Significance
Treatment of acute myelogenous leukemia (AML) has witnessed tremendous progress over the past three decades, owing mainly to the introduction of anthracycline/cytarabine-based protocols. However, a sizable proportion of patients with AML experience relapse or are refractory to standard therapy. Our study addressed a unique molecular/cellular mechanism of Homoharringtonine (HHT) among the known anticancer drugs. A different mode of action of HHT compared with the currently used main anti-AML therapeutics is its ability to directly target NKRF protein and the resultant down-regulation of the MYC gene, a driver of leukemogenesis. Our observation suggests that expression of MYC together with KIT could be a biomarker in patients with AML suitable for HHT treatment.
Keywords: Homoharringtonine, NF-κB repressing factor, acute myelogenous leukemia, MYC, KIT
Abstract
Homoharringtonine (HHT), a known protein synthesis inhibitor, has an anti-myeloid leukemia effect and potentiates the therapeutic efficacy of anthracycline/cytarabine induction regimens for acute myelogenous leukemia (AML) with favorable and intermediate prognoses, especially in the t(8;21) subtype. Here we provide evidence showing that HHT inhibits the activity of leukemia-initiating cells (Lin−/Sca-1−/c-kit+; LICs) in a t(8;21) murine leukemia model and exerts a down-regulating effect on MYC pathway genes in human t(8;21) leukemia cells (Kasumi-1). We discovered that NF-κB repressing factor (NKRF) is bound directly by HHT via the second double-strand RNA-binding motif (DSRM2) domain, which is the nuclear localization signal of NKRF. A series of deletion and mutagenesis experiments mapped HHT direct binding sites to K479 and C480 amino acids in the DSRM2 domain. HHT treatment shifts NKRF from the nucleus (including nucleoli) to the cytoplasm by occupying the DSRM2 domain, strengthens the p65–NKRF interaction, and interferes with p65-p50 complex formation, thereby attenuating the transactivation activity of p65 on the MYC gene. Moreover, HHT significantly decreases the expression of KIT, a frequently mutated and/or highly expressed gene in t(8;21) AML, in concert with MYC down-regulation. Our work thus identifies a mechanism of action of HHT that is different from, but acts in concert with, the known mode of action of this compound. These results justify further clinical testing of HHT in AML.
Acute myelogenous leukemia (AML) is a group of malignant clonal disorders of the hematopoietic system characterized by blockage of myeloid cell differentiation and accumulation of blast cells (1). At present, anthracyclines [e.g., doxorubicin (Doxo), daunorubicin (DNR), idarubicin] and cytarabine (Ara-C) remain in common use for induction therapy, presenting a complete remission (CR) rate of 50–75% in newly diagnosed patients (2). Compared with the marked efficacy of all-trans retinoic acid and arsenic trioxide (5-y disease-free survival rate >90%) in the treatment of acute promyelocytic leukemia (APL) (3), the 5-y survival rate of patients with AML was only 27.4% in 2008–2014 (4). The overall unsatisfactory outcomes of AML highlight the need for improved therapies.
Since 1980s, trials with Homoharringtonine (HHT)-based therapy have revealed the broad efficacy of the compound on a series of hematopoietic disorders, including a CR rate of 24% (65 of 274 patients) in AML (5), a complete hematologic remission rate of 72% (42 of 58 patients) in chronic myelogenous leukemia (6), and a CR rate of 25% (7 of 28 patients) in myelodysplastic syndrome (7) (SI Appendix, Fig. S1A). Mechanistic studies have shown that HHT is embedded in the A-site of the ribosomal subunit and interferes with the elongation process of protein biosynthesis, thereby blocking the transcription of short-half-life oncoproteins, such as MYC, CTNNB1, and MCL1, resulting in apoptosis of leukemic blasts (8, 9).
A recent randomized multicenter Phase III clinical trial reported that an HHT-based regimen (HAA regimen) yielded a significantly improved CR rate and event-free survival compared with a DNR/Ara-C (DA) regimen in de novo AML (non-APL) therapy, particularly in patients with favorable and intermediate-risk cytogenetics (10). Moreover, Zhu et al. (11) identified HAA as a potential treatment regimen for AML, especially for the t(8;21) subtype.
The AML t(8;21) subtype expresses the RUNX1-RUNX1T1 fusion protein and accounts for 10–15% of cases of AML (1). As an aberrant transcriptional factor, the RUNX1-RUNX1T1 protein reportedly activates MYC and subsequently induces self-renewal, cell proliferation, and survival in t(8;21) AML cells (12). In addition, a leukemogenic cooperation between RUNX1-RUNX1T1 and the KIT gene mutations and/or overexpression has been well documented in this type of leukemia (13–15). Although t(8;21) is considered a favorable subtype of AML, >40% of affected patients eventually relapse and die of this disease; therefore, novel, more effective treatments are needed.
While the preventive function of HHT in the initial elongation step of protein synthesis is well accepted (9, 16), the selective effects of this drug on t(8;21) AML suggest additional antileukemic mechanisms of action. In the present study, using t(8;21) AML as the study model, we have identified the direct binding protein of HHT and revealed a related mechanism underlying its therapeutic effects.
Results
In Vivo and in Vitro Antileukemic Effect of HHT in t(8;21) AML.
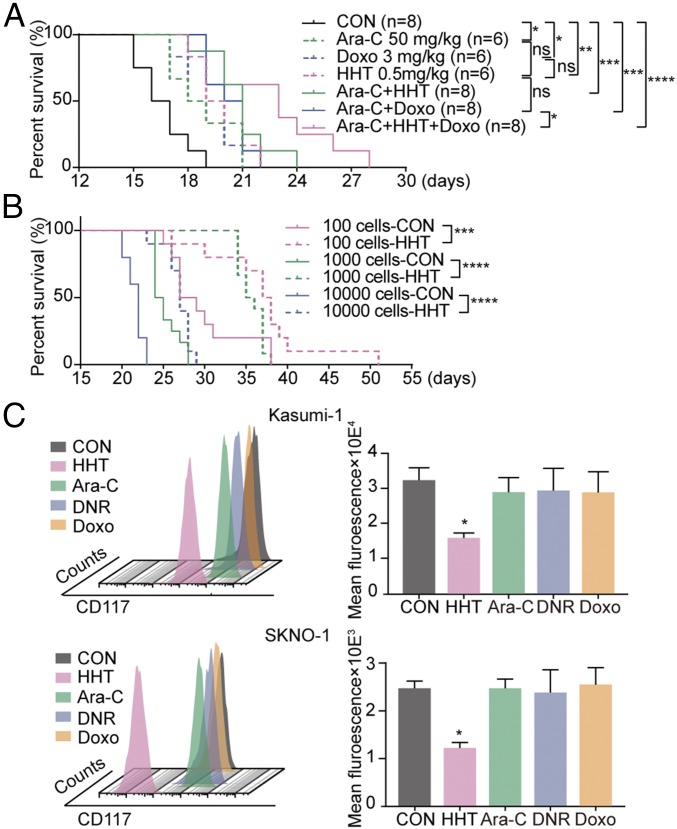
Several experimental systems were used to perform an in-depth analysis of HHT’s mode of action on t(8;21) AML. First, the effects of HHT and HHT-based regimens on t(8;21) AML were validated in a previously described RUNX1-RUNX1T1 and KITN822Kmut coexpression murine leukemia model (13). HHT significantly prolonged the median overall survival (19.5 d vs 16.5 d, P < 0.05) and enhanced the efficacy of a Doxo/Ara-C regimen (treatment vs control groups: 20.5 d vs 16.5 d, P < 0.001; HDA vs control: 23 d vs 16.5 d, P < 0.0001; HDA vs DA: 23 d vs 20.5 d, P < 0.05) (Fig. 1A). In addition, the body weight loss was similar in the mice receiving the HDA regimen and the control group (SI Appendix, Fig. S1B).
Fig. 1.
Therapeutic efficacy of HHT in vivo and in vitro. (A) Life span of t(8;21) AML mice treated with Ara-C, Doxo, HHT, or combinations of Ara-C+HHT, Ara-C+Doxo, and Ara-C+HHT+Doxo. (B) Life span of mice engrafted with 10,000, 1,000, or 100 LICs from vehicle-treated and HHT-treated t(8;21) AML donors. (C) CD117 fluorescence intensity of Kasumi-1 cells (Upper) and SKNO-1 cells (Lower) treated with HHT, Ara-C, Doxo, and DNR for 24 h. Data are mean ± SEM of three independent experiments. CON, control. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We then examined the effect of HHT on leukemia-initiating cells (LICs) (GFP+/Lin−/Sca-1−/c-kit+ bone marrow cells) in secondary limiting dilution transplantation experiments (SI Appendix, Fig. S1C). Compared with mice receiving LICs from vehicle-treated donors, mice receiving LICs from HHT-treated donors showed significantly prolonged survival in all three groups (27 d vs 22 d for the 10,000 cells group, P < 0.0001; 35.5 d vs 24.5 d for the 1,000 cells group, P < 0.0001; and 37.5 d vs 28 d for the 100 cells group, P < 0.001) (Fig. 1B).
Cell viability was significantly inhibited in a dose-dependent manner in both human RUNX1-RUNX1T1–expressing Kasumi-1 and SKNO-1 cell lines and the primary cells, with HHT half-maximal inhibitory concentration (IC50) values of 19.8 nM, 40.6 nM, and 17.0 nM, respectively (SI Appendix, Fig. S1D). Notably, HHT arrested the cell cycle at G0/G1 phase. This effect deviates from the effects of traditional chemotherapeutic drugs, such as the nucleoside analog Ara-C and the topoisomerase II inhibitor DNR or Doxo, which mainly induce cell cycle arrest in S phase or in S and G2/M phases, respectively (SI Appendix, Fig. S2). We also noticed a decrease in the intensity of cell surface CD117 (marker of KIT receptor tyrosine kinase) in both cell lines, a unique effect of HHT compared with Ara-C, DNR, or Doxo (Fig. 1C).
Responses of Distinct AML Cell Lines to HHT Correlate with the Expression of MYC and MYC Target Genes.
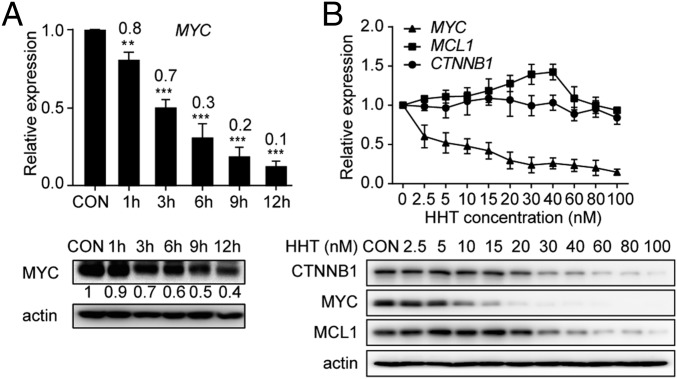
We postulated that transcriptome dynamics could provide clues for unravelling the therapeutic mechanism of HHT. Thus, gene expression profiling was performed in HHT-treated Kasumi-1 cells. Among 1,984 genes identified as significantly altered (P < 0.01) by a comparison analysis between cells with and without HHT treatment, the expression of MYC was the most significantly decreased in the HHT-treated samples (fold change, 15.03; adjusted P < 1 × 10−5) (SI Appendix, Fig. S3A). This finding was validated by quantitative RT-PCR and Western blot assays in HHT-treated Kasumi-1 cells (Fig. 2A).
Fig. 2.
Down-regulation of MYC expression under treatment with HHT. (A) mRNA (Upper) and protein (Lower) expression levels of MYC in Kasumi-1 cells treated with HHT. (B) mRNA (Upper) and protein (Lower) expression levels of MYC, CTNNB1, and MCL1 in Kasumi-1 cells treated with HHT. CON, control. Data are mean ± SEM of three independent experiments. **P < 0.01; ***P < 0.001.
We also measured the expression levels of classical HHT targets, including MYC, CTNNB1 and MCL1, using various HHT concentrations. We found that the mRNA levels of CTNNB1 and MCL1 were not affected, while a significant down-regulation of MYC transcriptional expression was observed in a time- and dose-dependent manner starting at a concentration of 2.5 nM (Fig. 2). At the protein level, the concentration of HHT that down-regulated MYC (10 nM) was much lower than the concentrations that down-regulated CTNNB1 and MCL1 (20 nM and 30 nM, respectively) (Fig. 2B).
The ENCODE ChIP-Seq Significance Tool was used to search for pathways/processes as possible targets of HHT (17). MYC was noted as a highly enriched transcriptional regulator in the promoters of genes altered by HHT. Gene Set Enrichment Analysis revealed that the HHT treatment of Kasumi-1 cells caused a significant decrease in the expression levels of MYC up-regulated targets previously identified from two independent datasets (SI Appendix, Fig. S3B) (18, 19); for example, BAX and CDK4 were down-regulated with HHT treatment (SI Appendix, Fig. S3C). Given that this phenomenon occurred within 24 h after onset of the drug’s effect, the down-regulation of MYC and the expression levels of its targeted genes might be a quick and critical event.
We next evaluated MYC expression in six AML cell lines—Kasumi-1, SKNO-1, NB4, HL-60, U937, and THP-1—and compared the results with their responses to HHT (SI Appendix, Fig. S4A). Notably, the transcriptional level of MYC was correlated with the sensitivity to HHT in these six cell lines (r = −0.8553, P = 0.0299) (SI Appendix, Fig. S4B).
Taken together, these results suggest that HHT could attenuate MYC and MYC-targeted gene expression in t(8;21) AML cells, and that the down-regulatory effect on MYC could be selectively correlated with the therapeutic efficacy of HHT.
HHT Recognizes MYC Transcriptional Regulator NKRF.
The selective effect of HHT on MYC expression provided a stimulus for further investigation of direct drug targets. We first incubated the Kasumi-1 cell lysates with biotin-labeled HHT (bio-HHT) and then performed electrospray-ionization quadrupole time-of-flight mass spectrometry (ESI-Q-TOF MS) with eluted proteins. Among 2,095 prey proteins detected in the bio-HHT group, 9 proteins were found to be enriched and associated with MYC expression (SI Appendix, Table S1). Among these nine proteins, six are located downstream of the MYC pathway (ABCD1, ABCD3, CLIC4, GLYR1, ASNS, and ABCF3) and three are upstream regulators of MYC expression (SRC, STAT1, and NKRF) (SI Appendix, Fig. S5). SRC and STAT1 are reportedly capable of up-regulating MYC mRNA levels, whereas NKRF is a known repressor of MYC expression via inhibition of p65 (20–22).
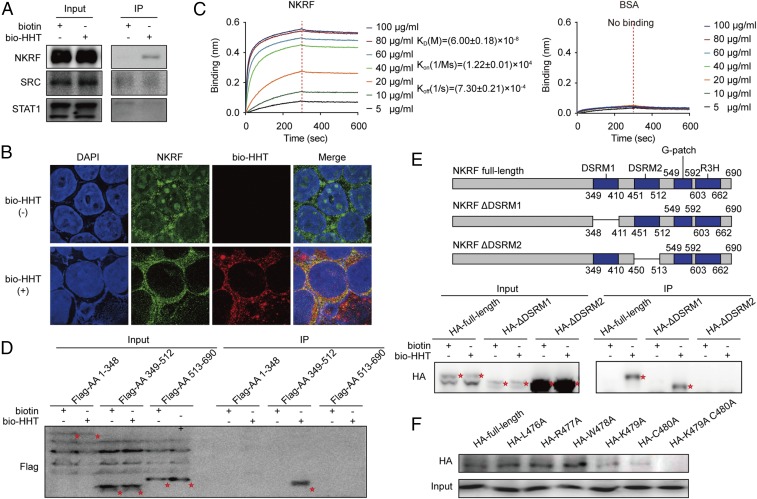
A streptavidin (SA) agarose affinity assay showed that only NKRF could directly interact with HHT in Kasumi-1 cells, while SRC and STAT1 could not (Fig. 3A and SI Appendix, Fig. S6). In an immunofluorescence assay, a typical nucleolar localization of NKRF was observed, as described previously (23), along with the presence of the protein in cytoplasm without drug treatment. Notably, on the effect of bio-HHT, NKRF staining almost disappeared from the nucleoli. Meanwhile, bio-HHT signals were partly colocalized with NKRF in the cytoplasm of Kasumi-1 cells (Fig. 3B). These results suggest that HHT not only might induce translocation of NKRF from nucleoli to cytoplasm, but also might directly interact with NKRF.
Fig. 3.
Mapping the direct binding site of NKRF protein to HHT. (A) Validation of the binding of SRC, STAT1, and NKRF with bio-HHT by SA agarose affinity assay. (B) Colocalization of NKRF with biotin-HHT analyzed by immunofluorescence staining. (Magnification: 630×.) (C) BLI analysis of the binding affinity between HHT and increasing concentrations of 6× His-NKRF. BSA served as a negative control. (D) Binding effect between bio-HHT and the full-length or truncated NKRF constructs (HA-labeled) detected by immunoprecipitation assay. Protein bands are highlighted by red asterisks. (E, Upper) Schematic representation of NKRF deletion mutants. (E, Lower) Binding effect between bio-HHT and the full-length or deleted NKRF mutants (HA-labeled) detected by immunoprecipitation assay. Protein bands are highlighted by red asterisks. (F) Effects of NKRF amino acid 479 and 480 substitutions on binding of NKRF to HHT.
To further confirm this interaction and quantitatively measure the binding affinity, recombinant His6-NKRF was prepared to examine its bio-HHT binding capacity using bio-layer interferometry (BLI). The results showed time- and concentration-dependent saturation with a KD of (6.00 ± 0.18) × 10−8 M (Fig. 3C), providing evidence that HHT could directly bind to NKRF.
Mapping of the HHT Direct Binding Site to K479 and C480 in the DSRM2 Domain of NKRF.
We next mapped the region(s) of NKRF protein that might be involved in the interaction with HHT. NKRF protein consists mainly of two double-stranded RNA-binding motifs (DSRMs), a G-patch domain and an R3H domain. Three flag-tagged, truncated NKRFs were expressed: amino acids 1–348, amino acids 349–512, and amino acids 513–690. Only the peptide with amino acids 349–512 containing two DSRMs bound with bio-HHT (Fig. 3D and SI Appendix, Fig. S7A). Furthermore, deletion of the DSRM1 domain in NKRF did not affect bio-HHT binding, whereas the truncated protein lacking DSRM2 completely lost its binding ability (Fig. 3E). BLI analysis also showed that the DSRM2 domain retained a strong affinity to bio-HHT, whereas the DSRM1 domain did not (SI Appendix, Fig. S7B). These data suggest that DSRM2 domain-containing amino acids (451–512) could harbor the HHT binding site.
A set of overlapping peptides (20–22 amino acids) was synthesized to narrow the binding site to a 10-amino acids peptide (SI Appendix, Fig. S8). By using a second set of shorter overlapping peptides (3–4 amino acids), the binding site was further narrowed to a stretch of amino acids 476–480 (SI Appendix, Fig. S9). By replacing the candidate amino acid with alanine (A) one by one and performing an SA agarose affinity assay, we finally identified that K479 and C480 might be the key binding amino acids. Indeed, the substitution of K479 or C480 diminished the binding considerably, whereas the substitution of both amino acids completely severed the binding (Fig. 3F).
HHT Down-Regulates MYC Expression by Binding NKRF and Interfering with p65 Translocation.
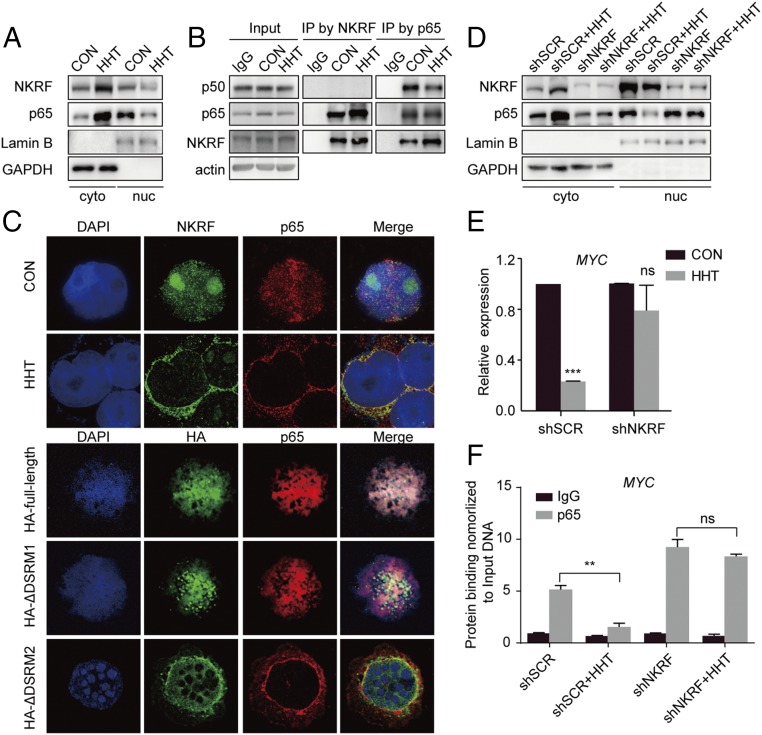
Based on the above results, we further hypothesized that HHT could modulate the biological effects of NKRF proteins. NKRF maintains NF-κB in an inactive state and represses activation of NF-κB downstream MYC expression (22). In this study, we used TNF-α to better analyze the results of p65 signaling activation. We found that in TNF-α–stimulated Kasumi-1 cells, the nuclear contents of p65 and NKRF decreased as NKRF accumulated in cytoplasm after HHT treatment (Fig. 4A). In addition, an increased p65–NKRF interaction and a decrease in p65-p50 heterodimers were observed under HHT treatment (Fig. 4B), while the p65 phosphorylation level showed no obvious change (SI Appendix, Fig. S10). These results indicate that HHT could promote formation of the p65-NKRF complex while interfering with p65-p50 heterodimerization without involvement of the classical posttranslational modification of p65.
Fig. 4.
Shift of p65 proteins from nucleus to cytoplasm on HHT treatment. (A) NKRF and p65 proteins shifted from the nucleus to cytoplasm under HHT treatment in TNF-α simulated Kasumi-1 cells. (B) Detection of binding effect of NKRF to p65 and p65 to p50 by immunoprecipitation assay. (C) Immunofluorescence staining of NKRF and p65 in TNF-α–stimulated Kasumi-1 cells treated with HHT compared with the control sample (Upper) and of HA and p65 in TNF-α–stimulated Kasumi-1 cells transfected with the HA-tagged full-length or deleted NKRF mutants (Lower). (Magnification: 630×.) (D) NKRF and p65 protein levels in nucleus and cytoplasm of shSCR- and shNKRF-transfected Kasumi-1 cells stimulated by TNF-α. (E) Quantification of MYC mRNA expression in shSCR- and shNKRF-transfected Kasumi-1 cells treated with HHT for 24 h. Data are mean ± SEM of three independent experiments. (F) Assessment of the amount of p65 on MYC gene promoter by ChIP assays in shSCR- and shNKRF-transfected Kasumi-1 cells stimulated by TNF-α. CON, control. Data are expressed as mean ± SEM. ns, P > 0.05; **P < 0.01; ***P < 0.001.
Immunofluorescence assays showed that NKRF partly colocalized with p65 signals, and that their colocalization signals under HHT treatment shifted from the nucleus and nucleoli to the cytoplasm (Fig. 4C, Upper), even under a low drug concentration (10 nM HHT) (SI Appendix, Fig. S11). The same translocation was also observed in Kausmi-1 cells when the DSRM2 domain, but not the DSRM1 domain, of NKRF protein was deleted (Fig. 4C, Lower).
To further determine the biological role of NKRF in the HHT-mediated antileukemia effect, endogenous NKRF was approximately 50% knocked down by short-hairpin RNA (shRNA) in Kasumi-1 and SKNO-1 cells (SI Appendix, Fig. S12). We found that NKRF knockdown diminished the HHT-induced p65 translocation (Fig. 4D). Meanwhile, HHT significantly lost its effect in down-regulating MYC (Fig. 4E). Our results support the idea that HHT might also target MYC at the transcriptional level by inhibiting the activity of p65-p50 (24). In fact, HHT treatment decreased p65 binding to the MYC promoter, whereas NKRF knockdown diminished this effect (Fig. 4F).
Notably, the suppressive effect of MYC protein on the effect of HHT was partially retained in cells in which NKRF expression was knocked down compared with the control setting (SI Appendix, Fig. S13), supporting the existence of the classical mechanism of HHT in inhibiting the protein synthesis of MYC.
HHT Decreases KIT Expression in Accordance with MYC Down-Regulation.
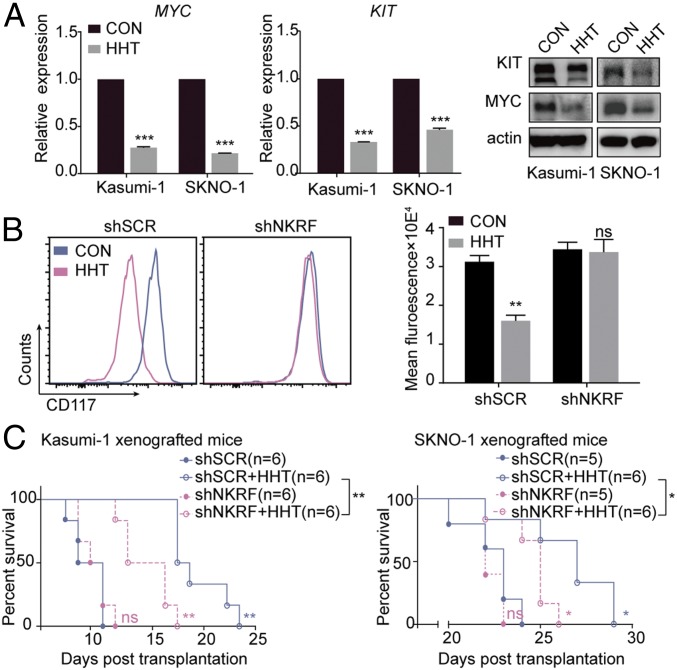
Our previous study reported that the overexpression of KIT could be detected in 81.3% of patients with t(8;21) AML (13). Here we observed a significant correlation (r = 0.7475, P < 0.0001) between the expression of MYC and KIT in 37 primary AML samples, especially in patients with t(8;21) AML (SI Appendix, Fig. S14 A and B, Left). This correlation was also verified in the analysis of 173 AML patients from The Cancer Genome Atlas database (r = 0.4599, P < 0.0001) (SI Appendix, Fig. S14B, Right). We also found that the mRNA and protein levels of KIT were down-regulated together with MYC in Kasumi-1 and SKNO-1 cells treated with HHT for 24 h (Fig. 5A).
Fig. 5.
Decrease in KIT expression on HHT treatment in accordance with down-regulation of MYC expression. (A) mRNA (Left and Middle) and protein (Right) expression levels of MYC and KIT in Kasumi-1 and SKNO-1 cells treated with HHT for 24 h. (B) CD117 fluorescence intensity in shSCR- and shNKRF- transfected Kasumi-1 cells treated with HHT for 24 h. (C) Life span of the mice xenografted with shSCR- or shNKRF-transfected Kasumi-1 cells (Left) and SKNO-1 cells (Right) and treated with HHT or vehicle control. CON, control. Data are mean ± SEM of three independent experiments. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
The association of KIT and MYC expression was further strengthened by the observation that in Kasumi-1 cells, exposure to JQ1, a small-molecule bromodomain inhibitor capable of repressing MYC expression (25), decreased the expression of not only MYC, but also of KIT (SI Appendix, Fig. S14C). Furthermore, the shRNA targeting MYC, when transfected into Kasumi-1 cells, showed the same effect (SI Appendix, Fig. S14D). We also observed that under HHT treatment and NKRF knockdown conditions, the repressive effect on CD117 intensity was markedly diminished (Fig. 5B), suggesting that the inhibition of HHT on KIT expression was dependent, at least in part, on MYC transcriptional level.
As expected, the effect of HHT on cell cycle blockage and cell proliferation inhibition was diminished in NKRF shRNA-transfected t(8;21) leukemia cell lines (SI Appendix, Figs. S15 and S16). In vivo, mitigation of the therapeutic effect of HHT through NKRF knockdown was also observed in t(8;21) AML mice. Two xenograft mouse models were established by engrafting NOD/SCID and B-NDG mice with Kasumi-1 and SKNO-1 cells transfected by scramble shRNA (shSCR) or shRNA targeting NKRF (shNKRF), respectively. In both models, HHT significantly extended the lifespan of mice injected with shSCR-transfected cells (median overall survival: 10 d vs. 17.5 d, P < 0.01 in Kasumi-1 xenografted mice and 23 d vs. 27 d, P < 0.05 in SKNO-1 xenografted mice), while the mice injected with shNKRF-transfected cells showed a shorter lifetime (P < 0.01 and < 0.05, respectively) (Fig. 5C).
Discussion
HHT, a traditional Chinese medicine-based agent, has been used for leukemia treatment for more than three decades (5–11), with well-recognized inhibitory effects on the expression of short-half-life proteins. Recently, more potential HHT target proteins have been reported, suggesting a broader mode of action of the drug (26, 27). In this study, we found that HHT inhibited the activity of LICs and improved survival in t(8;21) AML mice. Notably, HHT reduced both the mRNA and protein levels of MYC and also reduced the protein expression for CTNNB1 and MCL1 without affecting their mRNA (9). Moreover, the dosage of HHT needed to down-regulate MYC protein was much lower than that needed to repress the classical short-half-life proteins, such as CTNNB1 and MCL1 proteins. These results suggest the existence of unique mechanisms underlying the antileukemia effect of HHT in addition to the inhibition of protein synthesis. Indeed, we demonstrated that NKRF, an NF-κB–repressing factor, was targeted by HHT.
NKRF was first reported as a repressor of NF-κB (28) localizing in the cytoplasm, nucleoplasm, and nucleoli (23). Generally detected by a subunit of p65, NF-κB is constitutively activated in primitive human AML cells, and inhibition of NF-κB might induce leukemia-specific apoptosis (29). Initially, NKRF was found to inhibit the transcriptional activity of NF-κB by recognizing the specific negative regulatory elements (NREs: AATTCCTCTGA) at the promoters of NF-κB target genes (28). Further studies showed that NKRF can directly interact with NF-κB factors in a non-NRE manner to regulate several NF-κB target genes, such as MYC and MMP (22, 30). Consistent with these reports, we found that HHT is able to induce NKRF translocation from nucleoli to cytoplasm, possibly resulting from a protein configuration change on HHT binding to amino acids K479 and C480 in the DSRM2 domain of NKRF. In agreement with a report that NKRF does not bind to the MYC promoter (22), direct interaction was not observed in our ChIP-qPCR assay (SI Appendix, Fig. S17). Thus, we hypothesized that HHT could down-regulate MYC expression in a DNA-independent manner.
It was previously reported that amino acids 361–550 of NKRF containing the RNA-binding motifs might be its nuclear localization signal (23). We found that localization of the DSRM2-deleted NKRF mutant was changed from the nucleoli to the cytoplasm, mimicking the effect of HHT. Meanwhile, we observed that the interaction of NKRF and p65 was strengthened, while the formation of p65-p50 complex was weakened. The biological consequence of this event should be reduced binding of p65 to the promoter region of MYC and decreased transcriptional activity of MYC, as has been previously documented (24). In terms of the role of p65 in AML cell proliferation and the possibility of other functional roles of NKRF in addition to its suppression of the NF-κB pathway, the interaction of HHT with NKRF could lead to additional perturbation of AML cell growth and survival.
We further validate our findings by showing that NKRF knockdown diminished the inhibitory effect of HHT on MYC expression, and that NKRF-partially depleted leukemia cells were relatively resistant to HHT under both in vitro and in vivo conditions. However, NKRF knockdown could not fully deplete HHT induced down-regulation of MYC protein level and completely abrogate the effect in shNKRF mice. These results may be explained by the incomplete knockdown of NKRF or the existence of another mechanism, such as inhibition of HHT on MYC protein synthesis.
Our results show that in t(8;21) AML mice, the high-frequency aberrant KIT expression cooperated with RUNX1-RUNX1T1 in the development of full-blown AML and thus reduced the likelihood of a favorable prognosis (13–15). A previous study revealed a positive feedback loop between KIT mutation or overexpression and MYC mRNA level, leading to hematopoietic malignancies (31). In this study, we observed that high MYC mRNA levels in AML patient samples often coexist with the aberrant expression of KIT. We also showed that HHT could down-regulate endogenous MYC and KIT simultaneously.
In summary, our study reveals a mechanism of HHT that is distinct from inhibiting mRNA translation at the ribosome elongation process as described previously (8). This mechanism leading to suppression of AML cells might be unique and not shared with any other anticancer agents discovered so far. Thus, patients with MYC and KIT overexpression could achieve a favorable response to HHT treatment. HHT may optimize the current combination of chemotherapy for AML, especially in subgroups with an activated NF-κB-MYC pathway, through abrogating both the NKRF-MYC regulating axis and protein synthesis.
Materials and Methods
Reagents.
HHT (Hangzhou Minsheng Pharmaceutical) was dissolved in a 100 μM RPMI 1640 solution and stored at −20 °C. Ara-C, DNR (Pfizer), and Doxo (Shenzhen Main Luck Pharmaceuticals) were dissolved in 1 mM RPMI 1640 solution and stored at −20 °C. PE Mouse anti-human CD117 antibody was purchased from BD Pharmingen, and JQ-1 and MG132 were purchased from Selleck.
ESI-Q-TOF MS and SA Agarose Affinity Assay.
Bio-HHT was synthesized by WuXi PharmaTech. SA agarose beads (Thermo Fisher Scientific) were used to enrich HHT-binding proteins and then subjected to ESI-Q-TOF MS, as described in detail in SI Appendix.
BLI Assay.
The binding affinities of HHT and NKRF proteins were determined using the ForteBio Octet RED96 system equipped with a SA sensor as described previously (32) and detailed in SI Appendix.
Immunofluorescence Assay.
To determine the colocalization of NKRF with bio-HHT or NKRF with p65, cells were fixed and stained as described previously (26). Images were obtained using a confocal microscope with 0.25-μm z-stacking.
Animal Model.
The t(8;21) leukemia murine model harboring RUNX1-RUNX1T1 and KITN822K mutation was generated as described previously (13). Gradient-descent leukemia-initiating cells [GFP+/Lin−/Sca-1−/c-kit+ bone marrow cells from HHT or vehicle-treated t(8;21) AML mice] were sorted and injected into sublethally irradiated BALB/C mice. Two xenograft mouse models were established with Kasumi-1 shNKRF cells and SKNO-1 shNKRF cells. More details are provided in SI Appendix.
Cell assay, the plasmid construction, and information related to data deposit in the Gene Expression Omnibus database (33) are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Shi-Dong Wang and Dr. Qing-Hua Zhang (Wayen Biotechnologies) for their contributions to the proteome array experiments and data analysis and Dr. Lei-Miao Yin (Shanghai University of Traditional Chinese Medicine) for the ingenuity pathway analysis. This work was supported by Chinese National Key Basic Research Project 973 Grant 2013CB966800; Chinese Ministry of Health Grant 201202003; National Natural Science Foundation of China Grants 81470311, 81670137, and 81270619; the Samuel Waxman Cancer Research Foundation; the Shanghai Guangci Translational Medical Research Development Foundation; Shanghai Municipal Education Commission–Gao Feng Clinical Medicine Support Grant 20152501; and the Doctoral Innovation Fund from Shanghai Jiao Tong University School of Medicine.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE121175).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818539116/-/DCSupplemental.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Pulte D, Redaniel MT, Jansen L, Brenner H, Jeffreys M. Recent trends in survival of adult patients with acute leukemia: Overall improvements, but persistent and partly increasing disparity in survival of patients from minority groups. Haematologica. 2013;98:222–229. doi: 10.3324/haematol.2012.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ZY, Chen Z. Acute promyelocytic leukemia: From highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 4.Noone AM, et al. 2018 SEER cancer statistics review, 1975-2015. Available at https://seer.cancer.gov/csr/1975_2015/. Accessed October 1, 2018.
- 5.Grem JL, Cheson BD, King SA, Leyland-Jones B, Suffness M. Cephalotaxine esters: Antileukemic advance or therapeutic failure? J Natl Cancer Inst. 1988;80:1095–1103. doi: 10.1093/jnci/80.14.1095. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien S, et al. Homoharringtonine therapy induces responses in patients with chronic myelogenous leukemia in late chronic phase. Blood. 1995;86:3322–3326. [PubMed] [Google Scholar]
- 7.Feldman EJ, Seiter KP, Ahmed T, Baskind P, Arlin ZA. Homoharringtonine in patients with myelodysplastic syndrome (MDS) and MDS evolving to acute myeloid leukemia. Leukemia. 1996;10:40–42. [PubMed] [Google Scholar]
- 8.Kantarjian HM, et al. Homoharringtonine: History, current research, and future direction. Cancer. 2001;92:1591–1605. doi: 10.1002/1097-0142(20010915)92:6<1591::aid-cncr1485>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Lü S, Wang J. Homoharringtonine and omacetaxine for myeloid hematological malignancies. J Hematol Oncol. 2014;7:2. doi: 10.1186/1756-8722-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin J, et al. Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: A multicentre, open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2013;14:599–608. doi: 10.1016/S1470-2045(13)70152-9. [DOI] [PubMed] [Google Scholar]
- 11.Zhu HH, et al. Homoharringtonine, aclarubicin and cytarabine (HAA) regimen as the first course of induction therapy is highly effective for acute myeloid leukemia with t (8;21) Leuk Res. 2016;44:40–44. doi: 10.1016/j.leukres.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Steffen B, et al. AML1/ETO induces self-renewal in hematopoietic progenitor cells via the Groucho-related amino-terminal AES protein. Blood. 2011;117:4328–4337. doi: 10.1182/blood-2009-09-242545. [DOI] [PubMed] [Google Scholar]
- 13.Wang YY, et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: Implication in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad Sci USA. 2005;102:1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang YY, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA. 2011;108:2450–2455. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatlen MA, Wang L, Nimer SD. AML1-ETO–driven acute leukemia: Insights into pathogenesis and potential therapeutic approaches. Front Med. 2012;6:248–262. doi: 10.1007/s11684-012-0206-6. [DOI] [PubMed] [Google Scholar]
- 16.Gürel G, Blaha G, Moore PB, Steitz TA. U2504 determines the species specificity of the A-site cleft antibiotics: The structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol. 2009;389:146–156. doi: 10.1016/j.jmb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auerbach RK, Chen B, Butte AJ. Relating genes to function: Identifying enriched transcription factors using the ENCODE ChIP-seq significance tool. Bioinformatics. 2013;29:1922–1924. doi: 10.1093/bioinformatics/btt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuhmacher M, et al. The transcriptional program of a human B cell line in response to myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kharma B, et al. STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Res. 2014;74:6519–6530. doi: 10.1158/0008-5472.CAN-14-0847. [DOI] [PubMed] [Google Scholar]
- 21.Fox EM, et al. Signal transducer and activator of transcription 5b, c-Src, and epidermal growth factor receptor signaling play integral roles in estrogen-stimulated proliferation of estrogen receptor-positive breast cancer cells. Mol Endocrinol. 2008;22:1781–1796. doi: 10.1210/me.2007-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Z, et al. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niedick I, et al. Nucleolar localization and mobility analysis of the NF-kappaB repressing factor NRF. J Cell Sci. 2004;117:3447–3458. doi: 10.1242/jcs.01129. [DOI] [PubMed] [Google Scholar]
- 24.La Rosa FA, Pierce JW, Sonenshein GE. Differential regulation of the c-myc oncogene promoter by the NF-kappa B rel family of transcription factors. Mol Cell Biol. 1994;14:1039–1044. doi: 10.1128/mcb.14.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posternak V, Cole MD. Strategically targeting MYC in cancer. F1000 Res. 2016;5:408. doi: 10.12688/f1000research.7879.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y, et al. Small-molecule induction of phospho-eIF4E sumoylation and degradation via targeting its phosphorylated serine 209 residue. Oncotarget. 2015;6:15111–15121. doi: 10.18632/oncotarget.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T, et al. Homoharringtonine binds to and increases myosin-9 in myeloid leukaemia. Br J Pharmacol. 2016;173:212–221. doi: 10.1111/bph.13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nourbakhsh M, Hauser H. The transcriptional silencer protein NRF: A repressor of NF-κ B enhancers. Immunobiology. 1997;198:65–72. doi: 10.1016/s0171-2985(97)80027-7. [DOI] [PubMed] [Google Scholar]
- 29.Guzman ML, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98:2301–2307. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- 30.Reboll MR, et al. Mapping of NRF binding motifs of NF-kappaB p65 subunit. J Biochem. 2011;150:553–562. doi: 10.1093/jb/mvr099. [DOI] [PubMed] [Google Scholar]
- 31.Liu S, et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang HN, et al. Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc Natl Acad Sci USA. 2015;112:15084–15089. doi: 10.1073/pnas.1521316112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.