Significance
Angelman syndrome is a neurodevelopmental disorder caused by loss of function from the maternal allele of UBE3A, an imprinted gene. The paternal allele of UBE3A is silenced by a long noncoding antisense transcript in mature neurons. We have identified a boundary element that stops the transcription of the antisense transcript in human pluripotent stem cells and thus restricts UBE3A imprinted expression to neurons. We further determined that UBE3A imprinting requires both the loss of the boundary function and the sufficient expression of the antisense transcript to silence paternal UBE3A. These findings provide essential details about the mechanisms of UBE3A imprinting that may suggest additional therapeutic approaches for Angelman syndrome.
Keywords: genomic imprinting, Angelman syndrome, iPSC, antisense transcript, long noncoding RNA
Abstract
Angelman syndrome (AS) is a severe neurodevelopmental disorder caused by the loss of function from the maternal allele of UBE3A, a gene encoding an E3 ubiquitin ligase. UBE3A is only expressed from the maternally inherited allele in mature human neurons due to tissue-specific genomic imprinting. Imprinted expression of UBE3A is restricted to neurons by expression of UBE3A antisense transcript (UBE3A-ATS) from the paternally inherited allele, which silences the paternal allele of UBE3A in cis. However, the mechanism restricting UBE3A-ATS expression and UBE3A imprinting to neurons is not understood. We used CRISPR/Cas9-mediated genome editing to functionally define a bipartite boundary element critical for neuron-specific expression of UBE3A-ATS in humans. Removal of this element led to up-regulation of UBE3A-ATS without repressing paternal UBE3A. However, increasing expression of UBE3A-ATS in the absence of the boundary element resulted in full repression of paternal UBE3A, demonstrating that UBE3A imprinting requires both the loss of function from the boundary element as well as the up-regulation of UBE3A-ATS. These results suggest that manipulation of the competition between UBE3A-ATS and UBE3A may provide a potential therapeutic approach for AS.
Angelman syndrome (AS) is a rare neurodevelopmental disorder characterized by developmental delay, seizures, lack of speech, ataxia, and severe intellectual disability (1, 2). It is most frequently caused by mutation (3, 4) or deletion (5) of the maternally inherited allele of UBE3A. UBE3A is an imprinted gene. The paternally inherited allele is silenced in brain (6, 7). The silencing of UBE3A is caused by the expression of an opposing neuron-specific transcript antisense to UBE3A (UBE3A-ATS) (8). The regulation of UBE3A-ATS expression and the mechanism by which UBE3A-ATS represses UBE3A is of tremendous importance since activation of paternal UBE3A is a promising therapeutic strategy for AS (9–11).
UBE3A-ATS is part of the >600 kb SMALL NUCLEOLAR RNA HOST GENE 14 (SNHG14) long noncoding RNA, which initiates from SNRPN promoters on the paternally inherited chromosome (12). SNHG14 can be divided into two functional units based on tissue-specific transcription patterns in humans (13). The proximal portion of the SNHG14/SNRPN transcript includes two protein-coding mRNAs, SNURF and SNRPN; two newly described long noncoding RNAs with snoRNA 5′ ends and polyadenylated 3′ ends, termed SPAs (14); snoLNC RNAs (15); and the noncoding host gene for several C/D box small nucleolar RNAs (SNORD109A, SNORD107, SNORD108, and SNORD116) (12). The noncoding exons annotated as IPW were originally described as an independent gene encoding a polyadenylated noncoding RNA within the Prader-Willi syndrome (PWS) region (16). It is now known that they are exons in the proximal portion of SNHG14 (12). This portion of SNHG14, including all of the aforementioned transcripts and small RNAs, is ubiquitously transcribed in all tissues (12, 17, 18). The distal portion of SNHG14, which includes the noncoding host gene for additional small nucleolar RNAs (SNORD115 and SNORD109B) and the noncoding UBE3A-ATS, is transcribed almost exclusively in the brain (12, 13, 17, 19, 20). It is not known how the neuron-specific processing of SNHG14 occurs such that UBE3A-ATS expression and thus UBE3A imprinting, is restricted to neurons.
We previously found that UBE3A-ATS was expressed and UBE3A was imprinted in nonneuronal cells derived from a patient with an atypical deletion of a portion of the paternal SNRPN allele (21). Based on these results, we hypothesized that the imprinted expression of human UBE3A is restricted to neurons by a boundary element. Here, we use CRISPR/Cas9 technology in human AS induced pluripotent stem cells (iPSCs) and their neuronal derivatives to functionally define this boundary element and determine its role in mediating UBE3A imprinting.
Results
A Boundary Element Composed of IPW and PWAR1 Restricts UBE3A-ATS Expression to Neurons.
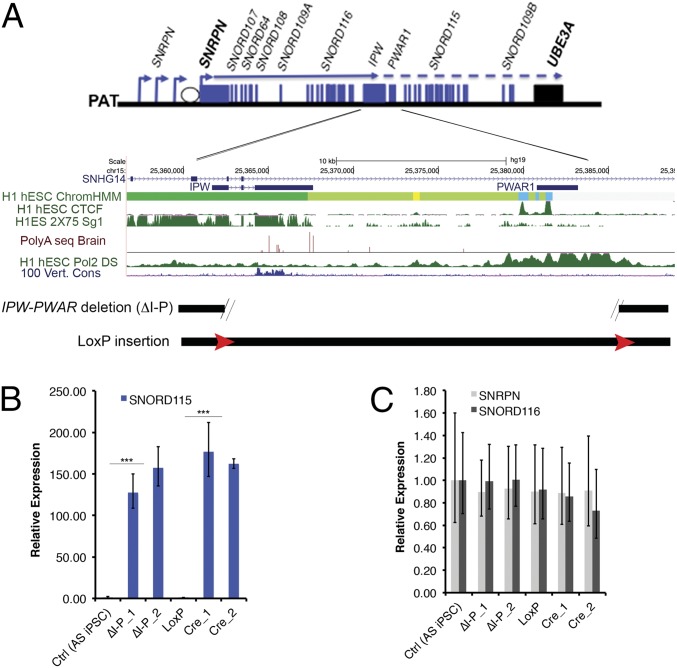
We previously reported that the distal portion of SNHG14 is expressed and UBE3A is imprinted in iPSCs derived from an individual with PWS due to an atypical paternal deletion (21). This unique paternal deletion demonstrated that imprinting of UBE3A can occur in nonneuronal tissues and that a boundary may restrict the expression of UBE3A-ATS and imprinting of UBE3A to neurons. The region separating the expressed proximal portion of the SNHG14 from the repressed distal portion includes a stretch of weak polyadenylation [poly(A)] sites within the last IPW exon (16, 22) and two divergently oriented CCCTC binding factor (CTCF) binding sites at PWAR1/PAR1 (heretofore referred to as PWAR1; Fig. 1A, ref. 23). PWAR1 was first described as an unspliced complementary DNA clone derived from a fetal brain library but is now interpreted to be an exon within SNHG14 (24). Poly(A) sites commonly mark the end of transcripts and signal transcriptional termination at the end of genes. CTCF is a structural protein with multiple potential functions, including insulating active and/or inactive chromatin domains and mediating long distance chromatin interactions. Publicly available RNA-seq data (https://encode.org/; ref. 25) showed that most of SNHG14 terminates at IPW where the poly(A) sites are located in most cell types. However, RNA polymerase II (RNAPII) was shown to accumulate further downstream within PWAR1 in human embryonic stem cells (ref. 26; https://encode.org/). These data led us to hypothesize that the two elements collectively efficiently terminate transcription of SNHG14 in nonneuronal tissues (Fig. 1A), thus, restricting imprinted UBE3A expression to neurons.
Fig. 1.
Deletion of a 24 kb region between IPW and PWAR1 leads to ectopic expression of SNORD115 in iPSCs. (A) A diagram of the SNRPN/SNHG14 transcriptional unit is shown (not to scale), followed by a more detailed view of the IPW-PWAR1 region including University of California, Santa Cruz (UCSC) Genome Browser data depicting genomic elements likely to contribute to the boundary function. Approximate deletion boundaries and loxP insertions are indicated at the bottom. (B) Reverse transcription– quantitative polymerase chain reaction (RT-qPCR) data quantifying SNORD115 in iPSCs. (C) RT-qPCR data quantifying SNORD116 and SNRPN in iPSCs. For both B and C, expression values relative to the control (Ctrl) (AS iPSC) sample are shown. Error bars reflect standard error of the mean (SEM) calculated from at least three replicate cultures from each sample. ΔI-P indicates IPW-PWAR1 deletion. ΔI-P_1 and ΔI-P_2 refer to independent clones generated using the same CRISPR constructs. LoxP indicates the floxed locus. Cre_1 and Cre_2 are independent clones harboring the Cre-mediated deletion. *** denotes significance at P < 0.005.
To test this hypothesis, we deleted a 24 kb region encompassing both IPW and PWAR1 in AS iPSCs. These iPSCs harbor a ∼5.5 Mb deletion of the maternally inherited allele of chromosome 15q11-q13, and thus enable us to easily focus on genes expressed from the paternal allele. A pair of CRISPRs designed to flank both IPW and PWAR1 were electroporated into AS iPSCs along with two single stranded oligonucleotides (ssODNs) designed to insert LoxP sequences at the CRISPR cut sites following homology directed repair. After screening 96 clones using a PCR strategy modified from Kraft et al. (27), we obtained seven deletion clones and one clone with LoxP inserted at both cut sites. The LoxP sites were subsequently recombined using Cre-recombinase to create the 24 kb deletion. We also obtained one clone in which the sequence intervening the two CRISPR cut sites was inverted (INV). Two clones harboring CRISPR-mediated deletions of IPW and PWAR1 (ΔI-P) and two clones from Cre-mediated recombination between LoxP sites (CreΔI-P) were chosen for further analysis. In iPSCs with both types of deletion, we detected the expression of SNORD115 (Fig. 1B), suggesting that the 24 kb region from IPW to PWAR1 prevents expression of the distal portion of SNHG14 in iPSCs. Deletion of this region did not affect the expression of SNRPN and the proximal portions of SNHG14 (Fig. 1C).
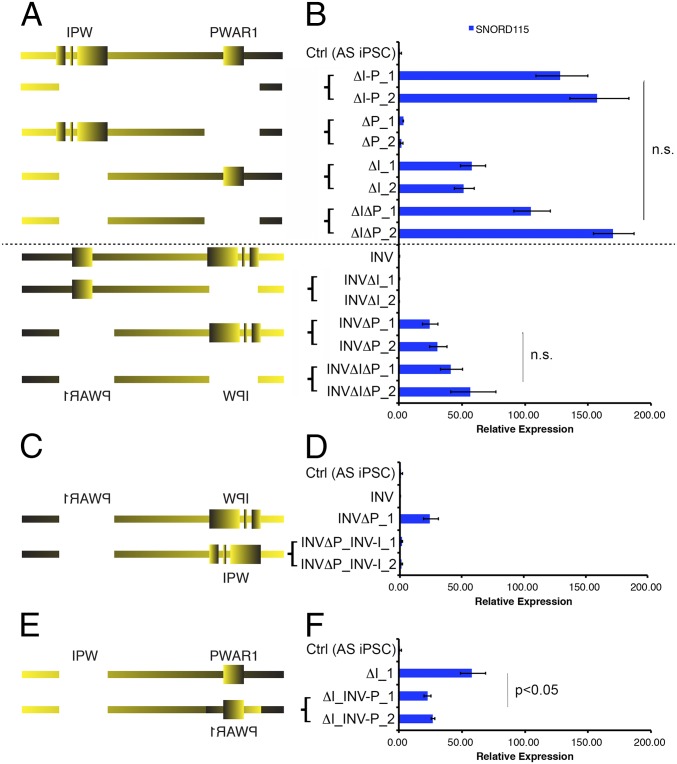
Both IPW and PWAR1 Contribute to Boundary Function.
To decipher individual contributions of IPW and PWAR1 to the boundary function, we deleted PWAR1 (ΔP) and IPW (ΔI) separately in AS iPSCs (Fig. 2A). In ΔP clones, we observed minimal expression of SNORD115. In ΔI clones, SNORD115 expression was detected at ∼50% of levels seen in ΔI-P clones. This suggested that the two components may work together to comprise full boundary function. Therefore, we deleted IPW and PWAR1 sequentially, (ΔIΔP) leaving the sequence between the two elements intact. The expression levels of SNORD115 in ΔIΔP clones were almost identical to those observed in ΔI-P clones (Fig. 2B). This confirmed that IPW and PWAR1 together are the pivotal elements providing boundary function between proximal and distal portions of SNHG14.
Fig. 2.
IPW and PWAR1 both contribute to boundary function. Diagrams of CRISPR-mediated deletions/inversions generated in unmodified AS iPSCs and INV AS IPSCs (A), INVΔP AS iPSCs (C), and ΔI AS iPSCs (E) are shown. RT-qPCR for SNORD115 in iPSCs with the corresponding deletion/inversion is shown in B, D, and F. ΔI-P_1 and ΔI-P_2 denote two independent clones generated using the same CRISPR constructs. Expression values relative to Ctrl (AS iPSC) are shown. Error bars reflect SEM calculated from, at least, three replicate cultures from each sample. n.s., not significant.
Poly(A)-dependent transcriptional termination requires proper orientation of the poly(A) sequence and downstream sequences required to bind cleavage stimulation factor and enhance poly(A)-dependent cleavage (28). Recent studies also suggest that the orientation of CTCF can influence its ability to form chromatin loops, although presumably, not all functions of CTCF require a specific orientation (29, 30). Paradoxically, when we inverted the 24 kb boundary in AS iPSCs (INV; Fig. 2B), we did not detect SNORD115 expression, suggesting that the boundary was still functional in the inverted orientation. To further understand this paradox, we deleted IPW and PWAR1 separately in the INV iPSCs. We did not detect SNORD115 when IPW was deleted in INV iPSCs (INVΔI; Fig. 2B). However, when PWAR1 was deleted in the INV iPSCs, SNORD115 was detected (INVΔP; Fig. 2B). Notably, SNORD115 expression in INVΔP lines is about 40% of that in ΔI-P lines (Fig. 2B). Sequential deletion of IPW and PWAR1 in the INV iPSCs (INVΔPΔI) resulted in a slight increase in SNORD115 expression but did not fully restore expression to the levels seen in ΔI-P or ΔIΔP iPSCs.
We took advantage of the fact that SNORD115 is expressed in ΔI and INVΔP iPSCs to individually test the directionality of IPW and PWAR1. We first restored IPW to its natural orientation in INVΔP iPSCs and found that SNORD115 expression was barely detectable (INVΔP_INV-I; Fig. 2 C and D), demonstrating that IPW can stop transcription in its natural orientation. Next, we inverted PWAR1 in ΔI iPSCs and found that SNORD115 expression was significantly reduced compared with the ΔI parent line, suggesting that the inverted PWAR1 gained a new function [(ΔI_INV-P); Fig. 2 E and F]. Together, these results suggested that both elements within the boundary require proper orientation to function appropriately.
Long-Distance Interactions Involving IPW and PWAR1.
IPW and PWAR1 constitute a strong chromatin boundary that may coincide with a putative topologically associated domain, based on published Hi-C data (31). To determine whether boundary function involves specific three-dimensional (3D) interactions, we first asked whether CTCF is bound to the PWAR1 region. CTCF is a structural protein that mediates chromatin loops and can separate chromatin boundaries. PWAR1 hosts a cluster of two divergent CTCF binding sites. We performed chromatin immunoprecipitation-sequencing (ChIP-seq) and ChIP-qPCR using antibodies against CTCF in iPSCs and iPSC-derived neurons with large deletions of maternal and paternal chromosome 15q11-q13. CTCF was bound at several sites across the imprinted domain on the paternally inherited allele in AS iPSCs, including the PWAR1 exon. However, the entire imprinted domain was largely devoid of CTCF binding in PWS iPSCs, which carry only a maternal allele of chromosome 15q11-q13 (SI Appendix, Fig. S1A). We identified allele-specific binding of CTCF at nine sites across the imprinted domain in iPSCs (SI Appendix, Fig. S1A and Table S4). CTCF binding outside of the imprinted domain was nearly identical in AS and PWS iPSCs (SI Appendix, Fig. S1A). Upon differentiation of AS iPSCs into neurons, CTCF binding at PWAR1 as well as several other sites was reduced (SI Appendix, Fig. S1 C and D). We observed retained CTCF binding in neurons at two different sites on the paternal allele, however (SI Appendix, Fig. S1B). CTCF binding at sites upstream of SNRPN and UBE3A promoters remained intact during the 10-wk time course of neural differentiation.
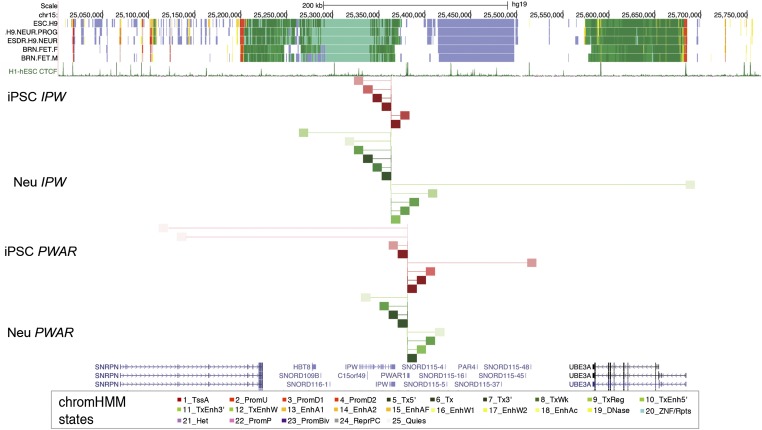
Next, we utilized circularized chromosome conformation capture followed by sequencing (4C-seq) to determine whether IPW and PWAR1 relied on specific long distance interactions to confer boundary function. The 4C enables the identification of all loci that interact with a specific viewpoint of choice. We performed 4C-seq using viewpoints located at IPW and PWAR1 in AS iPSCs and 10-wk neurons (Fig. 3). In iPSCs, the IPW viewpoint only showed significant interactions with PWAR1 and points upstream of it. In neurons, IPW interactions were mapped to points upstream and downstream, including the UBE3A promoter. Thus, IPW does not interact across the boundary in iPSCs but does in neurons where the boundary is dissolved. The CTCF binding sites at PWAR1 showed significant interactions with points upstream and downstream of the boundary in iPSCs. Points upstream that interact with the CTCF sites at PWAR1 include the upstream exons of SNRPN/SNHG14, which are annotated as strong enhancer or promoter states. Points downstream interacting with PWAR1 in iPSCs include a CTCF site at the distal end of SNORD115. In neurons, PWAR1 has few interactions and they are local. These data demonstrate that the 24 kb boundary restricts 3D interactions with IPW in iPSCs. Although 3D interactions with the CTCF sites at PWAR1 differ between iPSCs and neurons, they do not seem to be restricted by boundary function. In fact, 3D interactions with PWAR1 in iPSCs are more consistent with an interaction between the alternative upstream promoters of SNRPN/SNHG14 and the 3′ end of transcripts originating there.
Fig. 3.
Three-dimensional interactions with IPW and PWAR1. Analysis of 4C-seq data are shown along with chromatin state annotations from H9 hESCs, H9-derived neural progenitors, H9-derived neurons, and male/female fetal brain tissues from the Roadmap Epigenomics Project. CTCF binding sites and UCSC genes are shown for reference. The red lines and blocks refer to interactions in AS iPSCs, and the green lines and blocks refer to interactions in AS iPSC-derived neurons. The thin vertical lines at IPW and PWAR1 refer to the anchor point for 4C-seq. All interactions are significant (P < 0.001) with darker colors indicating decreased P value (higher significance).
UBE3A Imprinting Requires Sufficient Levels of UBE3A-ATS Expression.
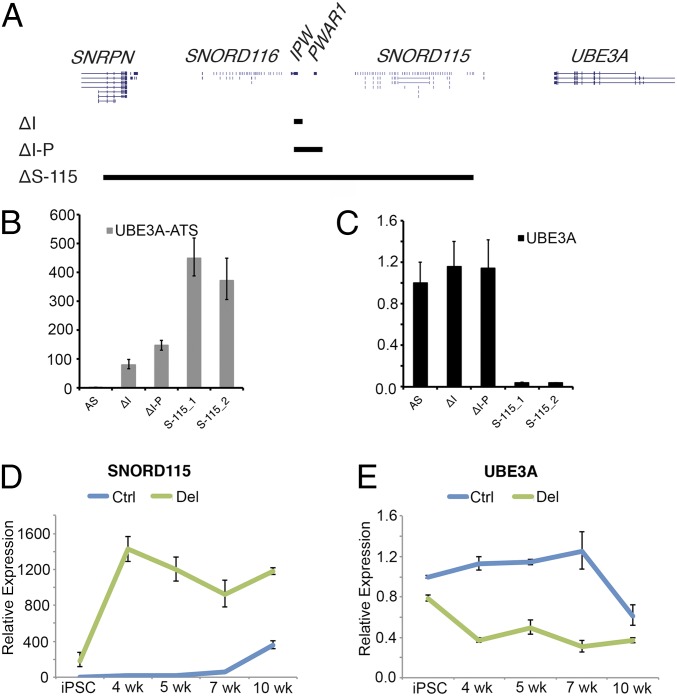
We previously reported imprinted UBE3A expression in an iPSC line that aberrantly expresses UBE3A-ATS due to an atypical PWS deletion. Based on these data, we predicted that UBE3A would be imprinted in iPSCs expressing SNORD115 and UBE3A-ATS. To our surprise, UBE3A imprinting was not observed in ΔI and ΔI-P clones where UBE3A-ATS is transcribed (Fig. 4B). Therefore, we tried to recapitulate our previous observation with the atypical PWS deletion in an AS iPSC line (21). We used CRISPR/Cas9 to remove a 303 kb region between SNRPN intron 1 and the last copy of SNORD115 (SNORD115-47) in AS iPSCs (ΔS-115; Fig. 4A). This deletion juxtaposes both canonical and upstream SNRPN/SNHG14 promoter(s) immediately upstream of UBE3A-ATS. Indeed, paternal UBE3A is completely repressed in iPSCs with this deletion (Fig. 4C) suggesting that increasing UBE3A-ATS transcription is necessary to imprint UBE3A.
Fig. 4.
Sufficient expression of UBE3A-ATS is required to imprint UBE3A. A diagram depicting relative sizes of ΔI, ΔI-P, and ΔS-115 deletions is shown in A. RT-qPCR for UBE3A-ATS and UBE3A are shown in B and C, respectively. RT-qPCR for SNORD115 and UBE3A is shown across a time course of neural development in AS iPSCs (Ctrl) and ΔI-P AS iPSCs (Del) in D and E, respectively. Expression values relative to the unedited AS sample are shown. The error bars reflect SEM calculated from, at least, three replicate cultures from each sample.
Since transcription of SNHG14 is normally increased during neurogenesis, we sought to determine whether an early increase in expression of UBE3A-ATS during neurogenesis would lead to premature imprinted UBE3A expression in neural derivatives of ΔI-P iPSCs, which lack the boundary. We differentiated AS and ΔI-P iPSCs into forebrain cortical neurons as previously described (32) and collected RNA samples during the time course of differentiation. We found that SNORD115 expression increases and UBE3A becomes silenced between weeks 7 and 10 of differentiation in AS iPSCs, consistent with our previously published observations (Fig. 4 D and E) (13, 19). The ΔI-P iPSCs showed a slight reduction of UBE3A expression compared with AS iPSCs. Within 4 wk of neural differentiation, SNORD115 expression in ΔI-P neural progenitors is increased to maximum levels, and UBE3A attains it lowest expression levels (Fig. 4 D and E). These data demonstrate that sufficient levels of UBE3A-ATS transcription are necessary to silence UBE3A and that the 24 kb boundary element also regulates the timing of UBE3A imprinting during neurogenesis.
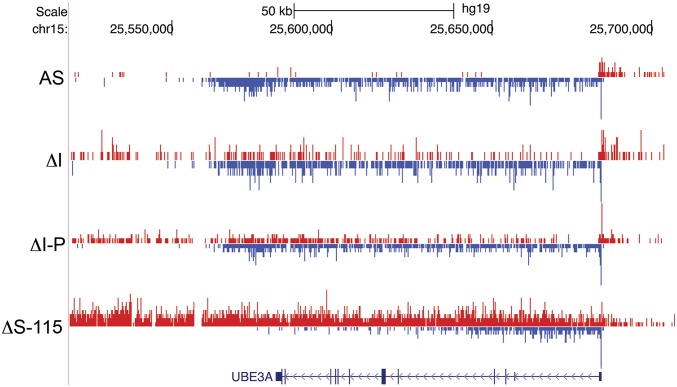
UBE3A-ATS is expressed in ΔI and ΔI-P iPSCs, but UBE3A is not imprinted. On the other hand, UBE3A-ATS is expressed, and UBE3A is imprinted in ΔS-115 iPSCs, enabling us to study AS iPSCs that imprint and do not imprint UBE3A. We sought to visualize and compare the interactions between UBE3A-ATS and UBE3A under these conditions. We performed precision nuclear run-on sequencing (PRO-seq) on these samples. PRO-seq determines the active sites of transcriptionally engaged RNAPII by mapping nascent transcription (33, 34). PRO-seq data from iPSC lines revealed plus-strand RNAPII density across UBE3A-ATS in ΔI, ΔI-P, and ΔS-115 iPSCs (Fig. 5). Minus-strand RNAPII density was seen across the entire UBE3A gene in all iPSCs, but the ΔS-115 iPSCs had robust PRO-seq density only in the first half of the gene (Fig. 5). These data suggest UBE3A imprinting coincides with reduction of the full-length transcript since polymerases do not appear to efficiently make it to the 3′ end of the gene.
Fig. 5.
Imprinting of UBE3A coincides with reduced RNAPII density across the 3′ half of UBE3A gene body. PRO-seq was used to map RNAPII density in AS, ΔI, ΔI-P, and ΔS-115 iPSCs. Plus-strand RNAPII density is shown in red. Minus-strand RNAPII density is shown in blue.
Discussion
Imprinted expression of UBE3A is restricted to neurons by the tissue-specific expression of UBE3A-ATS (8, 11, 35). UBE3A-ATS is at the 3′ end of SNHG14, which is the host gene for SNORD116 and SNORD115 as well as other noncoding RNAs (12). In humans, the proximal half of SNHG14 is expressed broadly in different tissue types, whereas the distal half, including UBE3A-ATS, is restricted to neurons (13, 17, 19). We used CRISPR/Cas9 to functionally define the boundary element that restricts UBE3A-ATS expression to neurons (Fig. 2). We found the boundary to be composed of two parts: one part includes poly(A) and conserved sequences in the last exon of IPW, whereas the other includes a cluster of CTCF sites in and around the exon annotated as PWAR1. Although both elements contribute to boundary function, IPW plays a larger role and is required to completely stop transcription in nonneuronal cells. IPW requires its natural orientation to stop transcription, suggesting that the poly(A) sites are important for boundary function. CTCF binds to the PWAR1 exon in iPSCs but not in neurons, suggesting that CTCF binding may contribute to boundary function as well. PRO-seq experiments demonstrate reduced RNAPII density downstream of PWAR1 in ΔI iPSCs, suggesting that these CTCF sites may pause RNAPII and facilitate RNAPII disengagement (SI Appendix, Fig. S2). CTCF has been previously shown to pause elongating RNAPII to influence alternative splicing (36). Interestingly, RNAPII is paused and/or disengaged near the first exons encoding SNORD115 in ΔI-P iPSCs by an as-yet-unknown mechanism. This suggests multiple redundancies may prevent UBE3A imprinting in this cell type. Based on these findings, we propose a simple model by which this bipartite boundary element stops transcription in most cell types. We propose that the poly(A) sites within IPW stop transcription via poly(A)-dependent cleavage, whereas CTCF binding at PWAR1 slows RNAPII enough to allow the XRN2 5′-3′ exonuclease to lead to termination in what is known as the “torpedo model” of transcription termination (37, 38).
It is not clear how the boundary function is lost during neurogenesis. The 4C-seq experiments demonstrate that 3D interactions with IPW are restricted to sites upstream in iPSCs but are bidirectional in neurons, consistent with a loss of boundary function during neurogenesis. CTCF binding within PWAR1 is present on the paternal allele in iPSCs but not in neurons. This loss of CTCF binding may contribute to the loss of boundary function in neurons. Consistent with this hypothesis, sites interacting with PWAR1 in neurons are limited to nearby loci are largely not bound by CTCF and overlap with several sites interacting with IPW.
We further speculate that loss of CTCF binding may contribute to reduced termination at IPW in neurons. CTCF is gradually lost from PWAR1 during the 10-wk course of neural differentiation (SI Appendix, Fig. S1C), correlating with full expression of UBE3A-ATS and imprinting of UBE3A (Fig. 4 D and E). iPSCs lacking the bipartite boundary imprint UBE3A precociously during neuronal differentiation, supporting the hypothesis that the boundary element also controls the developmental timing of UBE3A imprinted expression. An understanding of how IPW and PWAR1 independently contribute to the developmental timing of UBE3A imprinting may help determine how they facilitate boundary removal during neurogenesis. Paradoxically, deletion of PWAR1—including both CTCF sites—does not substantially decrease transcriptional termination in iPSCs (Fig. 1A). Perhaps this is due to the presence of additional elements capable of pausing RNAPII. Indeed, PRO-seq data reveal RNAPII pausing near the first exon of the SNORD115 cluster (SI Appendix, Fig. S2).
Finally, the surprising observation that UBE3A-ATS is expressed, but UBE3A is not imprinted in iPSCs with deletions of IPW or IPW plus PWAR1 (Fig. 4C) indicate that imprinted expression of UBE3A also requires sufficient expression of UBE3A-ATS in addition to the loss of boundary function. Indeed, a CRISPR-mediated deletion that increases UBE3A-ATS expression led to full repression of paternal UBE3A. PRO-seq experiments further demonstrated that UBE3A imprinting in these iPSCs coincided with reduced active RNAPII across the 3′ half of UBE3A (Fig. 5). These data further support the notion that UBE3A-ATS represses paternal UBE3A via transcriptional interference. If UBE3A imprinting occurs due to transcriptional interference, manipulation of UBE3A-ATS or UBE3A transcription may provide alternative therapeutic approaches for AS.
Materials and Methods
Cell Culture.
AS iPSC (AS del 1–0) and PWS iPSC (PWS del 1–7) lines were generated and maintained by mechanical passaging on mouse embryonic fibroblasts as previously described (13, 19).
CRISPR Genome Editing.
CRISPR guide RNA sequences were designed using CRISPR Genome Engineering Resource (https://zlab.bio/guide-design-resources) (39) and cloned into the px459 V2 vector (40, 41). The sequence of CRISPRs and ssODNs used in this paper are listed in SI Appendix, Table S2.
ChIP.
ChIP qPCR was performed using Millipore EZ-Magna ChIP G (17-409) following manufacturer’s instructions using 6–106 cells. SYBR green primers (SI Appendix, Table S1) were used for ChIP-qPCR. For ChIP-seq, library preparation and sequencing were performed by the Genomics Core in the Yale Stem Cell Center. FASTQ files were mapped and analyzed using Homer with the parameters described previously (42, 43). Full data are deposited in the Gene Expression Omnibus accession browser under the accession number GSE117283 (44).
4C-Seq.
The 4C-seq was carried out as described (45) using nuclei harvested from ∼26 iPSCs and iPSC-derived neurons. The NlaIII enzyme was used for the first digestion, and DpnII was used for the second digestion. Data were analyzed using the r3Cseq package (46).
PRO-Seq.
PRO-seq was carried out as described (33, 34, 47) using 1 × 106 permeabilized cells per iPSC line.
Detailed Materials and Methods are found in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Carissa Sirois and Dr. Marc Lalande for helpful discussions. The work was supported by the following funding sources: NIH Grant R01HD068730, Angelman Syndrome Foundation, and Connecticut DPH Stem Cell Research Program (Grant 12SCBUCHC) (to S.J.C.) and NIH Grant R35GM119465 (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Full data have been deposited in Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE117283).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815279116/-/DCSupplemental.
References
- 1.Williams CA, et al. Angelman syndrome 2005: Updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- 2.Williams CA. Neurological aspects of the Angelman syndrome. Brain Dev. 2005;27:88–94. doi: 10.1016/j.braindev.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 4.Matsuura T, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 5.Knoll JH, et al. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet. 1989;32:285–290. doi: 10.1002/ajmg.1320320235. [DOI] [PubMed] [Google Scholar]
- 6.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 7.Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- 8.Rougeulle C, Cardoso C, Fontés M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 9.Huang HS, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2011;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng L, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L, et al. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet. 2013;9:e1004039. doi: 10.1371/journal.pgen.1004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Runte M, et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 13.Chamberlain SJ, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, et al. Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Mol Cell. 2016;64:534–548. doi: 10.1016/j.molcel.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Yin QF, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 16.Wevrick R, Kerns JA, Francke U. Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]
- 17.Cavaillé J, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castle JC, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamberlain SJ, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galiveti CR, Raabe CA, Konthur Z, Rozhdestvensky TS. Differential regulation of non-protein coding RNAs from Prader-Willi syndrome locus. Sci Rep. 2014;4:6445. doi: 10.1038/srep06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martins-Taylor K, et al. Imprinted expression of UBE3A in non-neuronal cells from a Prader-Willi syndrome patient with an atypical deletion. Hum Mol Genet. 2014;23:2364–2373. doi: 10.1093/hmg/ddt628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutcliffe JS, et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 26.Birney E, et al. ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft K, et al. Deletions, inversions, duplications: Engineering of structural variants using CRISPR/Cas in mice. Cell Rep. 2015;10:833–839. doi: 10.1016/j.celrep.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Neve J, Patel R, Wang Z, Louey A, Furger AM. Cleavage and polyadenylation: Ending the message expands gene regulation. RNA Biol. 2017;14:865–890. doi: 10.1080/15476286.2017.1306171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Wit E, et al. CTCF binding polarity determines chromatin looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Guo Y, et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germain NDCP, et al. Gene expression analysis of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11-q13.1. Mol Autism. 2014;5:44. doi: 10.1186/2040-2392-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahat DB, et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 36.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 38.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352:aad9926. doi: 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cotney JL, Noonan JP. Chromatin immunoprecipitation with fixed animal tissues and preparation for high-throughput sequencing. Cold Spring Harb Protoc. 2015;2015:419. doi: 10.1101/pdb.err087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cotney J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chamberlain S, et al. 2018 A bipartite boundary element restricts UBE3A imprinting to mature neurons. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117283. Deposited July 18, 2018.
- 45.Wilderman A, VanOudenhove J, Kron J, Noonan JP, Cotney J. High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Reports. 2018;23:1581–1597. doi: 10.1016/j.celrep.2018.03.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thongjuea S, Stadhouders R, Grosveld FG, Soler E, Lenhard B. r3Cseq: An R/bioconductor package for the discovery of long-range genomic interactions from chromosome conformation capture and next-generation sequencing data. Nucleic Acids Res. 2013;41:e132. doi: 10.1093/nar/gkt373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.