. (F) A hypothetical model of NLRP6 inflammasome activation. In brief, full-length NLRP6 may be autoinhibited. On activation, it oligomerizes through both the NBD and the PYD, and the PYD filaments provide the platform for ASC recruitment and oligomerization through PYD–PYD interactions. The CARD in ASC then further oligomerizes and recruits caspase-1, driving caspase-1 dimerization and activation.
To gain more detailed understanding of full-length NLRP6 filament assembly, we collected a cryo-EM dataset on a Talos Arctica microscope equipped with a K2 Summit direct electron detector (Fig. 6B). The averaged power spectrum of full-length NLRP6 filaments is similar to that of NLRP6PYD filaments with a strong meridional reflection at 1/14 Å−1 (Fig. 6C and SI Appendix, Fig. S2A), indicating that the full-length NLRP6 filament diffraction was mainly from the PYD. Further 2D classification showed that the PYDs reside in the center of the NLRP6 filament surrounded by the NBD and LRR (Fig. 6D). The NBD-LRR region was blurry in the 2D average, likely due to lack of sufficient order (Fig. 6D). We further found that assembly of full-length NLRP6 filaments is concentration-dependent in vitro (Fig. 6E).
Discussion
NLRs share a similar domain arrangement and may possess an autoinhibited conformation in the absence of ligands in which the NBD and LRR are in a closed conformation, as shown by the crystal structure of NLRC4 (22). On ligand engagement, NLRs are presumed to assemble into ring-like oligomers through the NBDs when the NBD and LRR transit into an open conformation, as evidenced by structural studies of the NAIP/NLRC4 inflammasome (34–36). However, only limited details of the assembly of NLRPs are available at present. In this study, we used X-ray crystallography and EM to elucidate the assembly mechanism of NLRP6 (Fig. 6F).
We have shown that NLRP6PYD forms filaments that are able to nucleate ASCPYD filament assembly, and that NLRP6NBD enhances this ability of NLRP6PYD by lowering the concentration requirement by approximately 10-fold. Full-length NLRP6 also nucleates ASCPYD filament assembly but with a large Hill coefficient, which may indicate the need to overcome autoinhibition to gain the nucleating ability. Our data suggest that NLRP6PYD filament assembly cannot occur in the context of full-length, autoinhibited NLRP6, likely due to steric hindrance by the NBD and LRR. Only when these domains assume an open conformation can NLRP6PYD filament formation proceed within the full-length protein.
Indeed, when overexpressed, full-length NLRP6 assembles into filamentous structures with PYD filaments in the center and the NBD-LRR in the periphery, which may represent an open, active conformation. This open conformation is reminiscent of the overexpressed NAIP/NLRC4 complex in which the CARD forms the central filaments and the NBD-LRR forms the peripheral helical ring (35); however, the difference is that the NBD-LRR ring is more ordered in the NAIP/NLRC4 complex, while the central CARD filament appears as a column of density at low resolution. For NLRP6, the outer NBD-LRR region is not ordered as shown by 2D classes, which may be due to a flexible linker between PYD and the NBD-LRR. It is also possible that the NBD-LRR is not involved in self-association, but the biochemical data on its promotion of NLRP6PYD filament formation to nucleate ASCPYD filaments argue against this hypothesis. In contrast, our data support the hypothesis that the NBD associates in the outer ring and synergizes with the PYD to form the platform for ASC recruitment and oligomerization.
We further speculate that because the concentration of NLRP6 is likely limiting in cells, NLRP6 only forms short filaments in vivo instead of long filaments in vitro, and multiple such filaments can transmit the signal of NLRP6 activation to the polymerization and activation of the downstream ASC adaptor. ASC then acts as the supramolecular platform for recruitment and activation of caspase-1, allowing cells to make quick response to exogenous pathogen infections or endogenous danger signals.
Materials and Methods
NLRP6 Expression and Preparation.
Human NLRP6 PYD, PYD+NBD, and full-length proteins with different tags were expressed in E. coli or insect cells. Proteins were purified using Ni-NTA affinity resin followed by gel filtration chromatography.
EM Data Collection and Processing.
At the University of Massachusetts Cryo-EM Core Facility, the cryo grids were prepared using an FEI Vitrobot Mark IV and imaged using an FEI Titan Krios microscope operating at 300 keV with a K2 Summit camera to collect 723 movies of NLRP6PYD filaments at superresolution mode with a pixel size of 0.529 Å. The images were processed in RELION. Model building and refinement were performed in Coot and Phenix. The atomic coordinates and cryo-EM map have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6NCV) (37) and the EMData Bank, www.emdatabank.org (EMDB ID code 0438) (38).
Crystallization and Structure Determination of MBP-NLRP6PYD.
The best crystals were obtained by hanging drop vapor diffusion. Diffraction data were collected at beamline 24-ID-C of the Advanced Photon Source, and the structure was determined by molecular replacement. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6NDJ) (39).
FP Assay.
MBP-tagged ASCPYD was labeled with the fluorophore TAMRA, and filament formation was monitored by FP on removal of MBP by TEV protease in the absence and presence of various NLRP6 nucleators.
MD Simulations and Molecular Mechanics Calculation.
MD simulations were performed using the AMBER 14 suite of programs with the crystal and cryo-EM NLRP6PYD structures as initial models.
Supplementary Material
Acknowledgments
We thank Drs. Chen Xu and KangKang Song (University of Massachusetts Cryo-EM Core Facility) for data collection. This work was supported by National Institutes of Health Grants Al124491 and HD087988 (to H.W.), National Institutes of Health Grant R35GM122510 (to E.H.E.), Harvard Digestive and Disease Center Grant HDDC P30 DK034854 (to T.-M.F.), and a Cancer Research Institute Irvington Postdoctoral Fellowship (to C.S.). This work was based on research conducted at the Northeastern Collaborative Access Team beamlines, funded by the National Institute of General Medical Sciences from National Institutes of Health Grant P30 GM124165. The Eiger 16M detector on the 24-ID-C beamline is funded by National Institutes of Health Office of Research Infrastructure Programs High-End Instrumentation Grant S10OD021527. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates, structure factors, and cryo-EM map have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6NCV and 6NDJ) and the EMDataBank, www.emdatabank.org (EMDB ID code 0438).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817221116/-/DCSupplemental.
References
- 1.Lu A, Wu H. Structural mechanisms of inflammasome assembly. FEBS J. 2015;282:435–444. doi: 10.1111/febs.13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broz P, Dixit VM. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 3.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 5.Zanoni I, et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science. 2016;352:1232–1236. doi: 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 7.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 2018;557:62–67. doi: 10.1038/s41586-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu A, et al. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat Struct Mol Biol. 2016;23:416–425. doi: 10.1038/nsmb.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy M, Shapiro H, Thaiss CA, Elinav E. NLRP6: A multifaceted innate immune sensor. Trends Immunol. 2017;38:248–260. doi: 10.1016/j.it.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–830. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anand PK, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen GY, Liu M, Wang F, Bertin J, Núñez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Normand S, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci USA. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy M, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birchenough GM, Nyström EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–1542. doi: 10.1126/science.aaf7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 23.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.David L, et al. Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome. Proc Natl Acad Sci USA. 2018;115:1499–1504. doi: 10.1073/pnas.1721967115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu A, et al. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 2015;1:15013. doi: 10.1038/celldisc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288:13225–13235. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae JY, Park HH. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J Biol Chem. 2011;286:39528–39536. doi: 10.1074/jbc.M111.278812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinheiro AS, et al. Three-dimensional structure of the NLRP7 pyrin domain: Insight into pyrin-pyrin–mediated effector domain signaling in innate immunity. J Biol Chem. 2010;285:27402–27410. doi: 10.1074/jbc.M110.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinheiro AS, Eibl C, Ekman-Vural Z, Schwarzenbacher R, Peti W. The NLRP12 pyrin domain: Structure, dynamics, and functional insights. J Mol Biol. 2011;413:790–803. doi: 10.1016/j.jmb.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350:404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diebolder CA, Halff EF, Koster AJ, Huizinga EG, Koning RI. Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: Implications for NLR activation. Structure. 2015;23:2349–2357. doi: 10.1016/j.str.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350:399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 37.Shen C, Fu TM, Wu H. 2018 Cryo-EM structure of NLRP6 PYD filament. Protein Data Bank. Available at https://www.rcsb.org/structure/6NCV. Deposited December 12, 2018.
- 38.Shen C, Fu TM, Wu H. 2018 Cryo-EM structure of NLRP6 PYD filament. EMDataBank. Available at http://emsearch.rutgers.edu/atlas/0438_summary.html. Deposited December 12, 2018.
- 39.Shen C, Fu TM, Wu H. 2018 Crystal structure of human NLRP6 PYD domain with MBP fusion. Protein Data Bank. Available at https://www.rcsb.org/structure/6NDJ. Deposited December 13, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.