Significance
The spliceosome selects, and in the case of alternative splicing, contextually distinguishes between, splice sites with extremely high fidelity. However, when a splice site becomes mutated, as in many human diseases, the spliceosome can aberrantly select nearby “cryptic” splice sites, which produce nonfunctional protein. It is unclear how the spliceosome routinely selects against cryptic sequences. Here, we implicate core components of the spliceosome itself in its ability to select against cryptic splice sites and show that how the spliceosome selects against cryptic splice sites is different than how it selects between bona fide splice sites during alternative splicing. In addition to providing insight into an essential cellular process, our work is relevant to the development of therapeutics that act by modulating splice-site choice.
Keywords: cryptic splicing, alternative splicing, spliceosome, PRP8, CRISPR mutagenesis
Abstract
Pre-mRNA splicing must occur with extremely high fidelity. Spliceosomes assemble onto pre-mRNA guided by specific sequences (5′ splice site, 3′ splice site, and branchpoint). When splice sites are mutated, as in many hereditary diseases, the spliceosome can aberrantly select nearby pseudo- or “cryptic” splice sites, often resulting in nonfunctional protein. How the spliceosome distinguishes authentic splice sites from cryptic splice sites is poorly understood. We performed a Caenorhabditis elegans genetic screen to find cellular factors that affect the frequency with which the spliceosome uses cryptic splice sites and identified two alleles in core spliceosome component Prp8 that alter cryptic splicing frequency. Subsequent complementary genetic and structural analyses in yeast implicate these alleles in the stability of the spliceosome’s catalytic core. However, despite a clear effect on cryptic splicing, high-throughput mRNA sequencing of these prp-8 mutant C. elegans reveals that overall alternative splicing patterns are relatively unchanged. Our data suggest the spliceosome evolved intrinsic mechanisms to reduce the occurrence of cryptic splicing and that these mechanisms are distinct from those that impact alternative splicing.
Splicing is an essential step in gene expression. The spliceosome, a large, RNA-based molecular machine, identifies and catalyzes the removal of noncoding introns and joining of flanking coding exons of pre-mRNAs (1). Splicing is an inherently high-fidelity process, and errors in splicing have been linked to many human diseases (2). Introns contain specific consensus sequences at their 5′ and 3′ ends (5′ and 3′ splice sites, SSs) as well as at an internal branchpoint. The spliceosome recognizes and binds to these sequences, and proper SS recognition by the spliceosome is required for high-fidelity pre-mRNA splicing (1).
During cryptic splicing, the spliceosome selects a sequence element that resembles, but is not, a bona fide SS. This often occurs when a true SS has been mutated and nearby cryptic sites, usually defined by a 5′ GU or a 3′ AG, are selected. Use of a cryptic site can result in changes to a gene’s ORF and consequent disruption of gene expression. A small subset of core splicing factors have been implicated in cryptic splicing (3–5).
During assembly, the spliceosome undergoes considerable conformational rearrangements (1). Prp8 is the largest and most highly conserved protein in the spliceosome and has been directly implicated in splicing fidelity (6). The bulk of Prp8 surrounds and stabilizes the spliceosome’s catalytic core, contacting the catalytic U6 snRNA and pre-mRNA substrate (7, 8). One key rearrangement is the closure of the spliceosome’s catalytic core. In catalytic core closure, Prp8’s N-terminal and large domains come into close proximity, helping to organize and support the snRNAs and pre-mRNA for catalysis (9–13). There is evidence that the spliceosome’s catalytic core opens and closes repeatedly during a typical splicing cycle, and, importantly, “open” and “closed” forms of the spliceosome have been linked to changes in splicing fidelity (6, 14, 15). Cryptic splicing could result from local movements of the pre-mRNA within the catalytic core of the spliceosome while it is in an open form.
Here, we characterize two prp-8 alleles identified in a Caenorhabditis elegans screen designed to select for factors that alter cryptic splicing frequency. Our data suggest that stabilization of a more open form of the spliceosome’s catalytic core promotes cryptic splicing, and that cryptic splicing occurs independently of spliceosome selection between bona fide alternative sites, and by a different mechanism.
Results
Genetic Screen to Identify Alleles That Alter the Frequency of Cryptic Splicing.
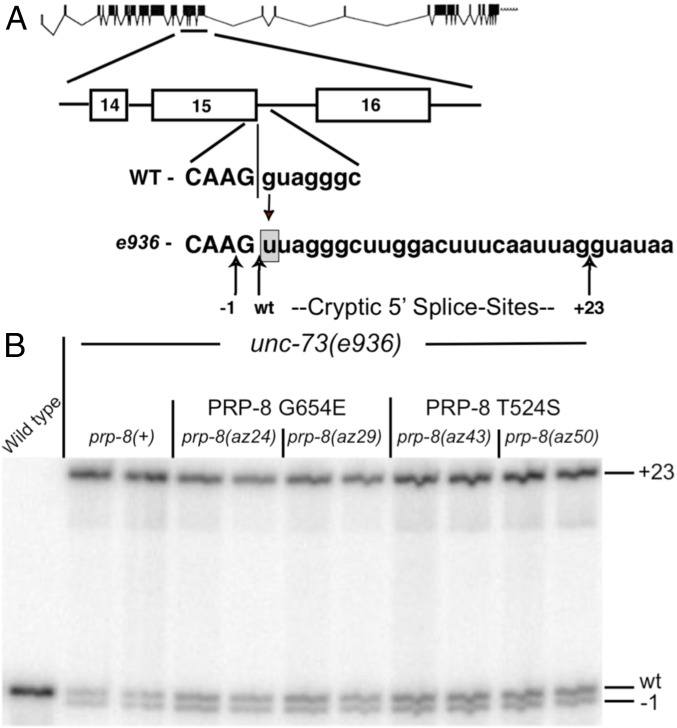
The C. elegans (worm) unc-73 gene encodes a protein required for proper axon guidance and cell migration. The unc-73(e936) allele bears a G → U mutation at the first nucleotide of intron 15, which disrupts splicing and causes uncoordinated locomotion. We previously reported the use of unc-73(e936) in a genetic screen to identify cellular factors that alter cryptic splicing patterns (16). In this approach, unc-73(e936) C. elegans are N-nitroso-N-ethylurea (ENU)-mutagenized and their F2 progeny screened for improved locomotion. Changes in locomotion often result from mutations in splicing factors that promote changes in splice-site choice (Fig. 1A), improving locomotion through increasing the level of in-frame unc-73 messages by promoting use of the wild-type 5′ SS (Fig. 1B and Table 1). No cryptically spliced e936 transcripts are subject to nonsense-mediated decay.
Fig. 1.
Two suppressor alleles map to prp-8. (A) Exon structure of unc-73 (32). The G → U substitution at the first base of intron 15 in the e936 allele, along with the cryptic SS activated by it, (−1, wt, +23) are indicated. (B) 32P-RT-PCR products from independent RNA extractions were separated on 6% polyacrylamide denaturing gels. Bands corresponding to use of the different cryptic SS indicated. Quantification presented in Table 1.
Table 1.
prp-8 alleles promote use of wild-type 5′ SS
| Genotype | +23, % | wt, % | −1, % |
| unc-73(e936);prp-8(+) | 73.7 | 14.8 | 11.5 |
| unc-73(e936);prp-8(+) | 74.3 | 13.5 | 12.2 |
| unc-73(e936);prp-8(az24) G654E | 53.6 | 24.2 | 22.2 |
| unc-73(e936);prp-8(az24) G654E | 53.2 | 23.1 | 23.7 |
| unc-73(e936);prp-8(az29) G654E | 53.4 | 27.0 | 19.6 |
| unc-73(e936);prp-8(az29) G654E | 55.1 | 25.7 | 19.1 |
| unc-73(e936);prp-8(az43) T524S | 52.2 | 28.3 | 19.6 |
| unc-73(e936);prp-8(az43) T524S | 53.8 | 28.4 | 17.8 |
| unc-73(e936);prp-8(az50) T524S | 56.9 | 26.3 | 16.8 |
| unc-73(e936);prp-8(az50) T524S | 56.3 | 25.0 | 18.7 |
Relative usage of each of the cryptic splice-sites for unc-73(e936) intron 15 in the gel shown in Fig. 1B.
We previously identified az24, a semidominant extragenic allele that altered cryptic-splicing frequency, but we had not identified the gene (16). By sequencing spliceosome genes in the 1.4-MB region of the genome to which we mapped az24, we identified G654E in the C. elegans homolog of Prp8 (PRP-8). To verify that prp-8(G654E) causes improved locomotion and changed cryptic splicing patterns seen in az24, we used CRISPR-cas9 to remake this mutation in wild-type worms and crossed this allele into the unc-73(e936) background (SI Appendix, Fig. S1). We call the CRISPR prp-8(G654E) allele az29. Both az24 and az29 improve worm locomotion, likely due to an increase in the frequency of in-frame splicing of the unc-73(e936) transcript (Fig. 1B and Table 1), leading to the production of more UNC-73 protein. We repeated the screen and identified a semidominant allele, az43, which mapped to prp-8(T524S). We used CRISPR-cas9 to generate prp-8(az50), a T524S amino acid substitution (SI Appendix, Fig. S1) designed to validate az43. Both the az43 and az50 T524S alleles affect e936 cryptic splice-site selection; they lead to increased usage of both the −1 and WT cryptic 5′ SS and lower the use of the +23 cryptic SS, increasing the abundance of mRNAs coding for full-length UNC-73 (Fig. 1B and Table 1).
prp-8 Alleles Map to Conserved Regions of PRP-8 That Surround and Stabilize Key Components of the Spliceosome’s Catalytic Core.
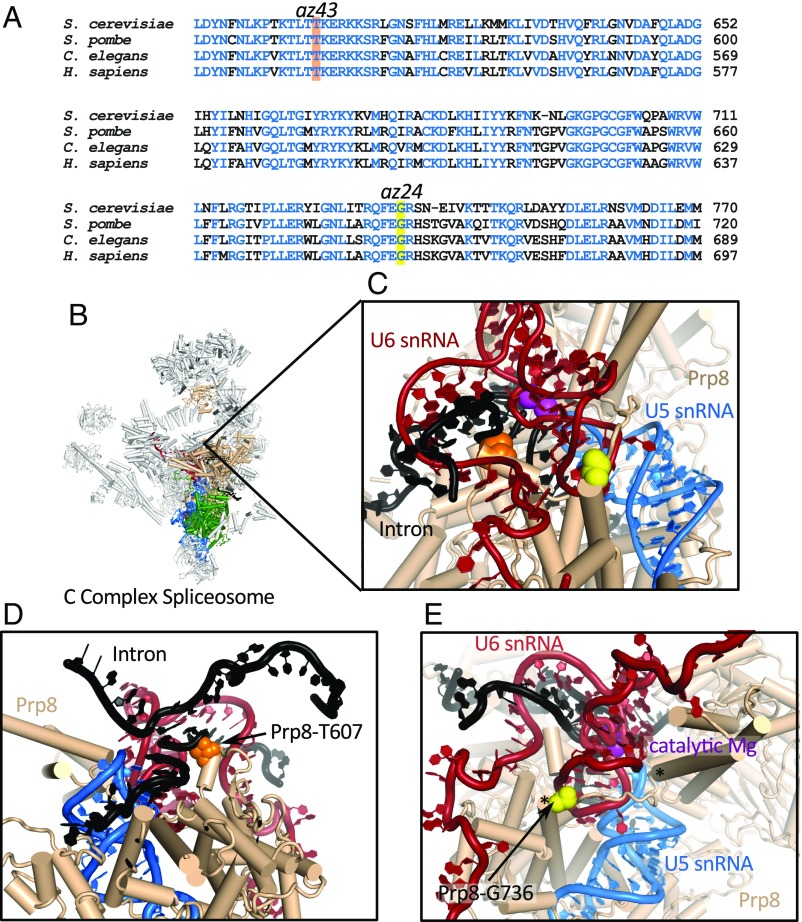
Prp8 surrounds and stabilizes the spliceosome’s catalytic core, directly contacting the catalytic U6 snRNA, pre-mRNA substrate, and the portion of U5 snRNA that positions the exons for ligation. It also makes extensive contacts with multiple spliceosomal proteins, including Snu114 (7, 9). az24 and az43 prp-8 alleles map to regions of Prp8’s N-terminal domain that are highly conserved in both sequence and structure (Fig. 2), consistent with being important for spliceosome function.
Fig. 2.
prp-8 alleles map to conserved regions of Prp8. (A) ClustalOmega (33) alignment of Prp8 sequences showing conservation (blue) in the region of az24 (yellow) and az43 (orange). (B) S. cerevisiae C complex structure (5LJ3) (7) highlighting Prp8’s (tan) central position, Snu114 (green), U5 snRNA (blue), and U6 snRNA (red). These interactions are virtually identical in all post B-complex structures. (C) Zoom to spliceosome’s catalytic core. Catalytic Mg2+, magenta. Positions of Prp8-T607 (orange) and Prp8-G736 (yellow) are shown. (D) Prp8-T607 (orange) contacts the intron (black) 5′ SS +3 phosphate. (E) Prp8-G736 (yellow) is part of a structured loop (edges indicated by asterisks) that forms during activation. This loop acts as a brace, supporting U6 snRNA.
Analysis of available spliceosome structures suggests that the az24 and az43 alleles act by destabilizing the spliceosome’s catalytic core. az43 maps to PRP-8 amino acid T524. In structures of the Saccharomyces spliceosome, this amino acid (T607 in S. cerevisiae and T532 in humans) interacts with the phosphodiester backbone of the pre-mRNA intron’s 5′ SS between the +2 and +3 positions (7, 8, 10). Thus, Prp8 serves as an additional contact stabilizing the 5′ SS within the spliceosome’s catalytic core (Fig. 2 C and D). Substitution of Ser for Thr in az43 removes a methyl group, decreasing the number of contacts supporting the 5′ SS.
The other allele, az24 (G736E in S. cerevisiae), maps to a loop in Prp8’s N-terminal domain (Fig. 2 C and E), which we refer to as the “740 loop.” This loop is only resolved in structures of activated spliceosomes (SI Appendix, Fig. S2), suggesting it is stabilized after the spliceosome’s catalytic core has closed (7, 9, 11, 17–19). The 740 loop braces the catalytic U6 snRNA internal stem loop, helping organize the spliceosome’s catalytic core (Fig. 2E). Mutation of a Gly to a Glu adds negative charge in proximity to negatively charged U6 snRNA, potentially destabilizing the spliceosome’s catalytic core.
Homologous S. cerevisiae prp8 Alleles Inform How Cryptic Splicing Frequency Is Altered.
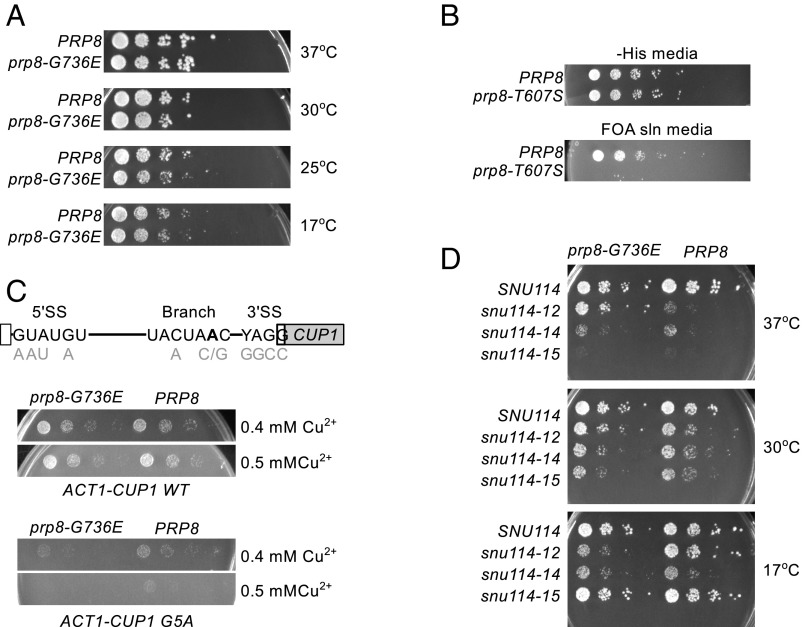
We created S. cerevisiae strains carrying homologous alleles to the az24 and az43 prp-8 alleles to take advantage of tools not available in C. elegans. We performed growth assays to determine whether prp8 alleles impact S. cerevisiae growth and/or survival. We were able to recover prp8-G736E(az24) S. cerevisiae, indicating that prp8-G736E(az24) does not affect viability (Fig. 3A). We compared the growth of prp8-G736E(az24) to that of PRP8. The strains grew identically (Fig. 3A), indicating that prp8-G736E(az24) does not affect S. cerevisiae growth. Strikingly, though az43 C. elegans show no gross morphological phenotypes: prp8-T607S(az43) is lethal in S. cerevisiae (Fig. 3B). Given that worm 5′ SS are more degenerate, it is surprising that removal of a 5′ SS backbone-stabilizing interaction would have a stronger effect in yeast. One explanation is that Prp8-T607S spliceosomes splice less efficiently due to problems stabilizing pre-mRNA. Because most S. cerevisiae introns are in essential transcripts, decreased efficiency may not be tolerable.
Fig. 3.
Effects of S. cerevisiae prp8 alleles. (A) prp8-G736E grows identically to wild-type S. cerevisiae. (B) FOA selection shows that, in the absence of wild-type PRP8, prp8-T607S is lethal. (C) Act-Cup analyses. (Top) Act-Cup reporter showing all intron substitutions. (Middle) prp8-G736E grows as PRP8 when challenged with a WT reporter. (Bottom) prp8-G736E growth on copper-containing media is compromised by G5A. (D) prp8-G736E exacerbates snu114-12 cold sensitivity while suppressing temperature sensitivity.
We used the well-characterized Act-Cup reporter to assess how S. cerevisiae prp8 alleles interact with different SS variants. In this system, the ACT1 intron has been fused to the copper detoxification protein CUP1, such that levels of CUP1 mRNA transcript and, thus, levels of Cup1 protein, are dependent upon splicing efficiency, which can be directly inferred by monitoring the growth of S. cerevisiae on copper-containing media (20). PRP8 and prp8-G736E(az24) S. cerevisiae were transformed with Act-Cup reporters (Fig. 3C), and the ability of prp8-G736E to splice these reporters was determined. We saw no change in general splicing efficiency, as indicated by the WT splicing reporter. The G5A reporter, which introduces a G → A substitution at position five of the 5′ SS, was the only reporter tested where prp8-G736E exhibited a phenotype (Fig. 3C). The negative effect on growth was slight but repeatable, consistent with RT-qPCR data showing a modest but not statistically significant decrease in the level of prp8-G736E G5A Act-Cup mRNA (SI Appendix, Fig. S3). The G5A 5′ SS mutation loosens contacts between pre-mRNA and the spliceosome, and a previously published prp8 allele implicated in spliceosome catalytic core stability is particularly sensitive to G5A (13). Together these data implicate Prp8 in the stability of the spliceosome’s catalytic core.
To gain more insight into the mechanisms underlying how Prp8 impacts cryptic SS selection, we used comparative growth assays to search for genetic interactions between prp8-G736E and other core spliceosome components implicated in catalytic core. We identified a genetic interaction between prp8-G736E and snu114-12. Snu114 is a GTPase that has homology with ribosomal translocation factor EF-G. Its GTPase activity correlates with several important steps in spliceosome assembly, catalysis, and disassembly (21). Snu114 physically and genetically interacts with Prp8 (22). The snu114-12 allele was designed to decrease Snu114’s rate of GTP hydrolysis and is temperature sensitive (22). prp8-G736E suppresses snu114-12 temperature sensitivity. prp8-G736E snu114-12 also exhibits cold sensitivity, a phenotype not present independently in prp8-G736E or snu114-12 (Fig. 3D and SI Appendix, Fig. S4). Together these data implicate Prp8 regulation by Snu114 in suppression of cryptic splicing.
Multiple C. elegans PRP-8 Amino Acid Substitutions at Position 654 Are Viable and Suppress Cryptic Splicing.
C. elegans PRP-8 amino acid G654 maps to the 740 loop, a region of PRP-8 that is unstructured during early assembly but later structures and interacts with the catalytic U6 internal stem loop (Fig. 2E and SI Appendix, Fig. S2). This intriguing structural transition, coupled with our yeast data, led us to test whether any additional amino acid substitutions at G654 are viable in C. elegans, and if such substitutions can also suppress cryptic splicing. To do this, we performed CRISPR mutagenesis. We used a 100-nt prp-8 repair oligonucleotide in which we substituted the G654 codon with three Ns to template random codons (SI Appendix, Fig. S5A). We recovered K, R, L, I, V, P, C, and E substitutions at position G654, as well as an 8-aa insertion, TKRRKSPS, between amino acids F652 and E653 (SI Appendix, Fig. S5B). All CRISPR mutant worms appear healthy, and no new recessive lethal alleles were recovered.
We crossed these alleles with unc-73(e936) to determine their effects on cryptic splicing frequency (Table 2). All CRISPR-generated prp-8 alleles showed changes in cryptic SS choice, increasing use of the WT unc-73(e936) exon 15 SS, although none to the same extent as the original G654E allele. Consistent with this molecular phenotype, using thrashing assays, a common assay of worm locomotion, we observed suppression of the unc phenotype to a degree consistent with the change in cryptic SS choice. The 8-aa 740 loop insertion allele also shows suppression at both the molecular and organismal level (Table 2). Together these data show that a wide variety of amino acids are tolerated at position G654 in C. elegans. We hypothesize that these changes to cryptic SS selection result from destabilization of the Prp8 740 loop.
Table 2.
Cryptic splicing measurements of the unc-73(e936) gene in the presence of the PRP-8 G654 substitution alleles
| Strain | Genotype | PRP-8 | “+23”, % | wt, % | “−1”, % | Thrashes/min |
| SZ181 | unc-73(e936); prp-8(+) | wt | 72.9, 73.7 | 12.1, 13.8 | 14.9, 12.5 | 10.2 ± 2.9 |
| SZ155 | unc-73(e936); prp-8(az29) | G654E | 54.3, 53.7 | 27.1, 24.9 | 18.6, 21.4 | 108.4 ± 9.9 |
| SZ255 | unc-73(e936); prp-8(az75) | G654K | 64.5, 62.6 | 19.5, 19.7 | 16.0, 17.7 | 50.8 ± 8.0 |
| SZ256 | unc-73(e936); prp-8(az76) | G654L | 56.7, 56.5 | 22.5, 23.7 | 20.8, 19.7 | 59.8 ± 12.7 |
| SZ257 | unc-73(e936); prp-8(az77) | G654P | 63.4, 62.0 | 20.1, 21.8 | 16.6, 16.2 | 41.4 ± 8.8 |
| SZ258 | unc-73(e936); prp-8(az79) | G654I | 59.3, 59.4 | 21.7, 21.8 | 19.0, 18.8 | 40.6 ± 10.1 |
| SZ259 | unc-73(e936); prp-8(az80) | G654C | 65.8, 62.6 | 20.4, 20.6 | 13.8, 16.7 | 33.4 ± 5.9 |
| SZ260 | unc-73(e936); prp-8(az82) | G654V | 57.4, 57.4 | 23.3, 23.3 | 19.3, 19.3 | 43.8 ± 5.5 |
| SZ261 | unc-73(e936); prp-8(az83) | 8aa ins. | 62.9,61.7 | 21.7, 23.4 | 15.4, 14.9 | 85.8 ± 15.9 |
| N2 | unc-73(+); prp-8(+) | wt | 100.0 | 133.4 ± 11.1 |
The last column is a thrash assay in which L4 worms were placed in a drop of M9 medium on a glass slide and the number of times that they bent their body across the central axis in 1 min was measured. Mean and SD of 10 independent trials listed.
Only Thr, Ser, and Val Can Be Tolerated at Position 524 in C. elegans.
We performed CRISPR-cas9 random mutagenesis experiments to produce the full range of possible T524 amino acid substitutions (SI Appendix, Fig. S5C). We recovered only T, S, and V as viable T524 substitutions (SI Appendix, Fig. S5D). Recovery of Thr and Ser are not surprising given that these are the wild type and the viable az43 alleles identified in our forward genetic screen. To test whether the T524V substitution in PRP-8 could also impact cryptic splicing frequency, we used genetic crosses to construct unc-73(e936); prp-8(T524V) animals. These double mutant animals were uncoordinated (no phenotypic suppression), slow to mature, and had few to no progeny, making it impossible to recover RNA for testing, unlike T524S C. elegans, which appear as wild type.
One striking observation from this CRISPR-based mutagenesis at T524 is that in the F1 progeny of the CRISPR-cas9 injected parents, we saw many dead and dying embryos and early-stage larvae. This is a phenomenon that we had not seen previously, and we did not observe any substitutions of T524 that were viable as heterozygotes but homozygous lethal. These results point to the possibility that other amino acid substitutions at T524 besides Ser and Val produce a dominant lethal form of PRP-8.
prp-8 Alleles Do Not Dramatically Impact Normal Alternative Splicing Patterns.
To determine whether prp-8 alleles promote global changes in alternative splicing of native genes, we performed high-throughput strand-specific mRNA sequencing of 100-nt paired-end reads generated from prp-8 C. elegans (SI Appendix, Table S1). We looked for changes in alternative splicing between libraries. We created de novo a dataset of alternative 5′ SS events, alternative 3′ SS events, and alternative cassette exon events. We next used MISO (23) to do pairwise comparisons for each SS choice event, comparing strains carrying the unc-73(e936) mutation against those with both unc-73(e936) and a prp-8 allele. Events in which MISO detected a change in percent spliced in (ΔPSI) ≥ 0.20 in each comparison (a change of ≥20%) were ranked by a total number of pairwise comparisons in which significant changes occurred (Table 3). This sum of significant changes for an event across all pairwise comparisons is the pairwise comparison sum, or pairSum. Table 3 shows the number of alternative splicing events detected for each pairSum for worms carrying each of the two prp-8 mutant alleles. We examined Sashimi plots for each flagged event along with BAM tracks to visually confirm them (Table 3) (23). The majority of events that passed visual confirmation were validated by 32P-RT-PCR (SI Appendix, Fig. S5).
Table 3.
Pairwise comparison sums of MISO analysis of alternative splicing changes
| pairSum | Alt 5′ splice-site events | Alt 3′ splice-site events | Alt cassette exons |
| PRP-8 G654E - Max pairwise comparison sum = 8 | |||
| 8 | 0/3 | 0/1 | 0/1 |
| 7 | 0/3 | 0/1 | 0/0 |
| 6 | 2/11 | 0/16 | 0/0 |
| PRP-8 T524S - Max pairwise comparison sum = 12 | |||
| 12 | 0/0 | 2/3 | 0/0 |
| 11 | 0/3 | 2/3 | 0/0 |
| 10 | 3/10 | 0/0 | 1/2 |
| 9 | 0/2 | 6/10 | 0/2 |
Number of potential events after inspection/number of potential initially flagged events. The bold column in the table represents the percentage of in-frame unc-73 messages in the animals.
Data from prp-8(G654E) worms indicated only two alternative 5′ splicing events (T12C9.7 and stip-1) (Table 4 and SI Appendix, Fig. S5 A and B). For the prp-8(T524S) mutant worms, we identified three alternative 5′ SS events, one alternative cassette exon event, as well as 10 alternative 3′ SS events (Table 5 and SI Appendix, Fig. S5 and Table S2). prp-8 T524S mutant altered 5′ SS usage for T12C9.7 in the opposite direction of G654E (Table 4 and SI Appendix, Fig. S5A). The five strongest changes in alternative 3′ SS in prp-8(T524S) worms were confirmed by 32P-RT-qPCR, without an obvious bias toward the upstream or downstream 3′ SS (Table 5 and SI Appendix, Fig. S5 C–G). Alternative 3′ SS choice events promoted by the prp-8 T524S mutation were unchanged or only minimally changed by the prp-8 G654E mutation, indicating these alleles have distinct effects on 3′ SS usage (Table 5 and SI Appendix, Fig. S5). We hypothesize that the changes in 3′ SS selection may be a consequence of altered interactions between Prp8 and the pre-mRNA branchpoint.
Table 4.
Alternative splicing events predicted from prp-8 G654E mRNA-Seq analysis
| Genotype | Up 5′SS use str. 1,2, % | |
| Gene & Sequence: T12C9.7; TTTATTTCTG▲GTT△gtgagaatttgttttatagttt | ||
| snrp-27(+); prp-8(+) | 18.65, 18.74 | |
| snrp-27(M141T); prp-8(+) | 97.55, 95.55 | |
| snrp-27(+); prp-8(G654E) | 35.61, 35.83 | |
| snrp-27(+); prp-8(T524S) | 1.41, 1.12 | |
| Gene & Sequence: stip-1; AGGATCAGGCGACATAT G▲ GTGGG△gtgagatcattgaaaa | ||
| snrp-27(+); prp-8(+) | 75.96, 75.74 | |
| snrp-27(M141T); prp-8(+) | 94.46, 95.54 | |
| snrp-27(+); prp-8(G654E) | 92.63, 91.88 | |
| snrp-27(+); prp-8(T524S) | 86.61, 87.07 | |
| Gene & Sequence: spat-2; AACAGCGTT△GTGAGG ATGGCCTTGCAAGAAGGG▲gtgcgggctta | ||
| snrp-27(+); prp-8(+) | 71.76, 72.07 | |
| snrp-27(M141T); prp-8(+) | 45.43, 42.86 | |
| snrp-27(+); prp-8(G654E) | 59.26, 61.84 | |
| snrp-27(+); prp-8(T524S) | 50.34, 51.64 | |
Sequences of the alternative spliced regions. 5′ SSs promoted in prp-8 mutant indicated by a filled triangle, partner 5′ SS indicated by an open triangle. Uppercase are sequences that can be exons; lowercase are always intronic. Below the sequence is quantitation of alternative 5′ splice-site usage by 32P-RT-PCR analysis. Genotypes are indicated. T12C9.7 was identified in the prp-8 G654E library, stip-1 in both prp-8 libraries, and spat-2 in the prp8 T524S library.
Table 5.
Analysis of alternative 3′ splicing events predicted from mRNA-Seq analysis of the prp-8 T524S mutant libraries
| Genotype | Up 3′ SS use strain 1,2, % | |
| Gene & Sequence: cdt-2 aaatcaaaaag▲ATTTCAG△TTTTTCCCGC | ||
| prp-8(+) | 18.24, 18.41 | |
| prp-8(G654E) | 27.65, 28.28 | |
| prp-8(T524S) | 51.51, 52.45 | |
| Gene & Sequence: T10E9.14 atattttgcag△TTTCAG▲GAAACTATCAAC | ||
| prp-8(+) | 16.76, 17.72 | |
| prp-8(G654E) | 19.34, 20.04 | |
| prp-8(T524S) | 5.44, 4.58 | |
| Gene & Sequence: F48D4.6 gttattcag▲TGTTCCAG△AAACAG△TGACGACTT | ||
| prp-8(+) | 54.90, 53.69 | |
| prp-8(G654E) | 49.98, 51.13 | |
| prp-8(T524S) | 79.53, 79.69 | |
| Gene & Sequence: W09G3.7 atcacgaatttag△CTTCCAG▲ATCGGTGGT | ||
| prp-8(+) | 84.59, 83.27 | |
| prp-8(G654E) | 81.61, 81.82 | |
| prp-8(T524S) | 62.01, 60.82 | |
| Gene & Sequence: dct-1 aacgcttcag△GTATGTCAG▲AATCGTGGGTGGAA | ||
| prp-8(+) | 62.48, 56.68 | |
| prp-8(G654E) | 48.90, 50.14 | |
| prp-8(T524S) | 44.33, 45.60 | |
Five of the 10 alternative 3′ splicing events were tested. Sequences of the alternatively spliced regions are shown, with the 3′ SSs promoted in the prp-8 mutant indicated by a filled triangle, and the partner 3′ SS by an open triangle (in F48D6.4 there are three alternative 3′ SSs). Uppercase indicates sequences that can be exons, while lowercase are always intronic. Below the sequence is quantitation of relative alternative 3′ SS usage by 32P-RT-PCR analysis (SI Appendix, Fig. S5). Genotypes are indicated.
We are confident in our analysis pipeline as this methodology previously yielded 26 high-confidence alternative splicing events for the snrp-27(az26) allele (24). While the snrp-27 and two prp-8 alleles have almost identical effects on unc-73(e936) cryptic 5′ SS choice, our sequencing analysis indicates that they can have differential effects on alternative splicing, suggesting that they may operate at distinct stages of splicing. In contrast to our previous studies, the small number of alternative splicing events that we observe suggest that these prp-8 mutations do not cause strong global changes in alternative splicing. We conclude that the prp-8 alleles lead to a small number of confirmable changes in the usage of SSs located in close proximity to each other (i.e., that look like cryptic sites), consistent with the spliceosome being in a more open conformation that may allow slippage between juxtaposed SSs.
Discussion
The remarkable number of high-resolution structures available paint a very clear picture of the relative conformations of the spliceosome’s catalytic core at multiple stages of the splicing cycle. These data illustrate how the spliceosome reorients during activation to close its catalytic core. However, such data present only static images of what is known to be an incredibly dynamic molecular machine. The spliceosome’s catalytic core is dynamic, changing its conformation from open to closed forms repeatedly during a typical splicing cycle (Fig. 4 A and B). Such dynamics are required to position pre-mRNA for catalysis, and spliceosome dynamics may be linked to high-fidelity splicing (6, 13, 14, 25).
Fig. 4.
Model. The spliceosome’s catalytic core can be found in both open and closed conformations. Prp8 is shown (tan blob) with the 740 loop (green), U6 snRNA (red), U5 snRNA (blue), pre-mRNA (black), and Mg2+ (magenta). (A) When the catalytic core is open, there is no pre-mRNA docked to U5 snRNA, catalytic Mg2+ are absent, and the 740 loop is unresolved. (B) Upon closure, the large domain of Prp8 rotates with respect to the Prp8 NTD (top of Prp8 moves toward bottom). The Prp8 740 loop forms and is resolved by cryoEM (SI Appendix, Fig. S2). The 740 loop supports the U6 internal stem loop, which when in a closed conformation, coordinates Mg2+. Pre-mRNA base pairs with the U5 snRNA. (C) We hypothesize prp-8 alleles negatively affect closure, disrupting pre-mRNA positioning and the 740 loop. prp8 spliceosomes would allow movement of pre-mRNA within their catalytic core, which could result in cryptic splicing.
Here, we present our analysis of two prp-8 alleles that alter the frequency of cryptic splicing in C. elegans. We show that improvement of unc-73(e936) locomotion is an exquisitely sensitive tool for identifying cellular factors involved in cryptic SS selection that act through a variety of mechanisms. Using a combination of structural and genetic approaches in S. cerevisiae, we suggest that these prp-8 alleles modulate cryptic splicing by negatively impacting catalytic core closure and/or stability during splicing (Fig. 4C). This may allow for cryptic SS to occupy the active site of the spliceosome even after initial definition of the 5′ SS. In the case of unc-73(e936), a less stable catalytic core allows an intron beginning with a nonconsensus U more frequent access to the spliceosome’s catalytic core, leading to an increase in in-frame messages and improved worm locomotion. We show that the prp-8 alleles identified in our screen only impact cryptic splicing and can promote small alterations in the ratio of alternative splicing for a small number of juxtaposed alternative 5′ or 3′ SS, but do not have global effects on alternative splicing. Together, our data directly link Prp8-mediated alteration of the spliceosome’s catalytic core to cryptic splicing. More tests are needed to address whether other methods of changing the spliceosome’s catalytic core also lead to changes in cryptic splicing, or if this mechanism is unique to Prp8. That prp-8 alleles do not greatly impact alternative splicing suggests that Prp8’s role in cryptic SS choice occurs independently of exon and/or intron definition, and that it likely occurs via a distinct mechanism.
Materials and Methods
Screen Design and Identification of Alleles That Alter Cryptic Splicing Frequency.
The ENU genetic screen to identify alleles of unc-73(e936) was adapted from a suppressor screen as described (16). az24 was isolated but not identified in ref. 16; az43 was identified in recent repeats of that screen.
CRISPR Mutagenesis.
CRISPR Cas9-directed mutagenesis to generate az29, az50, and T524 and G654 randomization alleles was performed using a co-CRISPR protocol as described (24). prp-8 repair oligos are described in SI Appendix, Table S3. Restriction digests of PCR products were used to track mutations (24, 26).
32P-RT-qPCR Analysis.
Starting with 4 µg of total mixed stage RNA, reverse transcription was performed (24). Primers are indicated in SI Appendix, Table S4. After 25–30 extension cycles (95 °C for 0.5 min, 59 °C for 0.5 min, 68 °C for 0.5 min), PCR products were recovered by phenol:CHCl3 extraction and ethanol precipitation. Ethanol precipitates were resuspended in 20 µL of formamide and 2 µL were separated on a 6% polyacrylamide urea TBE denaturing gel. Products were visualized with a Molecular Dynamics Typhoon PhosphorImager and quantified by ImageQuant (GE Healthcare Life Sciences).
S. cerevisiae Strain Construction and Growth Assays.
yMeg030 (Mat α prp8::LYS cup1::ura3-52 his leu lyse ura ade yCP50-PRP8), yMeg035 (MAT a prp8::LYS his leu lys ura yCP50/PRP8), and yTB108 (MAT α prp8::LYS snu114:KanMX6 his leu lys trp ura yCP50/PRP8 pRS316-SNU114) have been described (22, 27). pRS313-prp8-G736E was created using QuikChange. pRS313-prp8-T607S was created by gap repair of a synthesized fragment containing the T607S mutation (28). Plasmids are available from Addgene. Mutant strains were created by 5FOA shuffling (28). Growth assays were performed by serially diluting log phase cultures grown in rich media and spotting onto rich media. Growth was assessed at 37 °C (2 d), 30 °C (2 d), 25 °C (4 d), or 17 °C (6 d).
ACT-CUP Reporter Analyses.
PRP8 or prp8-G736E S. cerevisiae transformed with an Act-Cup reporter plasmid were grown as described (20, 28). Growth was assessed after 3 d at 30 °C. RT-qPCR analysis was performed with total RNA isolated by the hot phenol method from Act-Cup reporter strains. Specific primers used for qPCR have been described (13).
RNA-Seq Sample Preparation and Analysis.
RNA was isolated for library construction from mixed stage populations of C. elegans (September 2017 preps) or populations enriched for young adults (May 2016 preps) as described (24). FASTQ files and processed .gtf files are in Gene Expression Omnibus accession no. GSE113275. Two libraries from two independent RNA isolations were prepared per strain. Sequences trimmed to 80 bases were mapped to the C. elegans genome using STAR (29). We used a custom script to de novo identify all alternative 5′ SS and 3′ SS events separated by ≦ 50 nt (30) and AltEventFinder (31) to identify cassette exon alternative splicing events. We used MISO (23) to do pairwise comparisons of these libraries for all alternative splicing events. Events in which MISO detected a change in PSI of ≥0.20 were ranked by the total number of pairwise comparisons in which significant changes occurred. The sum of significant changes for an event across all pairwise comparisons is the pairSum. The delta PSI cutoff of 0.20 was chosen because of our previous experience and the issue of false flags that require individual inspection (Table 3) (24). A minimum change in alternative SS usage of 20% allowed us to separate true changes in alternative splicing. This approach may miss some subtle alternative splicing changes, but there is not a widespread destabilization of alternative SS selection.
Supplementary Material
Acknowledgments
We thank Sam Gu and Zhouli Ni of Rutgers University for help in getting the A.M.Z. laboratory started on CRISPR experiments in C. elegans; Manny Ares and Naomi Tesfuzigta for the idea for a single-site oligonucleotide-directed homologous repair genetic randomization experiment; Surabhi Jain and Ken Osterhoudt for help in genetic screening; Magda Konarska for key Prp8 cloning advice; and M. Jurica, A. de Bruyn Kops, M. Konarska, J. Arribere, and H. Madhani for helpful conversations. M.M. and C.G. were supported by the Agouron Institute and National Institute of General Medical Sciences Grant R01GM2119. C.G. is an American Cancer Society Research Professor of Molecular Genetics. Research in the A.M.Z. laboratory is supported by National Science Foundation Grant MCB-1613867. L.E.R. was a fellow of the University of California, Santa Cruz (UCSC) Postbaccalaureate Research Education Program (R25GM104552) funded by the NIH. A.R. is a fellow of the UCSC Initiative for Maximizing Student Diversity Program (R25GM058903).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE113275).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819020116/-/DCSupplemental.
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cvačková Z, Matějů D, Staněk D. Retinitis pigmentosa mutations of SNRNP200 enhance cryptic splice-site recognition. Hum Mutat. 2014;35:308–317. doi: 10.1002/humu.22481. [DOI] [PubMed] [Google Scholar]
- 4.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-box ATPases remodel pre-mRNA to activate alternative splice sites. Cell. 2016;164:985–998. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Q, et al. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev. 2016;30:2710–2723. doi: 10.1101/gad.291872.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayerle M, et al. Structural toggle in the RNaseH domain of Prp8 helps balance splicing fidelity and catalytic efficiency. Proc Natl Acad Sci USA. 2017;114:4739–4744. doi: 10.1073/pnas.1701462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhan X, Yan C, Zhang X, Lei J, Shi Y. Structure of a human catalytic step I spliceosome. Science. 2018;359:537–545. doi: 10.1126/science.aar6401. [DOI] [PubMed] [Google Scholar]
- 9.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016;353:904–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 10.Bertram K, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 11.Plaschka C, Lin P-C, Nagai K. Structure of a pre-catalytic spliceosome. Nature. 2017;546:617–621. doi: 10.1038/nature22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haselbach D, et al. Structure and conformational dynamics of the human spliceosomal Bact complex. Cell. 2018;172:454–464.e11. doi: 10.1016/j.cell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 13.MacRae AJ, et al. Prp8 positioning of U5 snRNA is linked to 5′ splice site recognition. RNA. 2018;24:769–777. doi: 10.1261/rna.065458.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 16.Dassah M, Patzek S, Hunt VM, Medina PE, Zahler AM. A genetic screen for suppressors of a mutated 5′ splice site identifies factors associated with later steps of spliceosome assembly. Genetics. 2009;182:725–734. doi: 10.1534/genetics.109.103473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen THD, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fica SM, et al. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542:377–380. doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai R, Yan C, Wan R, Lei J, Shi Y. Structure of the post-catalytic spliceosome from Saccharomyces cerevisiae. Cell. 2017;171:1589–1598.e8. doi: 10.1016/j.cell.2017.10.038. [DOI] [PubMed] [Google Scholar]
- 20.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartels C, Urlaub H, Luhrmann R, Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J Biol Chem. 2003;278:28324–28334. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- 22.Brenner TJ, Guthrie C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz Y, et al. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015;31:2400–2402. doi: 10.1093/bioinformatics/btv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahler AM, et al. SNRP-27, the C. elegans homolog of the tri-snRNP 27K protein, has a role in 5′ splice site positioning in the spliceosome. RNA. 2018;24:1314–1325. doi: 10.1261/rna.066878.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anokhina M, et al. RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J. 2013;32:2804–2818. doi: 10.1038/emboj.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paix A, Schmidt H, Seydoux G. Cas9-assisted recombineering in C. elegans: Genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res. 2016;44:e128. doi: 10.1093/nar/gkw502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayerle M, Guthrie C. Prp8 retinitis pigmentosa mutants cause defects in the transition between the catalytic steps of splicing. RNA. 2016;22:793–809. doi: 10.1261/rna.055459.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular and Cell Biology. Gulf Professional Publishing; Houston: 2004. [Google Scholar]
- 29.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragle JM, Katzman S, Akers TF, Barberan-Soler S, Zahler AM. Coordinated tissue-specific regulation of adjacent alternative 3′ splice sites in C. elegans. Genome Res. 2015;25:982–994. doi: 10.1101/gr.186783.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou A, et al. Alt event finder: A tool for extracting alternative splicing events from RNA-seq data. BMC Genomics. 2012;13(Suppl 8):S10. doi: 10.1186/1471-2164-13-S8-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steven R, et al. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- 33.Li W, et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.