Significance
Adult secondary hemophagocytic lymphohistiocytosis (sHLH) is a fulminant hyperinflammatory syndrome with mortality exceeding 80% in spite of state-of-the-art medical care. The pathogenesis of sHLH, unlike primary HLH, which is primarily genetic, is poorly understood. Here we describe a model of sHLH that recapitulates many aspects of human sHLH. We found that the hyperinflammation in this model is macrophage intrinsic and identified a unique transcriptional profile associated with sHLH macrophages that was also present in patients with sHLH. We also found that these hyperinflammatory macrophages were highly dependent on glycolytic metabolism. Consequently, we found that administration of the glycolysis inhibitor 2-deoxyglucose was sufficient to rescue animals with sHLH by significantly suppressing the inflammatory response.
Keywords: inflammation, sepsis, hemophagocytic lymphohistiocytosis
Abstract
Secondary hemophagocytic lymphohistiocytosis (sHLH) is a highly mortal complication associated with sepsis. In adults, it is often seen in the setting of infections, especially viral infections, but the mechanisms that underlie pathogenesis are unknown. sHLH is characterized by a hyperinflammatory state and the presence hemophagocytosis. We found that sequential challenging of mice with a nonlethal dose of viral toll-like receptor (TLR) agonist followed by a nonlethal dose of TLR4 agonist, but not other permutations, produced a highly lethal state that recapitulates many aspects of human HLH. We found that this hyperinflammatory response could be recapitulated in vitro in bone marrow-derived macrophages. RNA sequencing analyses revealed dramatic up-regulation of the red-pulp macrophage lineage-defining transcription factor SpiC and its associated transcriptional program, which was also present in bone marrow macrophages sorted from patients with sHLH. Transcriptional profiling also revealed a unique metabolic transcriptional profile in these macrophages, and immunometabolic phenotyping revealed impaired mitochondrial function and oxidative metabolism and a reliance on glycolytic metabolism. Subsequently, we show that therapeutic administration of the glycolysis inhibitor 2-deoxyglucose was sufficient to rescue animals from HLH. Together, these data identify a potential mechanism for the pathogenesis of sHLH and a potentially useful therapeutic strategy for its treatment.
Hemophagocytic lymphohistiocytosis (HLH) has classically been designated as primary or secondary. Primary HLH is thought to be inherited in an autosomal or X-linked manner with defects in genes responsible for CD8 and NK-mediated cytotoxicity (1). Secondary HLH (sHLH) has been thought to be acquired, although genetic susceptibility may also play a role by reducing the threshold of inflammatory activation or attenuating antiinflammatory negative feedback control (2). Infection is associated with roughly half of all secondary HLH cases while rheumatic diseases (where it is often referred to as macrophage activation syndrome) and neoplasm account for the rest (3, 4). Primary and secondary HLH are mechanistically distinguished by the dependence on intrinsic natural killer (NK) cell dysfunction in the former, but not the latter. Rather, NK cell dysfunction is thought to be a result of exuberant cytokine signaling in secondary HLH, although the pathogenesis of this entity is poorly understood, especially because of the lack of mouse models and the limitations of extrapolating mechanisms from clinical observations (2). Lymphocytic choriomeningitis virus (LCMV) infection in perforin-deficient mice, among other genetic models, closely model primary HLH (5), but a dearth of models exist for secondary HLH. As a result, mortality associated with sHLH can be as high as 88% despite modern medical care and a variety of new immuno- and chemotherapeutics (6–8)
To our knowledge, there is only one proposed animal model of sHLH. Behrens et al. (9) described a model of secondary HLH using repeated CpG injection (10). In their model, CpG, a toll-like receptor 9 (TLR9) agonist, was injected repeatedly intraperitoneally and mice subsequently developed signs and symptoms of nonlethal HLH over the course of 10 d. In their model, HLH-2004 criteria (11) were not met, although application of these criteria, which were developed for the primary HLH patients, is inherently problematic. Nonetheless, serum cytokine levels in general were not dramatically elevated and likely related to the lack of mortality and lack of robust hemophagocytosis observed. The model is in stark contrast with what is observed in the adult secondary HLH human population, where mortality is high and related to multiorgan dysfunction resultant from exuberant inflammation (6).
In this study, we report that sequential challenging of mice with viral TLR agonists followed by the TLR4 agonist lipopolysaccharide (LPS) recapitulated many of the aspects of sHLH. We found that the hyperinflammatory phenotype was macrophage intrinsic and we were able to recapitulate this in vitro. Transcriptional profiling revealed a unique profile, with enrichment of the SpiC transcriptional program, which was also present in sorted bone marrow cells from patients with sHLH, although SpiC was not necessary for the hyperinflammatory phenotype. Pathway enrichment analyses also revealed a significant down-regulation in genes involved in fatty acid/oxidative metabolism. sHLH macrophages displayed impaired mitochondrial respiration. Inhibition of glycolysis using 2-deoxyglucose (2-DG) significantly attenuated the magnitude of inflammation in vitro and in vivo and was sufficient to protect animals with sHLH from mortality.
Results
Viral Priming Leads to a Hyperinflammatory LPS Response in Vivo.
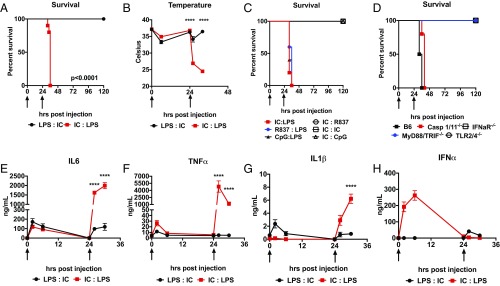
After first establishing lethal-dose zero (LD0) doses of Poly I:C and LPS (SI Appendix, Fig. S1 A and B), we found that sequentially challenging C57Bl6/J mice with LD0 dose of Poly I:C and then LD0 dose of LPS (IC:LPS), but not the reverse order, rapidly led to mortality in all animals (Fig. 1A), which was associated with a very rapid drop in their body temperature subsequent to LPS challenge (Fig. 1B). We had previously reported that glucose metabolism had an opposite effect on Poly I:C- and LPS-induced mortality and that treatment with 2-DG in the first 8 h subsequent to Poly I:C challenge led to high mortality (12). Therefore, we assessed blood glucose levels in IC:LPS-challenged animals, which were not significantly different (SI Appendix, Fig. S1C). We also challenged animals with 2-DG 24 h after priming with Poly I:C to exclude glucose utilization as the cause of IC:LPS mortality and did not note any enhancement in mortality (SI Appendix, Fig. S1D). We also excluded Poly I:C-induced deficiencies in circulating ketone bodies or FGF21 (SI Appendix, Fig. S1 E and F). We next examined a temporal requirement for inducing mortality subsequent to Poly I:C priming. We found that while prior exposure to Poly I:C 36 h before LPS challenge was sufficient to potentiate LPS mortality, this effect was lost when Poly I:C was introduced 48 h or longer before LPS (SI Appendix, Fig. S1G). To exclude the effect of genetic background and gender, we performed IC:LPS in BALB/c males and females and did not note a dependence on background or gender (SI Appendix, Fig. S1H).
Fig. 1.
Viral TLR agonism primes a hyperinflammatory LPS response. (A) Survival of 8-wk-old male C57Bl6/J mice treated with 5 mg/kg LPS i.p. or 10 mg/kg i.v. Poly I:C at the indicated times (arrows) in the indicated order (n = 10 mice per group). (B) Colonic temperatures of 8-wk-old male C57Bl6/J mice treated with LPS:IC or IC:LPS (n = 10 per group). (C) Survival of 8-wk-old male C57Bl6/J mice treated with 10 mg/kg Poly I:C, 5 mg/kg LPS, 10 mg/kg R837 i.p., 10 mg/kg CpG ODN1826 i.p. in the indicated order at the indicated times (arrows) (n > 8 per group). (D) Survival of 8-wk-old male C57Bl6/J, caspase 1/11 DKO, IFNaR KO, TLR2/4 DKO, or MyD88/TRIF DKO mice were challenged with IC:LPS (n > 8 per group). (E–H) Plasma IL-6, TNFα, IL-1β, or IFNα assessed following LPS and Poly I:C administered at the indicated time points (arrows) in the indicated order (n > 5 per group). ****P < 0.0001.
To test if any viral TLR agonist could prime LPS mortality, we subjected animals to sequential challenges with TLR3 (Poly I:C), TLR7/8 (R837), or TLR9 (CpG) agonists. While all viral agonists tested could prime LPS mortality, they could not prime mortality to each other (Fig. 1C). These data suggested that a conserved feature in viral inflammation was responsible for priming the LPS response.
We next examined the signaling pathways required for IC:LPS-induced mortality. Previous studies had demonstrated an important role for caspase 11 in the LPS response, and in particular in the intracellular LPS response, which could be primed by Poly I:C (13, 14). To test if caspase 11 was required for IC:LPS-induced mortality, we subjected caspase 1/11 double knockout mice to IC:LPS and did not observe any protection (Fig. 1D). While neither caspase 1 nor 11 were required for IC:LPS-induced mortality, mice lacking MyD88/TRIF or TLR2/4 were protected from IC:LPS (Fig. 1D). These data suggested that canonical TLR4 signaling was required for IC:LPS mortality. Since a variety of viral TLR agonists were capable of priming LPS, and type I IFN signaling has been shown to be important in the LPS response (15–17), we performed IC:LPS challenge in mice deficient in IFN α/β receptor 1 (IFNAR) and found that IFNAR was necessary for IC:LPS-induced mortality (Fig. 1D).
Finally, we compared the systemic inflammatory response between mice challenged with LPS:IC versus IC:LPS during primary and secondary challenge. We found that Poly I:C priming followed by LPS resulted in dramatically elevated levels of TNF, IL-1β, and IL-6 (Fig. 1 E–G) but not IFNα (Fig. 1H). Taken together, we found that sequential challenge with nonlethal doses of Poly I:C and LPS resulted in superinduction of inflammatory response and mortality.
Poly I:C Priming Followed by LPS Challenge Recapitulates Most Aspects of HLH.
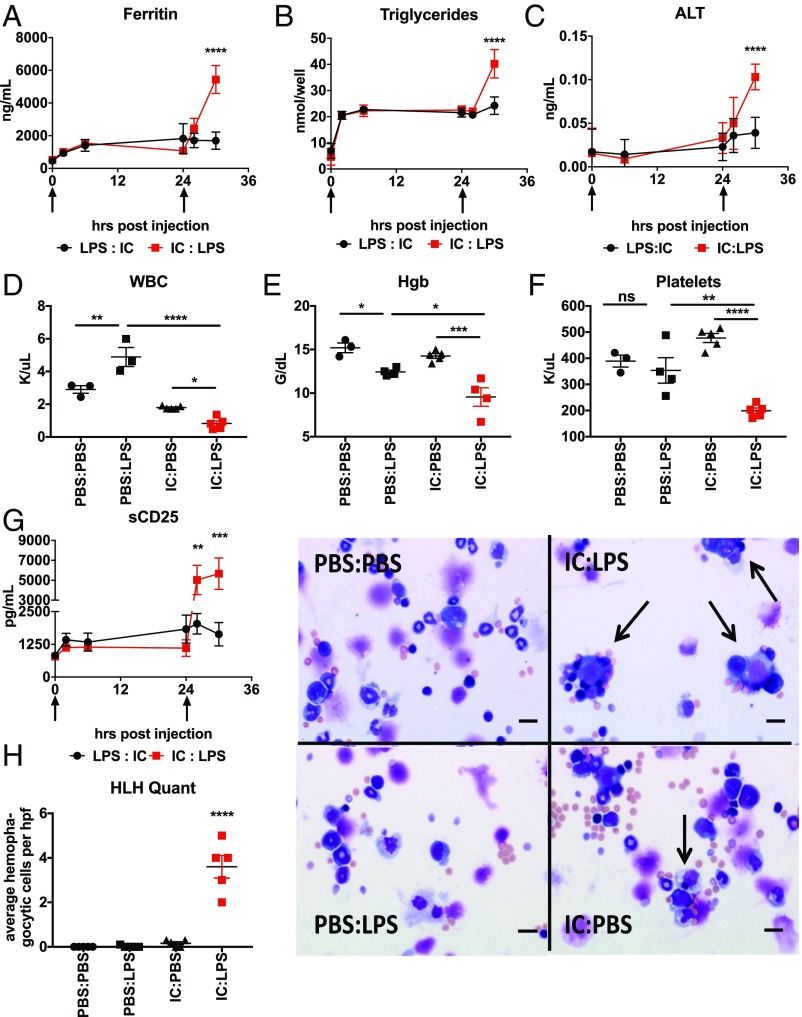
The cytokine storm induced by IC:LPS challenge was reminiscent of the hyperinflammatory state of HLH (18). To assess if other features of HLH were present in these mice, we examined plasma levels of ferritin (Fig. 2A), triglycerides (Fig. 2B), alanine aminotransferase (Fig. 2C), and soluble IL-2 receptor (Fig. 2G). Consistent with HLH, we observed robust increases in all three plasma biomarkers subsequent to LPS challenge in Poly I:C primed versus unprimed animals. Consistent with the lack of hyperinflammation and mortality, LPS:IC challenge did not have the same effect.
Fig. 2.
IC:LPS induces sHLH-like phenotype in mice. (A–C) Plasma ferritin (A), triglycerides (B), ALT (C) was assessed following LPS and Poly I:C administered at the indicated time points (arrows) in the indicated order (n = 5 per group). (D–F) Complete blood counts were performed on mice challenged sequentially at t = 0 and then t = 24 h with LPS, Poly I:C, or vehicle (PBS) in the indicated order. White blood cells (D), hemoglobin (E), and platelets (F) are displayed. (G) Soluble CD25 was assessed following LPS and Poly I:C administered at the indicated time points (arrows) in the indicated order (n = 5 per group). (H) Representative cytospin images of hemophagocytosis (arrows) from bone marrow aspirates harvested from mice 4 h after challenge with IC:LPS (Right) with quantification (Left). ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (Scale bars, 20 µm.)
One characteristic feature of HLH is hemophagocytosis, which is the ability of macrophages, usually seen in the bone marrow, liver, and spleen, to phagocytize leukocytes leading to consumptive peripheral cytopenias of rapid onset (19). To test if IC:LPS led to cytopenias, we performed complete blood cell characterization of animals challenged with IC:LPS and found that Poly I:C primed animals developed profound pancytopenia 6 h after LPS compared with unprimed animals (Fig. 2 D–F). Finally, we observed significantly more hemophagocytosis in cytospin preparations of bone marrow subsequent to IC:LPS compared with other stimulation conditions (Fig. 2G). Taken together, these data suggest that sequential TLR challenges in mice are sufficient to replicate most features of sHLH.
Influenza Infection Potentiates Mortality of Nonpathogenic Escherichia coli Peritonitis.
Having identified that Poly I:C priming caused a hyperinflammatory LPS response, we asked if our model could be recapitulated with live infections to assess the effect of hyperinflammation on pathogen control. We had previously shown in a different system that influenza induced a glucocorticoid response that attenuated the host response to the intracellular Gram-positive bacteria Listeria monocytogenes, which led to increased pathogen load and mortality (20). We also had previously shown that influenza infection followed by Legionella pneumophilia resulted in mortality due to defective tissue repair but without an effect on pathogen load or magnitude of inflammatory response (21).
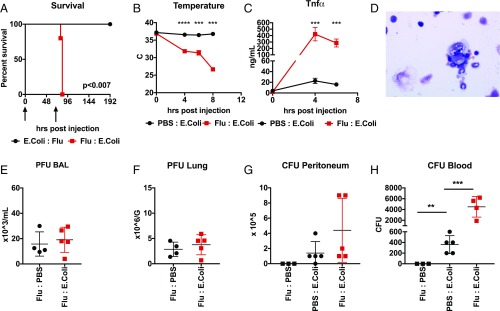
Because sequential IC:LPS challenge mimics viral–bacterial coinfections, we next examined whether IC:LPS response could be recapitulated using live infections. We chose to utilize the nonpathogenic DH5α strain of E. coli (22). We verified that high cfu i.p. inoculations of DH5α were unable to cause mortality (SI Appendix, Fig. S2A). To model viral priming, we infected mice with influenza virus intranasally and then, 3 d after infection, we challenged them with i.p. DH5α. We found that infection with influenza followed by DH5α resulted in 100% mortality (Fig. 3A). We also tested if other routes of entry of single-stranded RNA viruses would produce the same result. We found that i.v. LCMV infection could also potentiate DH5α mortality (SI Appendix, Fig. S2B). Consistent with our IC:LPS model, influenza-infected mice rapidly dropped body temperature after DH5α challenge (Fig. 3B) and exhibited greatly enhanced plasma cytokine levels (Fig. 3C) as well as evidence of hemophagocytosis in the bone marrow (Fig. 3D).
Fig. 3.
Influenza pneumonia primes mortality from nonpathogenic E. coli peritonitis. (A) Survival of 8-wk-old male C57Bl6/J mice (n = 10 per group) infected with influenza intranasally or DH5α E. coli i.p. in the indicated order at the indicted times (arrows). (B) Colonic temperatures of 8-wk-old male C57Bl6/J mice treated with influenza followed by E. coli or visa versa (n = 10 per group). (C) Plasma TNFα in mice treated with influenza or vehicle followed by E. coli (n = 5 per group). (D) Representative image of bone marrow aspirate of mice treated with influenza followed by E. coli. (Magnification: 100×.) (E and F) Plaque forming units of influenza from the bronchoalveolar lavage (E) or lung parenchyma (F) of mice treated with the indicated sequence of challenges 8 h after the second challenge. (G and H) Colony forming units of E. coli from the peritoneal lavage (G) or blood (H) of mice treated with the indicated sequence of challenges 8 h after the second challenge. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Interestingly, despite the enhanced TNF production, we found significantly higher DH5α bacterial burden in the blood of animals preinfected with influenza compared with controls (Fig. 3H). The amount of DH5α recovered from the peritoneal lavage also trended slightly higher (Fig. 3G). However, the amount of plaque forming units of influenza did not differ significantly between groups (Fig. 3 E and F). Taken together, these data indicate that secondary peritonitis, even with nonpathogenic bacteria, can cause a hyperinflammatory hemophagocytic phenotype akin to sHLH and suggest that despite hyperinflammation, pathogen control may be paradoxically impaired, as has been previously demonstrated in primary HLH (23, 24). Given our other previously reported coinfection models (20, 21), these data also indicate that certain pathogen pairs and not others may predispose to sHLH.
Hyperinflammation and Hemophagocytosis Are Macrophage Intrinsic.
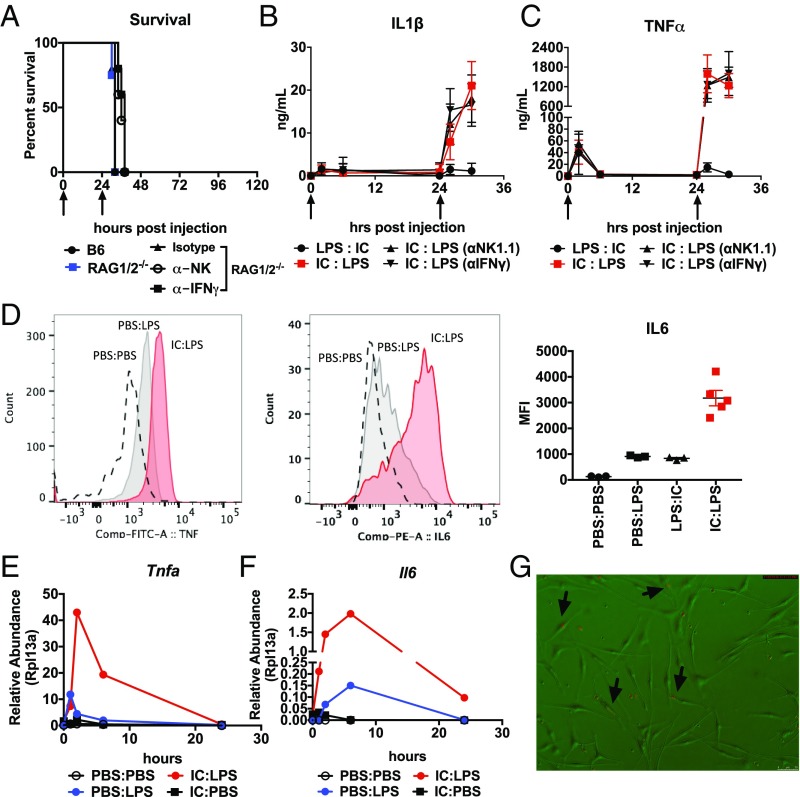
To dissect the mechanism of hyperinflammation in our IC:LPS model, we first asked if our model is dependent on NK cells or T cells, which are required in primary HLH. Thus, we tested IC:LPS challenge in RAG 1/2-deficient animals, as well as RAG 1/2-deficient animals in which NK cells had been depleted using the NK1.1 antibody (Fig. 4A). We found that T lymphocytes and NK cells were dispensable in mediating IC:LPS. Since IFNγ is a potent driver of macrophage activation and also implicated in the pathogenesis of primary HLH, we also tested whether or not IFNγ was required for IC:LPS response, which we tested by introducing IFNγ blocking antibodies into RAG 1/2-deficient mice (Fig. 4A). Concordant with the mortality data, RAG 1/2-deficient animals in which NK cells or IFNγ were depleted did not display any reduction in plasma cytokine levels (Fig. 4 B and C).
Fig. 4.
IC:LPS induced sHLH is macrophage intrinsic. (A) Survival of 8-wk-old male C57Bl6/J, RAG1/2 DKO, or RAG1/2 DKO treated with antibodies to NK1.1 or IFNγ challenged with IC:LPS (n > 8 per group). (B and C) Plasma IL-1β (B) and TNFα (C), following LPS and Poly I:C administered at the indicated time points (arrows) in the indicated order (n = 5 per group). (D) Representative histogram plots of ex vivo intracellular analyses of TNF (Left) and IL-6 (Center, quantified on the Right) of CD11b+ F480+ cells isolated from the liver (TNF) and bone marrow (IL-6) of mice challenged with indicated treatments (n > 3 per group). (E and F) Relative abundance of Tnfa (E) and Il6 (F) in BMDMs sequentially challenged with the indicated treatments (representative experiment, average of two experimental replicates per time point per group). (G) Representative image of BMDMs (bright field) challenged with IC:LPS incubated with CFSE-labeled (red) bone marrow cells (arrows indicate BMDMs which contain CFSE-labeled cells). (Magnification: 100×.)
To identify the source of the cytokine producing cell, we isolated leukocytes from IC:LPS-challenged animals and incubated them without any additional stimulus in the presence of Brefeldin and assessed the identity of cytokine producing cells. We found that CD11b+ F480+ cells in the liver robustly produced IL-6, and bone marrow CD11b+ F480+ cells robustly produced TNF (Fig. 4D). We also found that peritoneal macrophages were also robust producers of TNF (SI Appendix, Fig. S3E). We were unable to detect more CD11b+ F480+ cells in the peritoneum, bone marrow, liver, or spleen of animals challenged with IC:LPS, excluding the possibility that the hyperinflammation was also due to an increase in total macrophage numbers (SI Appendix, Fig. S3 A–D). We were also unable to robustly detect IL-6 in other cells within the liver, including in CD11b+ F480− cells (SI Appendix, Fig. S3F). Taken together, we hypothesized that the hyperinflammation in IC:LPS is largely macrophage intrinsic.
To further test this, we asked if we could induce hyperinflammatory response in isolated bone marrow-derived macrophages (BMDMs). We found that BMDMs displayed dramatically increased magnitude and accelerated kinetics of transcription of inflammatory genes subsequent to Poly I:C priming (Fig. 4 E and F). We found that secondary response genes (25, 26), such as IL-6 and MX1 were highly induced within 1 h of LPS stimulation when primed 24 h earlier with Poly I:C (Fig. 4F and SI Appendix, Fig. S3G). As with our in vivo model, we found that Poly I:C could not prime the Poly I:C response (SI Appendix, Fig. S3H), caspase 1/11 was dispensable, IFNAR was required for the hyperinflammatory response (SI Appendix, Fig. S3I), and the Poly I:C priming effect was lost after 36 h (SI Appendix, Fig. S3J). Although IFNAR was required, IFNβ was insufficient to prime LPS (SI Appendix, Fig. S3H). Finally, we found that only IC:LPS-treated macrophages, but not other treatment conditions, could phagocytize freshly prepared bone marrow cells that were labeled with carboxyfluorescein succinimidyl ester (CFSE) (Fig. 4G). Taken together, we demonstrate that the hyperinflammatory hemophagocytic phenotype of IC:LPS macrophages is cell intrinsic and has a transcriptionally distinct inflammatory profile compared with conventional LPS-stimulated macrophages.
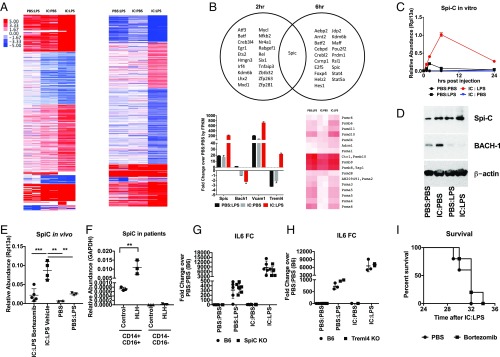
Macrophage SpiC Expression Is a Potential Biomarker of sHLH.
To characterize the transcriptional profile of IC:LPS macrophages, we performed RNA sequencing of macrophages stimulated with Poly I:C followed by LPS (IC:LPS), PBS followed by LPS (PBS:LPS), and Poly I:C followed by PBS (IC:PBS) at 2 and 6 h post LPS or PBS (Fig. 5A). We identified differentially expressed genes in IC:LPS compared with IC:PBS- and PBS:LPS-treated macrophages and focused our analyses on differentially expressed transcription factors. We identified one transcription factor specifically induced in IC:LPS macrophages, which was not present in PBS:LPS or IC:PBS at both time points and identified the E26 transformation-specific (ETS) family member SpiC as well as SpiC-regulated genes Vcam1, Bach1, and Treml4 (Fig. 5B). SpiC is the transcriptional master regulator of red-pulp macrophages (RPMs), which activates a transcriptional program that enables RPMs to phagocytose aged erythrocytes and metabolize iron and heme (27, 28). SpiC is thought to be negatively regulated by Bach1 which is proteasomally degraded in the presence of heme (28). Consistent with the proposed activation of SpiC in RPMs, IC:LPS macrophages displayed suppressed transcription of Bach1 and increased transcription of many proteasomal subunits (Fig. 5B). We confirmed SpiC induction in IC:LPS and characterized its kinetics by qRT-PCR (Fig. 5C). We also confirmed that SpiC protein was increased in IC:LPS and observed a concomitant decrease in Bach1 (Fig. 5D).
Fig. 5.
IC:LPS macrophages have a distinct transcriptional profile. (A) K-mean clustered heat map of uniquely differentially expressed genes in IC:LPS BMDMs (normalized to PBS:PBS, expressed as log2 fold change) at 2 h (Left) and 6 h (Right) in BMDMs following the indicated sequential stimulations. (B) Venn diagram of significantly up-regulated transcription factors (Top). Expression of Spic pathway genes (Bottom Left) and heat map of proteasome genes (Bottom Right). (C) qRT-PCR validation of Spic in BMDMs. (D) Representative Western blot of SpiC and BACH-1 in BMDMs 8 h after the indicated treatments. (E) Relative abundance of Spic in bone marrow cells harvested from mice 4 h after the indicated treatments. (F) Relative abundance of Spic in CD14+ CD16+ cells sorted from bone marrow aspirates of patients with sHLH. (G and H) Fold change of Il6 in BMDMs from SpiC KO (G) or Treml4 KO (H) or control animals 2 h after the indicated treatments. (I) Survival of mice challenged with IC:LPS treated with bortezomib or vehicle (n > 8 per group). **P < 0.01, ***P < 0.001.
Next, we asked if SpiC was also induced in vivo. We found that treatment with IC:LPS significantly increased SpiC transcription (Fig. 5E). Consistent with the proteasome requirement for SpiC activation, we found that treatment in vivo with the proteasome inhibitor bortezomib significantly inhibited induction of SpiC. Finally, we obtained bone marrow aspirates from patients with sHLH, sorted CD14+ CD16+ cells, and assessed SpiC transcriptional levels in these cells versus normal controls and found that SpiC was highly expressed in sHLH bone marrow aspirates (Fig. 5F). To test if SpiC or its transcriptional target Treml4, which has been implicated in feed-forward amplification of viral TLR responses (29), were required for the hyperinflammatory response to IC:LPS, we performed IC:LPS stimulation on SpiC-deficient and Treml4-deficient BMDMs and found that they were dispensable in the hyperinflammatory phenotype (Fig. 5 G and H). Consistent with the dispensability of SpiC, treatment with bortezomib in vivo had no effect on survival despite attenuation of SpiC expression (Fig. 5I). Together, these data demonstrate that SpiC and its transcriptional targets are enriched in IC:LPS macrophages in vitro and in vivo in mouse and humans, making it potentially a useful biomarker for sHLH, but that they likely do not participate in the pathogenesis of sHLH.
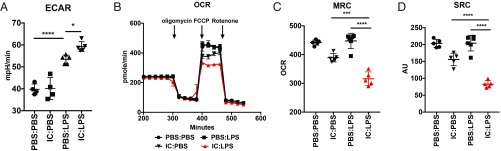
IC:LPS Macrophages Display an Altered Immunometabolic Profile.
Pathway analyses of up-regulated genes confirmed a hyperinflammatory phenotype in IC:LPS macrophages (SI Appendix, Fig. S4 A and B). Interestingly, pathway analyses of genes that were significantly down-regulated in IC:LPS BMDMs revealed significant enrichment in genes involved in OXPHOS and fatty acid metabolism (SI Appendix, Fig. S4 C and D), including the fatty acid transporter Cd36 and several genes involved in fatty acid oxidation, including Acaa2 and Hadh (SI Appendix, Fig. S4 E and F). Metabolic reprogramming of LPS-stimulated macrophages is well described (30, 31), and, given the enrichment in metabolic pathways in our transcriptome analyses, we asked if IC:LPS macrophages exhibited a different metabolic profile. Seahorse analyses of IC:LPS demonstrated that IC:LPS macrophages were significantly more glycolytic (Fig. 6A) and had significantly impaired OXPHOS metabolism (Fig. 6 B–D), consistent with the down-regulation of genes involved in fatty acid metabolism seen in our RNA-Seq data.
Fig. 6.
IC:LPS macrophages are metabolically distinct. (A) Glycolytic rate of BMDMs 2 h after the indicated treatments as determined by ECAR using Seahorse analyses. (B–D) OCR, maximal respiratory capacity (MRC), and spare respiratory capacity (SRC) of BMDMs 2 h after the indicated treatments as determined by Seahorse analyses. *P < 0.05, ***P < 0.001, ****P < 0.0001.
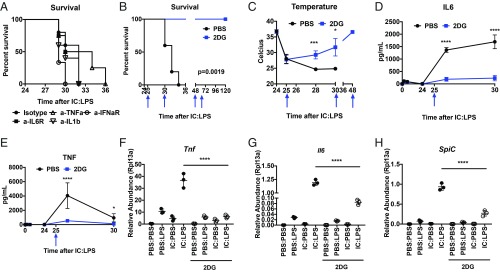
Given that IFNAR was required for mortality in IC:LPS (Fig. 1D) and the role of TNF, IL-1, and IL-6 in driving mortality in multiple models of inflammation (32, 33), we asked if cytokine blockade would be sufficient to rescue IC:LPS-challenged animals. While TNF blockade extended viability of IC:LPS-challenged animals, blocking TNF, IL-6, IL-1, or IFNAR did not significantly prolong survival (Fig. 7A). Proteasome inhibition (Fig. 5I) and IFNγ blockade (Fig. 4A) were also unable to confer protection. These data are consistent with the poor efficacy of cytokine blockade in patients with sHLH (34, 35). Given the glycolytic phenotype of IC:LPS-treated macrophages, we next asked if inhibition of glycolysis with the 2-DG can reduce inflammation and improve survival in our HLH model. Indeed, when we treated IC:LPS-challenged animals with 2-DG, we noted a dramatic improvement in survival (Fig. 7B). Consistent with prolonged survival, animals regained their body temperature (Fig. 7C).
Fig. 7.
Inhibition of glycolysis rescues animals from IC:LPS-induced sHLH. (A) Survival of animals challenged with IC:LPS treated with the indicated blocking antibodies 1 h and 6 h (if viable) after LPS (n > 8 per group). (B) Survival of animals challenge with IC:LPS treated with 2-DG or PBS at the indicated times (blue arrows) (n = 10 per group). (C) Colonic temperatures of animals challenged with IC:LPS treated with 2-DG or PBS at the indicated times (blue arrows) (n = 5 per group). (D and E) Plasma IL-6 (D) and TNFα (E) of animals challenged with IC:LPS treated with 2-DG or PBS at the indicated time (blue arrow) (n = 5 per group). (F–H) Relative abundance of Tnfa (F), Il6 (G), and Spic (H) of BMDMs 2 h after the indicated treatments. *P < 0.05, ***P < 0.001, ****P < 0.0001.
Previous studies had shown that deletion of PDK1, Hif1a, Glut1, or pharmacological inhibition of glycolysis with 2-DG reduced transcriptional induction of inflammatory genes subsequent to LPS stimulation (36–39). We thus asked if circulating inflammatory cytokines induced by IC:LPS were attenuated in vivo with 2-DG treatment. We observed that 2-DG significantly suppressed circulating TNFα and IL-6 (Fig. 7 D and E). Indeed, IC:LPS-challenged animals were surprisingly sensitive to 2-DG, consistent with our observation that the hyperinflammatory phenotype was macrophage intrinsic (Fig. 4) and dependent on glycolysis (Fig. 6). Consistent with our in vivo observation, 2-DG significantly attenuated Tnf and Il6 induction in BMDMs challenged with IC:LPS (Fig. 7 F and G). Spic induction was also attenuated with 2-DG (Fig. 7H). Taken together, these studies demonstrate that in our model of sHLH, macrophages develop a transcriptional repression of genes involved in fatty acid metabolism and preference for glycolysis, and that glucose utilization by these macrophages is required for the hyperinflammatory response in vitro and in vivo.
Discussion
sHLH is a mortal complication associated with infections with unknown etiology. In this study, we demonstrate that sequential activation first with viral TLR agonists followed by TLR4 agonist is sufficient to recapitulate most features of sHLH (Figs. 1–3). We found that this phenotype was macrophage intrinsic (Fig. 4). Transcriptional profiling of IC:LPS macrophages revealed a SpiC signature, which is normally observed only in red-pulp macrophages responsible for the phagocytosis of aged red blood cells, and we found that SpiC transcription was also present in bone marrow aspirates from patients with sHLH, the site where hemophagocytosis is most commonly observed (Fig. 5). The up-regulation of SpiC is likely due to phagocytosis of RBCs. sHLH macrophages also demonstrated a significant decrease in multiple genes involved in fatty acid metabolism, and metabolic analyses revealed a preference for glycolysis and a defect in mitochondrial respiration (Fig. 6). Finally, we found that these macrophages were dependent on glycolysis for the hyperinflammatory response and that administration of 2-DG was sufficient to rescue animals from sHLH (Fig. 7).
sHLH secondary to infection, like all sepsis syndromes, is likely a heterogeneous collection of diseases with many etiologies that lead to a similar phenotype. Genetic predisposition, especially in genes involved in the pathogeneses of primary HLH, likely plays a role in some cases (2). Repeated TLR9 stimulation produces a more protracted less fulminant disease course which is sometimes seen (9), and given our findings, it would be interesting to ask whether commensal Gram-negative bacteria are required for this phenotype. It is also interesting that chronic TLR7 activation, as is seen in yaa murine strains and TLR7 transgenic models (40–42), has not similarly been described to exhibit HLH-like phenotypes. In humans, sHLH can be seen in a variety of contexts, including in neoplasms, autoimmune diseases (primarily adult-onset Still’s disease and systemic lupus erythematosus), and infections. It remains to be seen if TLR pathways are the common upstream mechanism for disease pathogenesis in these various contexts and if the sHLH phenotypes that arise from these different contexts are distinct from each other.
We previously reported that influenza could potentiate a listerial infection (20) and promote L. pneumophila pneumonia (21), although in both cases the potentiation was not due to increased inflammation. Here we found that influenza infection could potentiate a nonpathogenic strain of E. coli but that this sequence caused a hyperinflammatory phenotype that closely resembles sHLH. We found that we could further reduce our model to the use of TLR agonists and showed that a variety of viral TLR agonists could create a maladaptive hyperinflammatory LPS response, consistent with the observation that type I IFN via IFNAR primes macrophage LPS responses (43). Many factors may account for these different outcomes in virally primed coinfection models, and it is likely that each coinfection exposes specific vulnerabilities in the host. Pathogen-specific immunity also likely plays an important role. Unlike E. coli, Listeria and Legionella are both pathogens that maintain a part of their lifecycle within host cells (44, 45) and have been shown to activate intracellular pathogen sensing pathways in addition to pathways activated by flagella and cell wall components that are distinct to these pathogens (46, 47). The portal of infection and sites of tissue injury also likely play an important role. What is interesting is that in all cases, viral priming of bacterial infections appears to be detrimental while the converse is not often observed except in cases of severe immunosuppression and viral reactivation, as is in the case of many human herpes viruses where its impact on outcomes is unclear (48, 49). Thus, coinfection models, although complex in their biology, may be a valuable tool to expose a finite set of host vulnerabilities that can then be better understood and translated into therapies.
The exact mechanism by which viral TLR agonism primes a hyperinflammatory TLR4 response remains to be elucidated. Unlike tissue resident macrophages that often express tissue-specific transcription factors (50, 51), it is possible that this maladaptive macrophage state is not coordinated by a distinct set of transcription factors. SpiC was a likely candidate since it was highly up-regulated in IC:LPS-challenged macrophages, coordinates the erythrophagocytic program in red-pulp macrophages (27, 28), and its downstream target Treml4 was recently implicated in feed-forward amplification of viral inflammation in myeloid cells (29). However, neither SpiC nor Treml4 was required for the IC:LPS phenotype (Fig. 5). At the same time, it may be a useful biomarker for diagnosis of sHLH, which is a difficult diagnosis to make based on a clinical diagnosis that incorporates a combination of nonspecific biomarkers of inflammation. It is likely that chromatin remodeling following viral TLR agonism plays a prominent role in the hyperinflammatory LPS response (26, 52–54), and more work remains to be done on dissecting the exact molecular mechanisms underlying IC:LPS response in macrophages. Finally, how IC:LPS-activated macrophages are capable of phagocytosing live cells remains an area of active investigation.
IC:LPS macrophages adopt a unique metabolic profile which we were able to exploit to rescue animals from sHLH (Figs. 6 and 7). It is now clear that specific metabolic programs fuel immune cell effector functions and that disruption of required metabolic pathways disrupts the ability for cells to perform these functions (30, 55, 56). We found that IC:LPS macrophages have altered mitochondrial respiration and are dependent on glycolysis. The administration of 2-DG was sufficient to rescue animals from sHLH-induced mortality and to attenuate the hyperinflammatory transcriptional program in vitro, suggesting that targeting glycolytic pathways may be a therapeutic approach to sHLH. It is interesting that the inhibition of glycolysis using 2-DG produces pleiotropic context-dependent outcomes. We have previously shown that while administration of 2-DG to animals challenged with Poly I:C or influenza resulted in driving the unfolded protein response toward neuronal cell death leading to enhanced mortality, administration of 2-DG to animals challenged with LPS or Listeria resulted in enhanced survival by mechanisms which are yet to be fully identified but independent of the magnitude of the inflammatory response (12). We have also recently shown using mouse models of malaria that 2-DG protected animals from cerebral malaria by affecting hemostasis while others have shown that it potentiates mortality in malarial models of fatal anemia (57, 58). It is likely that the composite outcome of 2-DG depends on the given set of biological processes that require glucose in any particular organismal state.
In summary, we report a murine model of sepsis, which highly resembles the infection-associated form of sHLH. We found that this phenotype was macrophage intrinsic and identified SpiC as a potentially useful biomarker for marking sHLH macrophages. Characterization of these macrophages revealed altered mitochondrial respiration and a reliance on glycolysis. Although our study was not designed to be a therapeutic study, our findings suggest that there may be a therapeutic role for the inhibition of glycolysis in sHLH, which remains a highly mortal infection-associated complication.
Methods
Animals.
Mice 8–10 wk of age were used. C57BL/6J, BALB/c, TLR2/4 double knockout and MyD88/TRIF double knockout mice, and IFNaR knockout mice were purchased from The Jackson Laboratory and bred at Yale University. SpiC knockout bone marrow cells were a kind gift from Malay Haldar, University of Pennsylvania, Philadelphia, PA. Treml4 knockout bone marrow cells were a kind gift from Terry Mean, Massachussets General Hospital, Harvard Medical School, Boston, MA. All animal experiments were performed according to institutional regulations upon review and approval of Yale University’s Institutional Animal Care and Use Committee.
Infections.
Influenza virus strain A/WSN/33 was a kind gift from the laboratory of Akiko Iwasaki, Yale University School of Medicine, New Haven, CT, and propagated using Madin-Darby canine kidney (MDCK) cells and titrated to determine concentration by plaque assay described below. For influenza infection, mice were anesthetized with a ketamine/xylazine mixture and indicated pfus of influenza in 30 µL PBS was administered intranasally dropwise. E. coli (DH5α) was purchased from Life Technologies and introduced at the indicated cfus intraperitoneally. For LCMV infections, mice were infected intraperitoneally with 2 × 105 pfu LCMV Armstrong.
Quantification of Influenza and DH5α.
To determine influenza viral titers in the lung, lung tissue was harvested, weighed, and disrupted by bead homogenization. For viral titers in the bronchoalveolar lavage (BAL) fluid, BAL was performed in euthanized animals via cannulation of the trachea, and lungs were lavaged with 1 mL of PBS. Virus titer was determined by infection of MDCK cells with titrated amounts of lung homogenate and BAL followed by addition of an agar overlay for 48 h after which cell monolayers were stained with crystal violet and plaque numbers were determined. To determine DH5α in the blood, peripheral blood was obtained by retroorbital collection and plated by serial dilution on LB agar plates. DH5α in the peritoneal lavage was determined by lavage with PBS and then serial dilution on LB agar plates.
Treatments.
LPS derived from E. coli 055:B5 (Sigma-Aldrich) diluted in PBS was injected intraperitoneally with the indicated doses. High molecular weight Poly(I:C) (InvivoGen) diluted in PBS was injected retroorbitally at the indicated doses. R837 (InvivoGen) was diluted in PBS and injected intraperitoneally at 10 mg/kg. CpG ODN 1826 (InvivoGen) was diluted in PBS and injected intraperitoneally at 10 mg/kg. Two-DG (Sigma) was reconstituted in water and injected intraperitoneally at the indicated times 200 mg/kg per dose. Bortezomib (Selleck, LLC) diluted in PBS was injected at 0.1 mg/kg intraperitoneally at the indicated times. Anti-NK1.1 antibody (PK136; Bio X Cell) diluted in PBS was injected at 100 μg per injection (100 μL) retroorbitally concurrently with each challenge. Anti-IFNγ antibody (XMG1.2, Bio X Cell) diluted in PBS was injected at 500 μg per injection (100 μL) retroorbitally concurrently with each challenge. Anti-IL6R (15A7, 10 mg/kg; Bio X Cell), anti-TNFα (XT3.11, 500 μg per injection; Bio X Cell), anti-IFNaR (MAR1-5A3, 10 mg/kg; Bio X Cell), anti-IL1β (B122, 500 μg per injection; Bio X Cell), and isotype antibody were diluted in PBS and injected retroorbitally.
Quantification of Plasma Cytokines, Biomarkers, and Complete Blood Count.
Plasma TNFα and IL-6 concentrations were assayed by sandwich ELISA using capture antibodies (eBioscience), biotin-conjugated detection antibodies (eBioscience and BD Biosciences, respectively), HRP-conjugated streptavidin (BD Biosciences), and TMB substrate reagent (BD Biosciences). Plasma IFNα and IL-1β concentrations were measured using kits according to the manufacturer’s protocols (eBioscience). Plasma Tropinin-I concentration (Life Diagnostics) and alanine aminotransferase (ALT) activity (Cayman Chemical) were assayed using kits according to manufacturers’ protocols. Plasma ferritin concentrations (Abcam) were determined using the kit according to the manufacturer’s protocols. sCD25 concentrations (MyBioSource) were determined using the kit according to the manufacturer’s protocols. For complete blood counts, 100 μL of whole blood was collected by the retroorbital route into EDTA (Gibco), and a Hemavet HV950FS (Drew Scientific) was utilized to analyze peripheral blood composition.
Cell Culture.
BMDMs were prepared from male animals 6–12 wk of age. Animals were killed by CO2 asphyxiation and femurs and tibias were isolated. The bone marrow cells were isolated and grown in macrophage growth medium [RPMI 1640 supplemented with 10% FBS (Gibco), 1% penicillin–streptomycin (Gibco), 2 mM l-glutamine (Gibco), 1 mM sodium pyruvate (Gibco), 0.01 M Hepes (AmericanBio), and 30% L929-conditioned media as a source of CSF-1] and plated on Petri dishes. Macrophage growth medium was supplemented on day 3. Cells were plated for use on day 6. For sequential stimuli, cells were first stimulated with 100 ng/mL Poly I:C (InvivoGen), 5 ng/mL LPS derived from E. coli 055:B5 (Sigma-Aldrich), or 100 units/mL recombinant IFNβ (R&D Systems). Twenty-four hours or at the indicated time after initial stimulation, the media were removed and cells were washed twice with warmed macrophage growth media, and then the media was replaced with Poly I:C or LPS. Two-DG was used at 2.5 mg/mL and added with the second stimulation.
For ex vivo cytokine analyses, peritoneal cells obtained from lavage, bone marrow cells obtained from the flushing of femurs and tibia, or leukocyte-enriched cells from liver from animals 2 h after animals received LPS or control, were seeded in triplicate in complete RPMI in the presence of GolgiPlug (BD Biosciences) and incubated for 2 h. Briefly, livers were mechanically disrupted and digested for 30 min in 2 mg/mL Collagenase IV (Worthington) and then centrifuged through 44%/66% Percoll gradient and cells at the interface were collected. Cells were then stained for extracellular markers. The following antibodies were utilized for FACS analyses: CD45 BUV395 (BD Biosciences), EMA (Thermo Fisher Scientific), F4/80 APC (BD Biosciences), and CD11b Pacific Blue (BD Biosciences). Intracellular staining was performed using the BD Fix/Perm kit and TNF FITC (BD Biosciences), IL6 PE (BD Biosciences). CD45+ cells within the singlet live gate, as defined by size, granularity, pulse width, and EMA negative cells were acquired for analyses. Samples were acquired on an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo (Tree Star Technologies).
For feeding of live cells to BMDMs, bone marrow cells were flushed from isogenic animals and cells were labeled with 3 μM CFSE and washed thoroughly. Target (10:1) to BMDM cells was added 2 h after stimulation and cells were imaged continuously using an AF6000 Modular System (Leica) in combination with a stage-top incubator INUBTFP-WSKM-F1 (Tokai Hit). Image acquisition, deconvolution, and maximum projection analysis were performed with the program LAS AF (Leica).
Human Samples.
Bone marrow aspirates from three patients with a diagnosis of sHLH were obtained from the Hematology Biorepository at Yale University (Stephanie Halene). All human samples were obtained after informed consent and procured according to institutional regulations upon review and approval of Yale University's Human Investigation Committee. Control bone marrow cells were purchased from Lonza. Samples were stained with CD14 APC and CD16 PE (BD Biosciences) and sorted by FACS for CD14+ CD16+ positive and negative cells. Cells were lysed with RLT and RNA was extracted as described above.
RNA Extraction and RNA Sequencing and Analyses.
For cell culture experiments, at the indicated times after stimulation, media were removed from the wells and cells were washed twice with PBS. RLT (Qiagen) was added to lyse cells and RNA extraction was performed using Qiagen RNAeasy kits as per manufacturer’s instructions. For RNA sequencing library preparation, on-column DNase digestion was performed as per manufacturer’s instructions. Sequencing libraries were constructed following Illumina Tru-seq stranded mRNA protocol. Paired-end sequencing performed with Next-seq 500 with 76-bp reads from each end. Illumina fastq files were downloaded from Illumina Basespace and aligned with Tophat2, transcripts were then assembled with Cufflinks, merged with Cuffmerge, quantified using Cuffquant, and normalized using Cuffnorm. The genes’ fragments per kilobase per million (FPKM) table from Cuffnorm was analyzed with Microsoft Excel to identify differentially expressed genes. All treatment conditions were normalized to the average of PBS:PBS control values. Unique IC:LPS expressed genes were determined by being greater than twofold different from both PBS:LPS and IC:PBS. Pathway enrichment analyses were done with MSigDB (software.broadinstitute.org/gsea/login.jsp), GO Enrichment Analyses (www.geneontology.org/page/go-enrichment-analysis), and KEGG Pathway Maps (https://www.genome.jp/kegg/kegg3a.html). The RNA sequencing data set can be found at the Gene Expression Omnibus data repository (59).
qRT-PCR.
RNA was purified from cells using QIAGEN RNeasy columns according to the manufacturer’s instructions. cDNA was generated with MMLV reverse transcriptase (Clontech) using oligo-dT6 primers (Sigma-Aldrich). qRT-PCR was performed on a CFX96 real-time system (Bio-Rad) using PerfeCTa SYBR Green SuperMix (Quanta Biosciences). Relative expression units were calculated as transcript levels of target genes relative to Rpl13a. Primers used for qRT-PCR are listed in SI Appendix, Table S1.
Western Blot.
Cells were lysed in RIPA buffer containing HALT protease and phosphatase inhibitor mixture (Thermo Fisher Scientific) according to the manufacturer’s protocols. Equal amounts of protein were loaded per well of a 10% Bis-Tris minigel (Invitrogen) and run in 1× Mops buffer (Invitrogen). Protein was transferred onto activated PVDF membrane (Millipore), then blocked in 5% milk in TBST (20 mM Tris, 150 mM NaCl, 0.05% Tween 20) for 30 min. Membranes were incubated with primary antibodies, anti-SpiC antibody (Abcam), anti-BACH1 antibody (R&D Systems), and anti–β-actin (Sigma Aldrich), overnight at 4 °C. Membranes were washed with TBST, then incubated with secondary antibodies for 1 h at room temperature. After washing, protein was visualized using enhanced chemiluminescence reagent (Thermo Fisher Scientific).
Metabolic Profiling.
FCCP, oligomycin, and rotenone were obtained from Sigma-Aldrich. Analysis of the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) was performed with a Seahorse XF96 Extracellular Flux Analyzer instrument in BMDMs. BMDMs were seeded overnight in quadruplicate at a density of 1 × 105 cells per well on a pretreated poly-d-lysine-coated 96-well polystyrene Seahorse plate in RPMI-1640 medium containing l-glutamine, 10% FCS, and 20% L929 supernatant and stimulated as described above. Before starting the assay, cells were washed and incubated in Seahorse assay medium supplemented with 10 mM glucose and 1 mM sodium pyruvate in a 37 °C incubator without CO2 for 45 min. Oligomycin (ATPase inhibitor, 1 μM), FCCP (0.2 μM), and rotenone (0.5 μM) were injected where indicated and the ECAR (mpH/min) and OCR (pmoles O2/min) were measured in real time.
Blood Glucose, Plasma Ketone Bodies, and Plasma-Free Fatty Acids.
Glycemia was measured by whole blood collection via the retroorbital plexus and assessed using a glucometer (OneTouch). Plasma was separated using lithium heparin-coated microcentrifuge tubes (BD Diagnostics). Plasma-free fatty acid was measured using a kit according to manufacturer’s instructions (Wako Diagnostics). Plasma beta-hydroxybutyrate was measured using a kit per manufacturer’s instructions (Cayman Chemical). Plasma L-type triglycerides were measured according to manufacturer’s instructions (Wako Diagnostics).
Histology.
All mice were anesthetized utilizing CO2 asphyxiation. Cytospin preparations were made according to the manufacturer’s instructions using a Cytospin Cytocentrifuge (Thermo Fisher Scientific). Slides were stained with hematocylin and eosin by standard methods and cells were visualized using a Zeiss Axio Imager A1 microscope, an AxioCam MRc5 camera, and AxioVision 4.8.3.0 imaging software (Carl Zeiss MicroImaging, Inc.) by a veterinarian (C.J.B.) trained in veterinary pathology with extensive mouse experience, who was blinded to experimental groups.
Quantification and Statistical Analyses.
Statistical information is indicated in the text or figure legends. Statistical analyses were performed using Prism 6.0 (GraphPad Software, Inc.). Where appropriate, Student’s t test or analysis of variance (ANOVA) was used. The log-rank Mantel–Cox test was used to compare Kaplan–Meier curves. A P value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank members of the R.M. laboratory for helpful discussions. This study was supported by the Howard Hughes Medical Institute, Else Kröner Fresenius Foundation, and the Blavatnik Family Foundation. A.W. was supported by NIH Grants (T32 AR07107-39 and K08 AI128745). Bone marrow aspirate samples were obtained from Dr. Stephanie Halene and the Yale Cooperative Center of Excellence in Hematology National Institute of Diabetes and Digestive and Kidney Diseases Grant U54DK106857.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE124765).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820704116/-/DCSupplemental.
References
- 1.Usmani GN, Woda BA, Newburger PE. Advances in understanding the pathogenesis of HLH. Br J Haematol. 2013;161:609–622. doi: 10.1111/bjh.12293. [DOI] [PubMed] [Google Scholar]
- 2.Brisse E, Wouters CH, Matthys P. Advances in the pathogenesis of primary and secondary haemophagocytic lymphohistiocytosis: Differences and similarities. Br J Haematol. 2016;174:203–217. doi: 10.1111/bjh.14147. [DOI] [PubMed] [Google Scholar]
- 3.Ramos-Casals M, Brito-Zerón P, López-Guillermo A, Khamashta MA, Bosch X. Adult haemophagocytic syndrome. Lancet. 2014;383:1503–1516. doi: 10.1016/S0140-6736(13)61048-X. [DOI] [PubMed] [Google Scholar]
- 4.Atteritano M, et al. Haemophagocytic syndrome in rheumatic patients. A systematic review. Eur Rev Med Pharmacol Sci. 2012;16:1414–1424. [PubMed] [Google Scholar]
- 5.Jordan MB, Hildeman D, Kappler J, Marrack P. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood. 2004;104:735–743. doi: 10.1182/blood-2003-10-3413. [DOI] [PubMed] [Google Scholar]
- 6.Otrock ZK, Eby CS. Clinical characteristics, prognostic factors, and outcomes of adult patients with hemophagocytic lymphohistiocytosis. Am J Hematol. 2015;90:220–224. doi: 10.1002/ajh.23911. [DOI] [PubMed] [Google Scholar]
- 7.Parikh SA, Kapoor P, Letendre L, Kumar S, Wolanskyj AP. Prognostic factors and outcomes of adults with hemophagocytic lymphohistiocytosis. Mayo Clin Proc. 2014;89:484–492. doi: 10.1016/j.mayocp.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Hayden A, Park S, Giustini D, Lee AY, Chen LY. Hemophagocytic syndromes (HPSs) including hemophagocytic lymphohistiocytosis (HLH) in adults: A systematic scoping review. Blood Rev. 2016;30:411–420. doi: 10.1016/j.blre.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Behrens EM, et al. Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J Clin Invest. 2011;121:2264–2277. doi: 10.1172/JCI43157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard-Guyonvarc’h C, et al. Unopposed IL-18 signaling leads to severe TLR9-induced macrophage activation syndrome in mice. Blood. 2018;131:1430–1441. doi: 10.1182/blood-2017-06-789552. [DOI] [PubMed] [Google Scholar]
- 11.Henter JI, et al. HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48:124–131. doi: 10.1002/pbc.21039. [DOI] [PubMed] [Google Scholar]
- 12.Wang A, et al. Opposing effects of fasting metabolism on tissue tolerance in bacterial and viral inflammation. Cell. 2016;166:1512–1525.e12. doi: 10.1016/j.cell.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 14.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 15.Yoo CH, et al. Interferon β protects against lethal endotoxic and septic shock through SIRT1 upregulation. Sci Rep. 2014;4:4220. doi: 10.1038/srep04220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huys L, et al. Type I interferon drives tumor necrosis factor-induced lethal shock. J Exp Med. 2009;206:1873–1882. doi: 10.1084/jem.20090213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dejager L, et al. Pharmacological inhibition of type I interferon signaling protects mice against lethal sepsis. J Infect Dis. 2014;209:960–970. doi: 10.1093/infdis/jit600. [DOI] [PubMed] [Google Scholar]
- 18.Weaver LK, Behrens EM. Hyperinflammation, rather than hemophagocytosis, is the common link between macrophage activation syndrome and hemophagocytic lymphohistiocytosis. Curr Opin Rheumatol. 2014;26:562–569. doi: 10.1097/BOR.0000000000000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoller EE, et al. Hemophagocytosis causes a consumptive anemia of inflammation. J Exp Med. 2011;208:1203–1214. doi: 10.1084/jem.20102538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamieson AM, Yu S, Annicelli CH, Medzhitov R. Influenza virus-induced glucocorticoids compromise innate host defense against a secondary bacterial infection. Cell Host Microbe. 2010;7:103–114. doi: 10.1016/j.chom.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamieson AM, et al. Role of tissue protection in lethal respiratory viral-bacterial coinfection. Science. 2013;340:1230–1234. doi: 10.1126/science.1233632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chart H, Smith HR, La Ragione RM, Woodward MJ. An investigation into the pathogenic properties of Escherichia coli strains BLR, BL21, DH5alpha and EQ1. J Appl Microbiol. 2000;89:1048–1058. doi: 10.1046/j.1365-2672.2000.01211.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhao XW, et al. Defects in neutrophil granule mobilization and bactericidal activity in familial hemophagocytic lymphohistiocytosis type 5 (FHL-5) syndrome caused by STXBP2/Munc18-2 mutations. Blood. 2013;122:109–111. doi: 10.1182/blood-2013-03-494039. [DOI] [PubMed] [Google Scholar]
- 24.Sieni E, et al. Familial hemophagocytic lymphohistiocytosis: When rare diseases shed light on immune system functioning. Front Immunol. 2014;5:167. doi: 10.3389/fimmu.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohyama M, et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haldar M, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156:1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez-Ortiz ZG, et al. The receptor TREML4 amplifies TLR7-mediated signaling during antiviral responses and autoimmunity. Nat Immunol. 2015;16:495–504. doi: 10.1038/ni.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langston PK, Shibata M, Horng T. Metabolism supports macrophage activation. Front Immunol. 2017;8:61. doi: 10.3389/fimmu.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norelli M, et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat Med. 2018;24:739–748. doi: 10.1038/s41591-018-0036-4. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto H, et al. The clinical importance of a cytokine network in the acute phase of sepsis. Sci Rep. 2018;8:13995. doi: 10.1038/s41598-018-32275-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wohlfarth P, et al. Interleukin 1 receptor antagonist anakinra, intravenous immunoglobulin, and corticosteroids in the management of critically ill adult patients with hemophagocytic lymphohistiocytosis. J Intensive Care Med. 2017:885066617711386. doi: 10.1177/0885066617711386. [DOI] [PubMed] [Google Scholar]
- 35.Schulert GS, Grom AA. Macrophage activation syndrome and cytokine-directed therapies. Best Pract Res Clin Rheumatol. 2014;28:277–292. doi: 10.1016/j.berh.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan Z, et al. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol. 2015;194:6082–6089. doi: 10.4049/jimmunol.1402469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semba H, et al. HIF-1α-PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nat Commun. 2016;7:11635. doi: 10.1038/ncomms11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freemerman AJ, et al. Metabolic reprogramming of macrophages: Glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J Biol Chem. 2014;289:7884–7896. doi: 10.1074/jbc.M113.522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian S, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci USA. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deane JA, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 44.Shin S, Roy CR. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 2008;10:1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 45.Freitag NE, Port GC, Miner MD. Listeria monocytogenes–From saprophyte to intracellular pathogen. Nat Rev Microbiol. 2009;7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagmann CA, et al. RIG-I detects triphosphorylated RNA of Listeria monocytogenes during infection in non-immune cells. PLoS One. 2013;8:e62872. doi: 10.1371/journal.pone.0062872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massis LM, Zamboni DS. Innate immunity to legionella pneumophila. Front Microbiol. 2011;2:109. doi: 10.3389/fmicb.2011.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heininger A, et al. Cytomegalovirus reactivation and associated outcome of critically ill patients with severe sepsis. Crit Care. 2011;15:R77. doi: 10.1186/cc10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walton AH, et al. Reactivation of multiple viruses in patients with sepsis. PLoS One. 2014;9:e98819. doi: 10.1371/journal.pone.0098819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.T’Jonck W, Guilliams M, Bonnardel J. Niche signals and transcription factors involved in tissue-resident macrophage development. Cell Immunol. 2018;330:43–53. doi: 10.1016/j.cellimm.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 52.Lee J, et al. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 54.Saeed S, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buck MD, et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: Insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang A, et al. Glucose metabolism mediates disease tolerance in cerebral malaria. Proc Natl Acad Sci USA. 2018;115:11042–11047. doi: 10.1073/pnas.1806376115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cumnock K, et al. Host energy source is important for disease tolerance to malaria. Curr Biol. 2018;28:1635–1642.e3. doi: 10.1016/j.cub.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 59. Wang, et al. (2019) Genome-wide RNA sequencing of sequential TLR agonist stimulation in C57Bl6/J macrophages. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124765. Deposited January 7, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.