Significance
Understanding the dynamics of competing species is essential for explaining the origin and maintenance of species diversity. However, ecologists have typically ignored the potential for rapid evolution to alter the contemporary population dynamics of competing species. By disrupting the ability of aquatic plants to evolve in response to interspecific competition, we show that competition drives evolutionary change and this evolutionary change simultaneously feeds back to alter the abundance of competing species over just a few generations. Rather than increasing niche differences as classic theory predicts, evolution causes population trajectories to converge by changing the competitive hierarchy. Our results suggest that understanding how species diversity is maintained requires explicitly accounting for the effects of rapid evolution on competitive population dynamics.
Keywords: biodiversity, eco-evolutionary dynamics, competition, character displacement, diversity maintenance
Abstract
Increasing evidence for rapid evolution suggests that the maintenance of species diversity in ecological communities may be influenced by more than purely ecological processes. Classic theory shows that interspecific competition may select for traits that increase niche differentiation, weakening competition and thus promoting species coexistence. While empirical work has demonstrated trait evolution in response to competition, if and how evolution affects the dynamics of the competing species—the key step for completing the required eco-evolutionary feedback—has been difficult to resolve. Here, we show that evolution in response to interspecific competition feeds back to change the course of competitive population dynamics of aquatic plant species over 10–15 generations in the field. By manipulating selection imposed by heterospecific competitors in experimental ponds, we demonstrate that (i) interspecific competition drives rapid genotypic change, and (ii) this evolutionary change in one competitor, while not changing the coexistence outcome, causes the population trajectories of the two competing species to converge. In contrast to the common expectation that interspecific competition should drive the evolution of niche differentiation, our results suggest that genotypic evolution resulted in phenotypic changes that altered population dynamics by affecting the competitive hierarchy. This result is consistent with theory suggesting that competition for essential resources can limit opportunities for the evolution of niche differentiation. Our finding that rapid evolution regulates the dynamics of competing species suggests that ecosystems may rely on continuous feedbacks between ecology and evolution to maintain species diversity.
Classic theory suggests that natural selection arising from interspecific competition should generate phenotypic differences between species that weaken interspecific competition, favoring species coexistence (1–5). However, while this eco-evolutionary process has become central to explanations for diversity (6, 7), empirical evidence for such feedbacks between ecology and evolution remain equivocal for three reasons (8). First, evolution in response to interspecific competition is most commonly inferred from post hoc observational evidence that morphological differences between species are larger when species occur together (in sympatry) vs. apart (in allopatry) (8–10), with the strongest evidence coming from natural experiments (11, 12). While a few experimental studies demonstrate trait change in response to interspecific competition (13, 14), the causal influence of competition, the repeatability of the evolutionary change, and the speed of evolution relative to the rate of competitive population dynamics are often unknown (8).
Second, and maybe more importantly, while the feedback from evolutionary change to the population dynamics of the competing species is essential for contemporary evolution to affect diversity maintenance, this feedback is rarely empirically quantified (8, 15, 16). In lieu of direct empirical evidence, population dynamic consequences are typically assumed following the common theoretical expectation that evolved trait and behavioral differences should promote coexistence by increasing niche differences (7, 8, 11, 17–19). However, as emphasized in recent reviews, the assumption that evolution should increase niche differences may not always be justified (16, 20). Developments in species coexistence theory demonstrate an equally important role for differences between species in competitive ability in determining competitive outcomes (21–23). Indeed, theory suggests that evolution of competitive ability may be more likely when opportunities for the evolution of niche differentiation are limited, as occurs when species cannot substitute other resources for the ones used by their competitors (24, 25). In general then, it remains unclear how competitive population dynamics should change as a consequence of evolution, and the resulting eco-evolutionary feedbacks could be more complex than is generally appreciated (16, 20).
A third reason for poorly resolved empirical evidence for competitive, eco-evolutionary feedbacks relates to the concurrent nature of the ecological and evolutionary changes. If ecological and evolutionary processes simultaneously feed back on one another, this dynamic cannot be resolved by assuming a separation of timescales, and quantifying the population-dynamic consequences of past evolutionary change (15, 26). These three limitations have motivated recent calls for combining experimental evolution approaches (8) more typical of studies in laboratory microbial systems (27) with the tools of quantitative coexistence theory (16) to understand how rapid evolution in response to competition affects species coexistence in more natural systems.
Here, we demonstrate how eco-evolutionary feedbacks influence coexistence by experimentally manipulating the ability of aquatic plant species to evolve in response to interspecific competition while simultaneously quantifying their multigenerational competitive population dynamics. Our approach allows us to address three questions: (i) Does interspecific competition cause evolutionary change on ecological timescales? (ii) Is this evolutionary change consistent and large enough to alter the contemporary dynamics of the competing species? And, (iii) how does this feedback influence coexistence via the evolution of niche differences and/or species’ competitive abilities? To answer these questions, we studied two species of floating, aquatic plants—Lemna minor and Spirodela polyrhiza. Both species have fast life cycles with asexual reproduction every 3–7 d and ∼20 generations per growing season (28), providing an ideal system for understanding how eco-evolutionary feedbacks affect the contemporary dynamics of competing species.
We imposed two selection treatments on multigenotype populations of the two species competing in replicate experimental ponds in the field (Materials and Methods). In the “heterospecific selection” treatment, the two species competed with each other in competitive arenas and were free to evolve in response to interspecific competition. In the “conspecific selection” treatment, the two species also competed with each other, but in this treatment we prevented evolution in response to interspecific competition. We did so by replacing all individuals in these competitive arenas every 2 wk with the same number of individuals of each species, but drawn from multigenotype populations growing and evolving in single-species monocultures in the same ponds. Thus, our experimental manipulation preserves the ongoing effects of interspecific competition on the population sizes of the two species, but prevents evolution in response to interspecific competition, and thereby prevents this evolution from affecting the ecological dynamics of the competing species (29). Importantly, the species in both treatments were able to evolve to other biotic and abiotic selection pressures arising naturally in the field during the experiment. To quantify eco-evolutionary trajectories, we combined assessments of genotypic and phenotypic change with surveys of multigenerational competitive population dynamics. Finally, we did additional competition experiments using the evolved populations to quantify how evolution affects ecological dynamics by altering niche and competitive-ability differences.
Results and Discussion
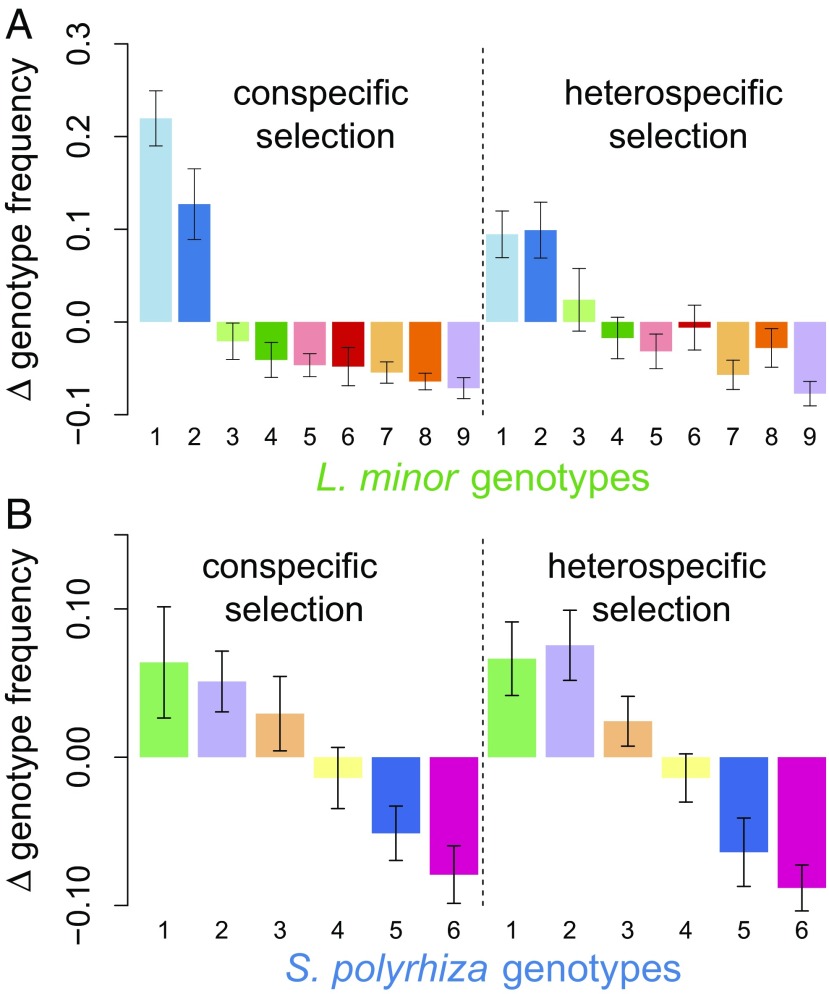
Interspecific competition drove rapid evolutionary change (Fig. 1). Specifically, selection in response to conspecific vs. heterospecific competitors generated differences in the genotypic composition of the evolved populations of L. minor [permutational multivariate analysis of variance (PERMANOVA): F(1,12) = 2.80, P = 0.019], but not of S. polyrhiza [F(1,12) = 0.1, P = 0.99; SI Appendix, Fig. S1 and Table S1]. The different evolutionary trajectories for L. minor were driven most strongly by changes in the frequency of a single genotype (genotype 1 in Fig. 1). Selection for this genotype was positive in both treatments, but significantly less so when in competition with heterospecific competitors [t(12) = 4.94, P < 0.001; SI Appendix, Fig. S2]. This genotype had the most extreme phenotypic trait values in the population (SI Appendix, Fig. S3), suggesting that interspecific competition was an agent of directional selection on L. minor. Importantly, differences in genotypic evolution caused differences in phenotypic evolution between treatments (described further below).
Fig. 1.
Competition between species drives evolutionary change. Bars show the mean change in frequency of each genotype of (A) L. minor and (B) S. polyrhiza in the conspecific and heterospecific selection treatments. The change in genotype frequency is the change from the beginning of the experiment in June until the sampling date in August. Each color and number represents a different genotype. Errors are SEMs.
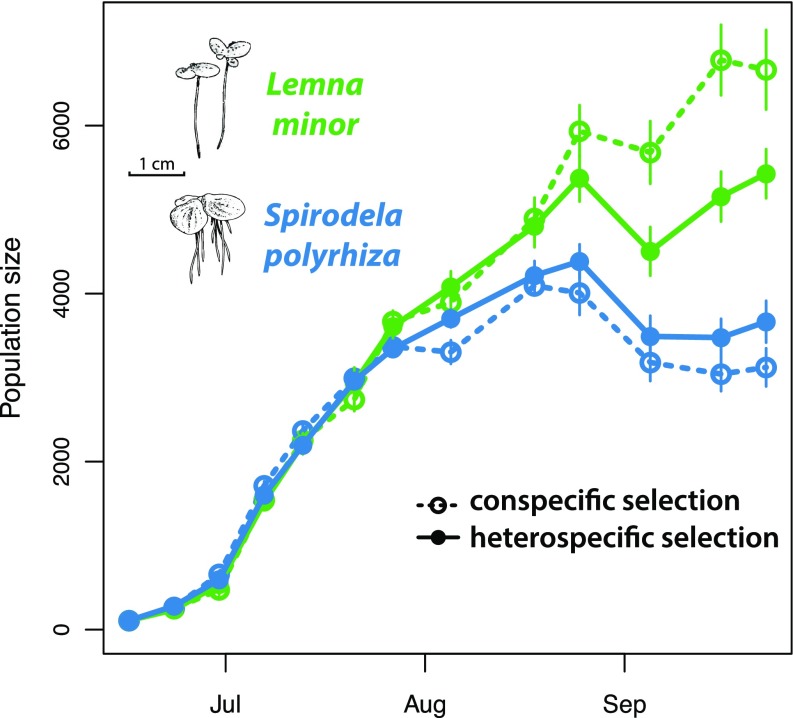
The evolutionary change was sufficiently large and rapid to affect the concurrent ecological dynamics of the competing species (Fig. 2). After a period of rapid growth from low density by both species, L. minor became numerically dominant in the conspecific selection treatment, with nearly twice as many individuals as S. polyrhiza (Fig. 2 and SI Appendix, Fig. S4). By contrast, competitor abundances were more even in the heterospecific selection treatment, with significantly lower final abundances of L. minor and significantly higher final abundances of S. polyrhiza (Fig. 2 and SI Appendix, Fig. S4 and Table S2; likelihood-ratio tests comparing population trajectories, L. minor, = 29.54, P < 0.001; S. polyrhiza, = 8.02, P = 0.018). Indeed, over the last third of the experiment, the average population size of L. minor was between 15 and 20% lower in the heterospecific selection treatment [F(1,11.92) = 8.16, P = 0.015; Fig. 2].
Fig. 2.
Evolution in response to competition alters the population dynamics of competing species. Population trajectories of the competing species, L. minor (green) and S. polyrhiza (blue), when they were able to evolve in response to interspecific competition (heterospecific selection, solid line) or unable to evolve to interspecific competition (conspecific selection, dashed line). Errors are SEMs.
Following coexistence theory, the observed changes in dynamics could be caused by the evolution of increased niche differences, a decrease in the competitive ability of L. minor relative to S. polyrhiza, or a combination of these effects (16, 20). Evaluating these scenarios requires quantifying niche and competitive-ability differences in each treatment. Expressions for these quantities can be derived from the mutual invasibility criterion of coexistence (21), and their values estimated based on a parameterized model of competitive population dynamics describing the species’ interaction (23). We parameterized an appropriate competition model using data from a separate set of field competition experiments, which were required to disentangle the effects of intraspecific and interspecific competition on dynamics (Materials and Methods). These experiments involved measuring the population growth of individuals from the evolved populations of each species in each treatment, competing against a density gradient of conspecifics and (separately) heterospecifics from the same treatment (ref. 23 and SI Appendix, Figs. S5 and S6 and Table S3). We then used the parameter estimates from the competition model to quantify niche and competitive-ability differences in each treatment.
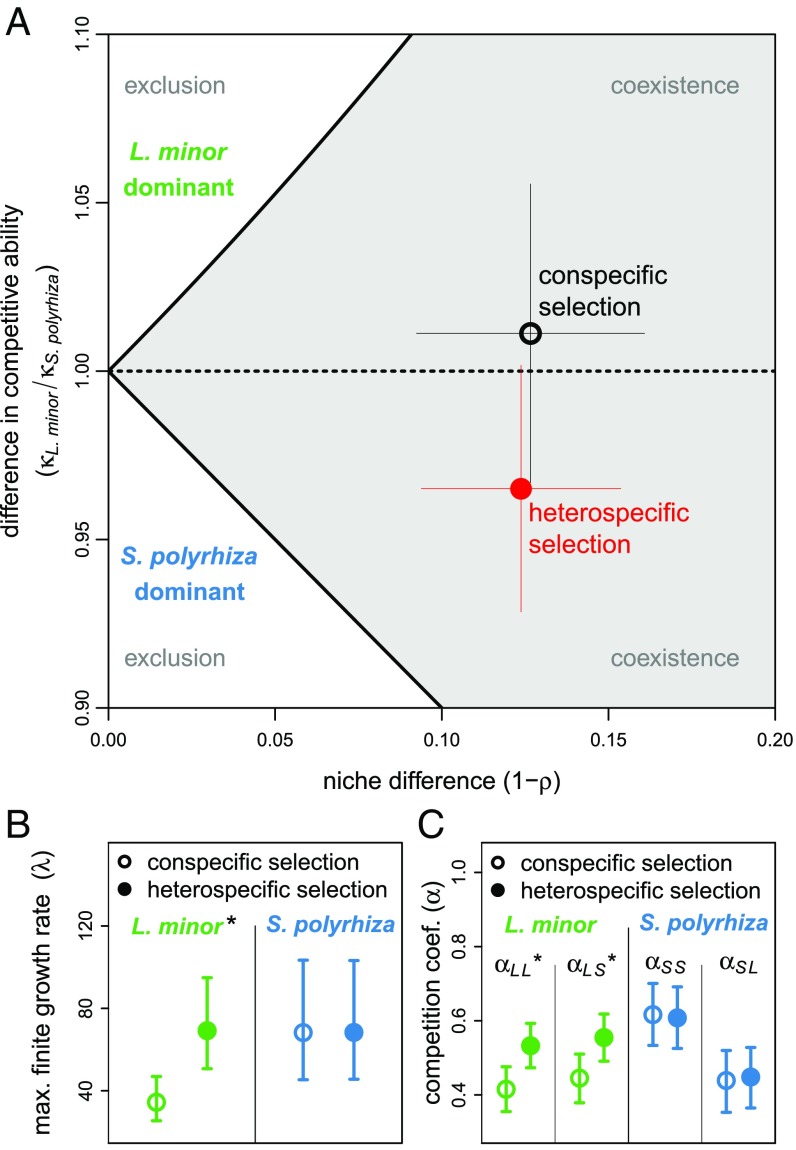
Results show that, while the predicted coexistence outcome did not change between treatments, the more even abundances of the competing species under heterospecific selection were more consistent with a change in species’ competitive abilities than with an increase in niche differences (Fig. 3). Specifically, we found little difference between treatments in the estimated niche difference, especially compared with the decrease in competitive ability of L. minor relative to S. polyrhiza (Fig. 3A). To evaluate the likelihood that these two alternative pathways contributed to the more even population abundances observed in our main experiment, we used Monte Carlo simulations to draw 106 possible combinations of the competition model parameters for each species in each treatment, based on the uncertainty in the original parameter estimates. We then calculated equilibrium population abundances, and niche and competitive-ability differences for each parameter combination within this set. For the parameter combinations that gave more even population abundances in the heterospecific vs. conspecific selection treatment—a situation that matches the observed abundances that we are aiming to explain (Fig. 2)—the competitive ability of L. minor decreased relative to S. polyrhiza in 96.4% of cases. By contrast, more even abundances were equally likely to be associated with increases or decreases in niche differences (46.3% and 53.7% of cases, respectively), suggesting little consequence of evolved changes in niche differences as a driver of the observed dynamics.
Fig. 3.
The niche and competitive-ability differences between L. minor and S. polyrhiza after evolution in response to conspecific and heterospecific selection. (A) Stable coexistence occurs when the stabilizing niche difference () exceeds the magnitude of the competitive-ability difference , shown by the gray shaded region defined by the inequality (Materials and Methods). (B) The effects of the selection treatments on species’ maximum finite rate of growth . (C) The effects of the selection treatments on intraspecific () and interspecific () competition coefficients. Lines in A show 1 SD. Lines in B and C show 95% confidence intervals. The asterisk (*) indicates that the difference is significant (SI Appendix, Table S3).
The lower competitive ability of L. minor occurred despite this species evolving a greater maximum finite rate of growth in the heterospecific selection treatment (Fig. 3B). All else being equal, a higher maximum finite rate of growth will increase competitive ability (23). However, the positive effect of this demographic change was more than counteracted by a large decrease in the ability of L. minor to maintain offspring production under crowded conditions [i.e., evolution caused L. minor’s sensitivity to competition (23) to increase because of increases in this species’ response to both intraspecific () and interspecific () competition; Fig. 3C]. While existing theory accommodates the possibility of evolutionary changes in competitive ability (24), it does not generally consider separate (and contrasting) effects of evolution on these different components of competitive ability. Regardless of the specific evolutionary constraints driving these effects, the findings exposed here by combining experimental evolution with quantitative coexistence theory highlight the potential for complex evolutionary responses to competition.
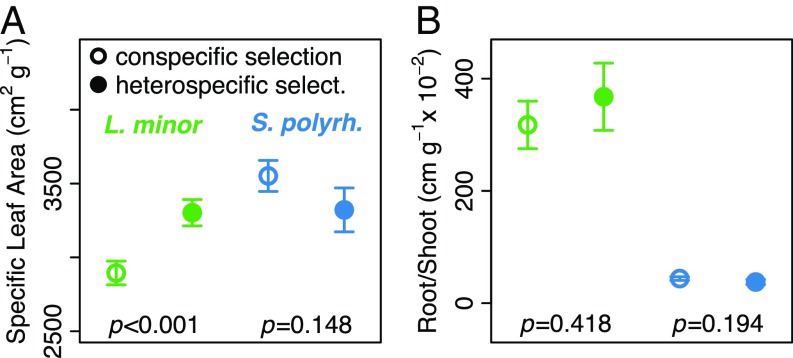
Finally, we asked whether phenotypic traits differed between heterospecific and conspecific competitive environments in a manner consistent with the evolved competitive dynamics. S. polyrhiza traits did not differ between treatments (Fig. 4), consistent with the lack of genetic differences between treatments (Fig. 1). By contrast, L. minor had greater specific-leaf area under heterospecific selection [Fig. 4A; F(1,12) = 20.98, P < 0.001]. This increase is consistent with the large decrease in the heterospecific selection treatment of the L. minor genotype that had the lowest specific-leaf area (genotype 1 in Fig. 1; SI Appendix, Fig. S3). Importantly, for each species, the differences in specific-leaf area between treatments in the field were consistent with those found under common-garden conditions in the laboratory (SI Appendix, Fig. S7), confirming that the observed differences in the field had a genetic component.
Fig. 4.
The effects of the conspecific and heterospecific selection treatments on phenotypic trait evolution. (A) Specific-leaf area. (B) Root length-to-frond mass ratio. Errors are SEMs.
Traditionally, the evolution of traits in response to interspecific competition would be interpreted as evidence for changes in niche differences (7, 8, 11, 17–19). However, given that our demographic analyses suggest that competitive ability and not niche differences evolved in response to interspecific competition (Fig. 3), trait change in our study is more consistent with a relationship between trait evolution and changes in competitive ability. Indeed, higher specific-leaf area tends to be associated with higher growth rate (30) and greater sensitivity to resource depletion (31), both of which are consistent with the evolved changes in L. minor’s maximum finite rate of growth (Fig. 3B) and sensitivity to competition (Fig. 3C), respectively. The differences in trait values at the individual level also had some important implications for the differences in the structure of the evolving populations between treatments. While the population size of L. minor was lower in the heterospecific selection treatment (Fig. 2), as was total biomass [F(1,12) = 10.32, P = 0.008; SI Appendix, Fig. S8], the total frond area of L. minor did not differ between treatments [F(1,12) = 1.33, P = 0.271; SI Appendix, Fig. S8], because the area of individual fronds was greater under heterospecific selection [F(1,12) = 5.84, P = 0.033; SI Appendix, Fig. S9]. By contrast, the increase in population size of S. polyrhiza in the heterospecific selection treatment translated directly into an increase in both the total (population-level) area [F(1,12) = 12.01, P = 0.005] and total (population-level) biomass of fronds [F(1,12) = 17.11, P = 0.001; SI Appendix, Fig. S8] in the competitive arenas.
In sum, our results suggest that evolution in response to interspecific competition resulted in more even population sizes of the competitors (Fig. 2) and did so via evolved changes in competitive ability rather than the evolution of greater niche differences (Fig. 3). In general, evolved changes in competitive ability are predicted by theory when species cannot substitute other resources for the ones used by their competitors (i.e., the same resources are essential for both competing species; refs. 24 and 25), conditions that may characterize our system. In the experimental ponds, as in nature, L. minor and S. polyrhiza likely compete most strongly for space, light, and essential nutrients, resources that may not be substitutable. The degree of limitation by these factors is unknown, and even space may be less limiting than expected as plants can overlap on the surface of the water before populations equilibrate. Nonetheless, a potential explanation for our results is that competition for these nonsubstitutable resources constrained opportunities for the evolution of niche differences, while allowing changes in the efficiency with which shared resources are exploited (24, 25). Our system is unlikely to be unusual in this respect, because competition for nonsubstitutable resources is likely to structure other functionally sessile and/or autotrophic communities, including terrestrial plant and marine benthic communities (32, 33). While our experiment likely captures nonspatial opportunities for niche differences that occur in natural ponds—and captured sufficient opportunities for niche differentiation to allow coexistence in both treatments (Fig. 3A)—it is possible that more complex spatially varying environments would provide additional opportunities for the evolution of niche differences, as well as competitive abilities, in response to interspecific competition (12, 34, 35).
Our study has a number of limitations to consider when relating our findings to the evolution of competing species more generally. First, our results reflect the effects of selection on standing genetic variation. While our study does not, therefore, account for evolutionary dynamics arising from de novo mutations (27), selection on standing genetic variation is likely to be a more important driver of evolution rapid enough to alter concurrent ecological dynamics (36). Second, the competing species in our experiment began with a random selection of nine and six genotypes of L. minor and S. polyrhiza, respectively. Our results suggest that even with these low levels of genotypic variance, evolution in response to competition can alter competitive dynamics. Nevertheless, it is possible that higher levels of genotypic diversity or different combinations of genotypes may have generated different results, including the evolution of niche differentiation. Future work quantifying competition at the genotype level may be able to quantify levels of additive genetic variance in niche and competitive-ability traits, and thus quantify the potential for evolution along these axes as a function of genotypic diversity. Third, because the individuals in our study are clonal, traits are perfectly linked. Nonetheless, it is unclear how recombination would affect the speed and direction of evolution in our system. Fourth, while our experiment allowed us to attribute the differences between treatments to the influence of interspecific competition, it is unknown how the contribution of evolution in both treatments in response to selection pressures other than competition affected the observed dynamics of the competing species. Understanding these effects would be a worthy goal for future work (37). Finally, phenotypic plasticity, including via maternal effects, could have contributed to our results. While we cannot rule this possibility out, additional experiments testing for these effects demonstrate that plasticity tends to increase the overall competitive performance of L. minor in heterospecific competitive environments, and so, if anything, is likely to have counteracted the overall decrease in competitive performance we observed in our evolution experiment (SI Appendix, Fig. S10).
For much of the last century, most ecologists have treated the maintenance of species diversity as a purely ecological process (21, 38, 39). In addition, while evolution was commonly thought to shape the traits of competing species, the simultaneous feedbacks between these ecological and evolutionary processes, as expected from theory, have been difficult to empirically evaluate. Only by experimentally disrupting the eco-evolutionary feedback through altering species’ abilities to evolve in response to their competitive environment were we able to directly show that evolution concurrent with ecological dynamics strongly affects the population dynamics of competing species over just a few generations. The outcome of this approach, coupled with the principles of quantitative species coexistence theory, gives a unique process-level view of the eco-evolutionary dynamics expected to shape contemporary patterns of biodiversity. Our results suggest that understanding competitive population dynamics—a cornerstone of ecological knowledge—may require accounting for the simultaneous influence of rapid evolutionary change.
Materials and Methods
Species, Collection, and Culturing.
Lemna minor and Spirodela polyrhiza are small, globally distributed, floating, aquatic plants belonging to the Lemnoideae subfamily of the Araceae family (28). The plants are morphologically simple, composed of a floating frond with small rootlets attached to the underside (Fig. 2). Flowering is rare, and instead, reproduction occurs every 3–7 d via asexual budding of daughter fronds. Populations often contain multiple genotypes (40). L. minor and S. polyrhiza likely compete most strongly for space, light, and essential nutrients. Populations vary greatly in density, and where nutrient availability is high, they can grow at high densities in overlapping layers. Our experiment used six genotypes of S. polyrhiza and nine genotypes of L. minor (SI Appendix, Tables S4 and S5). Genotypes were collected from ponds in central Europe in 2015, and two genotypes of S. polyrhiza from the same region were obtained from the collection of M. Huber and S. Xu (then at the Max Planck Institute for Chemical Ecology, Jena, Germany). To distinguish between genotypes, we developed microsatellite markers for each species (SI Appendix, Table S6). To generate sufficient numbers of individuals to initiate the experiment, we cultured each genotype for 8 wk in a greenhouse, in large (36 64 32 cm), green, plastic tubs containing ∼45 L of tap water and a layer (∼1–3 cm) of general purpose potting soil (GO/ON flower soil with 100–300 mg/L N, 150–450 mg/L P2O2, and 1,200–2,000 mg/L K2O).
Field Setup.
The experiment was done in 13, 1,260-L green, fiberglass cattle tanks (140 100 90 cm), which were regularly arranged in a field at the University of Zurich (47.3743°N, 8.5510°E). In May 2016, we distributed 80 L of general purpose potting soil (details as above) across the bottom of each tank, and then each was two-thirds filled with tap water (∼840 L). One liter of pond water (containing plankton) and three to five snails collected from nearby natural ponds were then added to each experimental pond. Diverse and abundant zooplankton, algal, and insect communities were observed in the experimental ponds during the experiment. To mimic the shaded conditions under which the plants commonly occur, each pond was shaded with two layers of 45% shade cloth. Competitive population dynamics and evolution of the two plant species occurred in small competitive arenas in each pond. Competitive arenas were white plastic containers (122-mm diameter, 1,100 mL) attached to a wooden frame floating on the surface of the water in each pond, with a single frame supporting a 5 6 array of 30 containers. Each container was attached to the frame with ∼3 cm of the container protruding above the surface of the water. The bottom of each container was punctured, allowing exchange of water and plankton between container and pond.
Experimental Manipulation.
There were two treatments in the main experiment, one in which both species competed and were able to respond to selection imposed by interspecific competition (heterospecific selection), and one in which both species competed but were unable to respond to selection imposed by interspecific competition (conspecific selection). There was one replicate of each treatment in each of the 13 ponds. Both treatments were initialized by placing 108 fronds of each species into each of two competitive arenas within each pond. At the beginning of the experiment, there were 12 individuals of each genotype of L. minor in each replicate of each treatment, and there were 12 individuals of three genotypes of S. polyrhiza and the remaining three genotypes had 24 individuals each (we initially thought each of the latter clones were two separate clones but genotyping subsequent to the establishment of the experiment revealed them to be a single genotype, hence their higher initial density). The two-species experimental communities were established in June 2016, and the component populations were allowed to grow and compete.
To prevent evolution in response to interspecific competition in the conspecific selection treatment, we replaced all individuals of both species in this treatment every 2 wk with individuals from multigenotype but single-species source populations that were not subject to selection from their heterospecific competitor, but were subject to selection from conspecific competitors. For each replicate, the single-species source populations were initiated at the same time, in the same pond, with the same genotypes, each with the same number of individuals as in the two-species communities, but with only one species per container. Thus, these single-species populations, which were used as a source of individuals for the conspecific selection treatment (described next), were free to evolve to the same abiotic and biotic conditions as the populations in the heterospecific selection treatment, but they were unable to evolve in response to competition from heterospecific individuals, to which they were not exposed.
To execute a single experimental replacement, we counted and then discarded all individuals of both species from a replicate of the conspecific selection treatment. We then replaced these individuals with same number of individuals of each species in that replicate, but using individuals from a single-species source population for each species from the same pond. We repeated this procedure every 2 wk for all replicates. There were 11 single-species source populations for each species in each pond, giving us a fresh source population for each replacement. Because the replacement method retains the number of individuals of each species in each replicate of the conspecific selection treatment, the manipulation preserves the ongoing effects of interspecific competition on the sizes of the populations of the two species in this treatment. However, the manipulation prevents evolution in response to interspecific competition, and therefore prevents such evolution from affecting the dynamics of the interaction. In the heterospecific selection treatment, the populations of the two species were able to compete and evolve in response to one another. The populations in both treatments were physically mixed during the weekly photographic censuses (described below). Single-species source populations were physically mixed with a plastic fork as a procedural control. We randomized the position of the treatments and single-species source populations within each pond.
Quantifying Evolutionary Change.
Evolution—the change in genotype frequency over multiple generations—was quantified by genotyping between 24 and 32 individuals of each species in each replicate sampled 50 d (August 4, 2016) after the experiment was initiated. In total, we genotyped 1,280 individuals using four microsatellite markers for each species (SI Appendix, Table S6) in a single multiplex procedure (SI Appendix). To determine whether there were differences between treatments in genotypic composition, we used PERMANOVA (41). For each species, we first described compositional differences between populations across both treatments using Euclidean dissimilarity matrices. We used a symmetric distance measure (Euclidean distance) because including shared absences of genotypes provides important information about the effects of our treatments. We then implemented PERMANOVA using the “adonis” function in the “vegan” package in R (42), including pond as a random factor (41). Genotypic compositional differences were visualized using principal-coordinates analysis (41). We further assessed for consistency in the direction of evolutionary change with univariate analyses on the numerically dominant genotype (genotype 1 in Fig. 1). We first assessed the probability that this genotype consistently increased in frequency in both treatments using exact binomial tests. We then assessed if the magnitude of the change in the frequency differed between treatments using a paired t test.
Quantifying and Analyzing Population Trajectories.
We photographed all individuals in each replicate of each treatment approximately every week (8.6 ± 2.9 d) for the duration of the experiment. So that all individuals could be distinguished in each photograph, we carefully removed all individuals from each competitive arena with a plastic fork and placed them in larger containers for photographing before returning them to their original competitive arena within each pond. Individuals of both species were then manually counted in the photographs using ImageJ (43). To test for differences in the population trajectories between treatments, we fit a two-parameter logistic function to phenomenologically describe the time-series data for each species in each treatment. The function took the form , where , the population size of species i, was modeled as a function of time, t, and where a is the predicted asymptotic population size and b is the population growth rate. This model was fit to the time-series data using the “nlme” package in R. We included pond as a random effect and used an autoregressive correlation error structure to account for repeated counts over time. To determine whether the trajectories differed between treatments, we used likelihood-ratio tests to compare full models including treatment effects with reduced models where treatment effects were excluded. As an additional test for whether the selection treatments caused differences in population sizes, we compared the mean population size for each species between treatments across the last five census dates. These tests were done using linear mixed-effects models with treatment as a fixed factor and pond as a random effect.
Additional Competition Experiments and Competition Model.
To quantify niche and competitive-ability differences between the species in each treatment, we parameterized a two-species competitive population dynamics model describing the interaction between the evolved populations of L. minor and S. polyrhiza. The aim here was not to describe the population trajectories in the two treatments (as described above), but rather to understand if the differences in dynamics between treatments could be explained by evolved differences in niche differences and/or in species competitive abilities. Parameterizing an appropriate competition model requires estimating the maximum finite rates of growth, and the per capita strength of intraspecific and interspecific competition, but estimating these quantities from observed population trajectories in mixture (i.e., in our main experiment) is difficult to do with confidence. Therefore, we identified and parameterized an appropriate competition model using data from a separate set of competition experiments, done under the same field conditions, using individuals sampled from the evolved populations of each species in the main experiment. These separate competition experiments began on August 10, 6 d after the genotype sampling. To initiate the experiments, a small number of individuals of both species were haphazardly sampled from each replicate of each treatment in the main experiment. Fronds collected from different replicates of the same treatment were then combined, providing the material for the separate competition experiments. We took this approach because parameterizing the competition model separately for each replicate of the main experiment was not feasible.
The separate competition experiments involved measuring population growth in response to a range of densities of conspecific and heterospecific individuals (23). To enable accurate estimates of the competition model parameters while limiting the total number of individuals required in these experiments, we did these competition trials in smaller competitive arenas (open-ended pipes, 46-mm diameter), attached vertically to the inside wall of one of the same type of container used in the main experiment, which was itself attached to a wooden frame floating within one of five experimental ponds. These five ponds were maintained from the beginning of the season to have similar conditions as the 13 experimental ponds used in the main experiment. To quantify the strength of intraspecific competition, we measured population growth of S. polyrhiza at densities of 1.6, 2.1, 4.9, 10.3, 30.9, 50.9, 76.9, and 96.9 individuals⋅cm−2, and of L. minor at 1.6, 2.7, 7.6, 15.2, 52.5, 89.4, 126.7, and 163.5 individuals⋅cm−2. To quantify the strength of interspecific competition, we measured population growth of individuals of the focal species at low density (1.6 cm−2) competing against their heterospecific competitor at each of the densities listed above. These competitor densities were chosen because they covered a range of densities up to levels beyond the maximum densities observed in the main experiment (111.89 fronds⋅cm−2), as has been shown to be optimal for accurately estimating model parameters (44). Note also that the lowest densities (1.6 fronds⋅cm−2) still contained 27 individuals, allowing for sufficient representation of each of the genotypes at even the lowest densities. The treatments containing only low densities of conspecifics were replicated three times, and there was one replicate of each of the remaining density combinations. The treatment densities were randomly distributed across the five ponds. Photographs of all individuals in all treatments were taken 2 wk after the experiment was initiated. Individuals in the photographs were then counted to estimate population growth over 2 wk, the same duration as the evolution manipulation in the main experiment. Data from these competition experiments were fit to the Law–Watkinson competition model (45), which takes the following form:
| [1] |
where Ni,t is the population size of species i at time t, and Ni,t+1 is the population size of species i 2 wk later. The parameter is the maximum finite rate of growth, and the parameters and are the intraspecific and interspecific competition coefficients, respectively. Data from the competition experiments were fit to this model using nonlinear least-squares regression using the “nls” function in R. We included the selection treatments as a factor in our model fits, allowing separate estimates of each of the competition model’s parameters in each treatment. We used likelihood-ratio tests to compare full models allowing a separate estimate of each parameter for each treatment, with reduced models allowing only a single estimate of each parameter across both treatments. For details of the competition model assumptions, see SI Appendix.
Quantifying Niche and Competitive-Ability Differences.
Using our parameter estimates for the Law–Watkinson competition model, we can define expressions that quantitatively describe the niche difference, which stabilizes species coexistence, and differences between species in their competitive abilities, which promote competitive exclusion (21). Both these quantities are based on the mutual invasibility criterion of species coexistence (SI Appendix). Following the methods of earlier studies (21, 23), the niche overlap for the Law–Watkinson model is , and the stabilizing niche difference is . This expression quantifies the degree to which the per capita strength of intraspecific competition (the denominator) exceeds the per capita strength of interspecific competition (the numerator). The difference between species in their competitive abilities in the Law–Watkinson model is (23). This expression determines who will win in competition in the absence of niche differences. The first term of this ratio quantifies the difference between the species in their productivity in the absence of competition, and the second term quantifies the difference between species in their sensitivity to competition from both heterospecific and conspecific competitors (23). Coexistence occurs when (21). For more information on the derivation of these quantities from the mutual invasibility criterion, and their relationship to population abundances, see SI Appendix.
We used our estimates for the parameters in the Law–Watkinson model to estimate niche (1 − ) and competitive-ability differences () in both selection treatments. To estimate the SD of these composite variables, and to evaluate the likelihood that niche vs. competitive-ability differences were responsible for the differences in dynamics between treatments, we used error propagation methods based on Monte Carlo simulations, using the “propagate” package in R (SI Appendix).
Trait Measurements.
We sampled 25–60 individuals of each species from each replicate of the two treatments in September 2016. We photographed these fronds and quantified total frond area in each sample using ImageJ. We quantified area per frond by dividing the total frond area by the number of fronds in the sample. We measured dry mass by first removing all roots and turions (dormant resting stages in S. polyrhiza) and then dried the remaining fronds at 70 °C for 24 h before weighing. We assessed root length as the longest root of a single haphazardly chosen cluster of fronds from each replicate. With these data, we calculated for each replicate and species, specific-leaf area and the ratio of root length to dry mass. Population-level estimates of frond area and biomass were calculated by multiplying the individual-level estimates of these variables by the population sizes of the species on the final census date. We compared the value of each individual-level trait, and area and biomass traits at the population level, for each species between treatments using linear mixed-effects models, with treatment as a fixed effect and pond as a random effect. To determine whether the observed phenotypic changes had a genetic component, morphological traits of individual clones were also assessed after growth in common-garden conditions in the laboratory (SI Appendix).
Supplementary Material
Acknowledgments
We thank Ariane Le Gros, Laura Stefan, Cyrill Hess, Renato Guidon, Andrea Reid, and other members of the Plant Ecology Group for assistance; and Sabine Güsewell, Jacob Usinowicz, and Marti Anderson for analytical advice. We thank Josh van Buskirk and the University of Zurich for use of their field infrastructure. We thank Walter Lämmler, Shuqing Xu, and Meret Huber for providing clones and advice. M.M.T. was supported by the ETH Zürich Center for Adaptation to Changing Environments.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data Deposition: Data relating to this work have been deposited on figshare (doi: 10.6084/m9.figshare.7599095.v1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816298116/-/DCSupplemental.
References
- 1.Macarthur RH, Levins R. The limiting similarity, convergence, and divergence of coexisting species. Am Nat. 1967;101:377–385. [Google Scholar]
- 2.Slatkin M. Ecological character displacement. Ecology. 1980;61:163–177. [Google Scholar]
- 3.Taper ML, Case TJ. Quantitative genetic models for the coevolution of character displacement. Ecology. 1985;66:355–371. [Google Scholar]
- 4.Abrams PA. Character displacement and niche shift analyzed using consumer-resource models of competition. Theor Popul Biol. 1986;29:107–160. doi: 10.1016/0040-5809(86)90007-9. [DOI] [PubMed] [Google Scholar]
- 5.Doebeli M. An explicit genetic model for ecological character displacement. Ecology. 1996;77:510–520. [Google Scholar]
- 6.Pfennig DW, Pfennig KS. 2012. Evolution’s Wedge: Competition and the Origins of Diversity (Univ of California Press, Berkeley, CA), 303 p.
- 7.Schluter D. The Ecology of Adaptive Radiation. Oxford Univ Press; New York: 2000. [Google Scholar]
- 8.Stuart YE, Losos JB. Ecological character displacement: Glass half full or half empty? Trends Ecol Evol. 2013;28:402–408. doi: 10.1016/j.tree.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Brown WL, Wilson EO. Character displacement. Syst Zool. 1956;5:49–64. [Google Scholar]
- 10.Schluter D, McPhail JD. Ecological character displacement and speciation in sticklebacks. Am Nat. 1992;140:85–108. doi: 10.1086/285404. [DOI] [PubMed] [Google Scholar]
- 11.Grant PR, Grant BR. Evolution of character displacement in Darwin’s finches. Science. 2006;313:224–226. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- 12.Stuart YE, et al. Rapid evolution of a native species following invasion by a congener. Science. 2014;346:463–466. doi: 10.1126/science.1257008. [DOI] [PubMed] [Google Scholar]
- 13.Schluter D. Experimental evidence that competition promotes divergence in adaptive radiation. Science. 1994;266:798–801. doi: 10.1126/science.266.5186.798. [DOI] [PubMed] [Google Scholar]
- 14.Zuppinger-Dingley D, et al. Selection for niche differentiation in plant communities increases biodiversity effects. Nature. 2014;515:108–111. doi: 10.1038/nature13869. [DOI] [PubMed] [Google Scholar]
- 15.Schoener TW. The newest synthesis: Understanding the interplay of evolutionary and ecological dynamics. Science. 2011;331:426–429. doi: 10.1126/science.1193954. [DOI] [PubMed] [Google Scholar]
- 16.Germain RM, Williams JL, Schluter D, Angert AL. Moving character displacement beyond characters using contemporary coexistence theory. Trends Ecol Evol. 2018;33:74–84. doi: 10.1016/j.tree.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Dayan T, Simberloff D. Ecological and community-wide character displacement: The next generation. Ecol Lett. 2005;8:875–894. [Google Scholar]
- 18.Pfennig KS, Pfennig DW. Character displacement: Ecological and reproductive responses to a common evolutionary problem. Q Rev Biol. 2009;84:253–276. doi: 10.1086/605079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beans CM. The case for character displacement in plants. Ecol Evol. 2014;4:852–865. doi: 10.1002/ece3.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lankau RA. Rapid evolutionary change and the coexistence of species. Annu Rev Ecol Evol Syst. 2011;42:335–354. [Google Scholar]
- 21.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–366. [Google Scholar]
- 22.Adler PB, Hillerislambers J, Levine JM. A niche for neutrality. Ecol Lett. 2007;10:95–104. doi: 10.1111/j.1461-0248.2006.00996.x. [DOI] [PubMed] [Google Scholar]
- 23.Hart SP, Freckleton RP, Levine JM. How to quantify competitive ability. J Ecol. 2018;106:1902–1909. [Google Scholar]
- 24.Abrams PA. Alternative models of character displacement and niche shift. 1. Adaptive shifts in resource use when there is competition for nutritionally nonsubstitutable resources. Evolution. 1987;41:651–661. doi: 10.1111/j.1558-5646.1987.tb05836.x. [DOI] [PubMed] [Google Scholar]
- 25.Fox JW, Vasseur DA. Character convergence under competition for nutritionally essential resources. Am Nat. 2008;172:667–680. doi: 10.1086/591689. [DOI] [PubMed] [Google Scholar]
- 26.Smallegange IM, Coulson T. Towards a general, population-level understanding of eco-evolutionary change. Trends Ecol Evol. 2013;28:143–148. doi: 10.1016/j.tree.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 27.Le Gac M, Plucain J, Hindré T, Lenski RE, Schneider D. Ecological and evolutionary dynamics of coexisting lineages during a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA. 2012;109:9487–9492. doi: 10.1073/pnas.1207091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landolt E. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae), Volume 2. The Family of Lemnaceae—A Monographic Study, Volume 1. ETH Zürich; Zürich: 1986. p. 566. [Google Scholar]
- 29.Pimentel D, Feinberg EH, Wood PW, Hayes JT. Selection, spatial distribution, and the coexistence of competing fly species. Am Nat. 1965;99:97–109. [Google Scholar]
- 30.Kunstler G, et al. Plant functional traits have globally consistent effects on competition. Nature. 2016;529:204–207. doi: 10.1038/nature16476. [DOI] [PubMed] [Google Scholar]
- 31.Wright IJ, et al. The worldwide leaf economics spectrum. Nature. 2004;428:821–827. doi: 10.1038/nature02403. [DOI] [PubMed] [Google Scholar]
- 32.Tilman D. Plant Strategies and the Dynamics and Structure of Plant Communities. Princeton Univ Press; Princeton: 1988. [Google Scholar]
- 33.Buss LW. Competition and community organization on hard surfaces in the sea. In: Diamond J, Case TJ, editors. Community Ecology. Harper and Row; New York: 1986. pp. 517–536. [Google Scholar]
- 34.Hart SP, Usinowicz J, Levine JM. The spatial scales of species coexistence. Nat Ecol Evol. 2017;1:1066–1073. doi: 10.1038/s41559-017-0230-7. [DOI] [PubMed] [Google Scholar]
- 35.Tan J, Rattray JB, Yang X, Jiang L. Spatial storage effect promotes biodiversity during adaptive radiation. Proc Biol Sci. 2017;284:2017.0841. doi: 10.1098/rspb.2017.0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Strauss SY. Ecological and evolutionary responses in complex communities: Implications for invasions and eco-evolutionary feedbacks. Oikos. 2014;123:257–266. [Google Scholar]
- 38.Gause GJ. The Struggle for Existence. Williams and Wilkins; Baltimore: 1934. [Google Scholar]
- 39.Hutchinson GE. The paradox of the plankton. Am Nat. 1961;95:137–145. [Google Scholar]
- 40.Cole CT, Voskuil MI. Population genetic structure in duckweed (Lemna minor, Lemnaceae) Can J Bot. 1996;74:222–230. [Google Scholar]
- 41.Anderson MJ. 2017 Permutational multivariate analysis of variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online. Wiley Online Library, eds Balakrishnan N, et al. Available at https://onlinelibrary.wiley.com/doi/10.1002/9781118445112.stat07841. Accessed August 04, 2018.
- 42.R Core Team 2018. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 3.5.1.
- 43.Rasband WS. 1997–2016. ImageJ (NIH, Bethesda, MD)
- 44.Inouye BD. Response surface experimental designs for investigating interspecific competition. Ecology. 2001;82:2696–2706. [Google Scholar]
- 45.Law R, Watkinson AR. Response-surface analysis of two-species competition: An experiment on Phleum arenarium and Vulpia fasciculata. J Ecol. 1987;75:871–886. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.