Significance
Plants are exposed to conflicting stresses simultaneously in nature. As stress responses are costly, plants likely coordinate these responses to minimize fitness costs. The nature and extent to which plants employ inducible mechanisms to cope with combined physical and biological stresses remains unknown. We identify a genetic mechanism by which leaves of distinct ages differentially control stress-response cross-talk. At the organism level, this mechanism balances stress-response trade-offs to maintain plant growth and reproduction during combined stresses. We also show that this leaf age-dependent stress-response prioritization influences the establishment of plant-associated leaf bacterial communities. This study illustrates the importance of active balancing of stress-response trade-offs for plant fitness maintenance and for interaction with the plant microbiota.
Keywords: combined stress, phytohormone, plant fitness, microbiota, stress trade-off
Abstract
In nature, plants must respond to multiple stresses simultaneously, which likely demands cross-talk between stress-response pathways to minimize fitness costs. Here we provide genetic evidence that biotic and abiotic stress responses are differentially prioritized in Arabidopsis thaliana leaves of different ages to maintain growth and reproduction under combined biotic and abiotic stresses. Abiotic stresses, such as high salinity and drought, blunted immune responses in older rosette leaves through the phytohormone abscisic acid signaling, whereas this antagonistic effect was blocked in younger rosette leaves by PBS3, a signaling component of the defense phytohormone salicylic acid. Plants lacking PBS3 exhibited enhanced abiotic stress tolerance at the cost of decreased fitness under combined biotic and abiotic stresses. Together with this role, PBS3 is also indispensable for the establishment of salt stress- and leaf age-dependent phyllosphere bacterial communities. Collectively, our work reveals a mechanism that balances trade-offs upon conflicting stresses at the organism level and identifies a genetic intersection among plant immunity, leaf microbiota, and abiotic stress tolerance.
In nature, plants encounter and must appropriately respond to diverse stresses to survive and reproduce. Different stress signaling pathways are connected, providing regulatory potential to maximize fitness (1). For instance, plants exposed to abiotic stresses such as high salinity and drought often display reduced immune activity (2). As stress responses are costly, prioritization in stress responses would allow plants to allocate more resources to abiotic stress responses and increase plant fitness in the absence of biotic stress (3). However, this prioritization does not explain how plants maintain fitness when simultaneously exposed to biotic and abiotic stresses.
Phytohormones play critical roles in stress responses and prioritization (4). In an evolutionarily conserved mechanism, phytohormone signaling mediated by abscisic acid (ABA) promotes abiotic stress tolerance and suppresses signaling of the biotic stress-related phytohormone salicylic acid (SA). Consequently, plant immunity is lowered during abiotic stresses such as drought and salt stress (5). In Arabidopsis thaliana, ABA suppresses expression of the SA biosynthesis gene SA INDUCTION DEFICIENT 2 (SID2) (5). ABA application causes proteasome-mediated degradation of the SA receptor NONEXPRESSOR OF PR GENE 1 (NPR1) (6). Molecular cross-talk between ABA and SA provides a plausible mechanism for prioritization of abiotic over biotic stress responses. Such a mechanism would provide plants with adaptive potential when abiotic stresses are major threats. However, it is unknown whether this hormonal cross-talk is still advantageous when plants simultaneously encounter biotic and abiotic stresses.
In natural soils, healthy plants are colonized by taxonomically structured microbial communities, collectively called the plant microbiota (7), which contribute to plant performance under adverse environmental conditions (8–11). SA modulates colonization of the A. thaliana root microbiota by specific bacterial taxa, although this was not accompanied by a detectable impact on host survival of the tested SA biosynthesis and signaling mutants (12, 13). In field experiments, drought induced a relative enrichment of multiple lineages of Actinobacteria and Chloroflexi in the rice root microbiota, whereas, in sorghum roots, relative enrichments of Actinobacteria and Firmicutes were observed (14–17). Whether and how cross-talk between biotic and abiotic stress response pathways impacts the assembly of the plant microbiota remains unknown.
Age and developmental stage are important factors influencing stress responses in animals and plants. For instance, as A. thaliana plants age, SA-mediated immunity is enhanced (18). Plants also display age-dependent responses at the organ level. Young A. thaliana rosette leaves exhibit greater SA accumulation and SA-mediated immunity in comparison with older rosette leaves (19). Evidence for leaf age-dependent variation also exists for abiotic stress (20, 21). Based on the optimal defense theory (ODT), in defending themselves against herbivores, plants prioritize tissues, such as young leaves, that are more valuable for the whole plant (22). However, whether leaf age-dependent variation in stress responses is a simple prioritization analogous to the ODT or an active strategy to increase plant fitness is not understood.
Here we show that biotic and abiotic stress responses are differentially prioritized in a leaf age-dependent manner in A. thaliana to maintain fitness under combined stresses. Abiotic stresses dampen immunity in old rosette leaves, whereas the SA signaling components PBS3 and NPR1 protected young rosette leaves from ABA-mediated immune suppression. pbs3 mutant plants exhibited enhanced abiotic stress tolerance but showed compromised fitness maintenance under combined biotic and abiotic stresses. Defining a hitherto uncharacterized link between stress signaling cross-talk and microbiota structure, PBS3 is indispensable for the proper establishment of salt stress- and leaf age-dependent leaf bacterial communities. We propose that balancing leaf-age dependent cross-talk between SA and ABA signaling is a critical determinant of plant performance during combined stresses.
Results
Leaf Age Controls ABA–SA Cross-Talk Independently of Vegetative Phase Change.
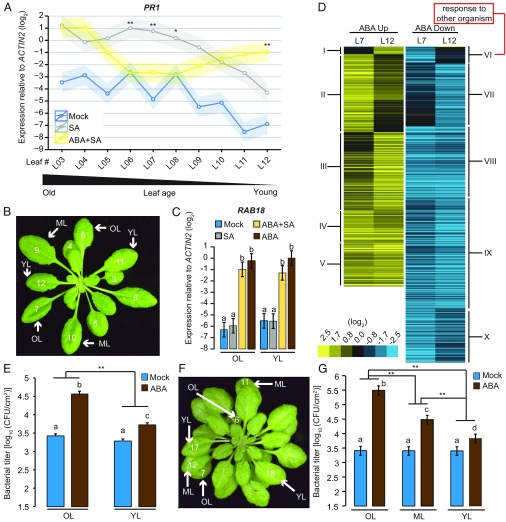
To gain insights into mechanisms underlying the cross-talk between ABA and SA signaling, we investigated the impact of ABA on the SA response in A. thaliana. We found that pretreatment with ABA unexpectedly blocked SA-mediated induction of the SA-response marker gene PATHOGENESIS RELATED PROTEIN 1 (PR1) (5) only in a subset of rosette leaves (L06 to L08; numbers refer to the positions of rosette leaves as determined by their order of appearance), but enhanced PR1 expression in young rosette leaves (L12; Fig. 1 A and B), indicative of a leaf age-specific effect of ABA on the SA response. In contrast, expression of the ABA-response marker gene RESPONSE TO ABA 18 (RAB18) (23, 24) was independent of leaf age (Fig. 1C and SI Appendix, Fig. S1A).
Fig. 1.
Leaf age effects of ABA on SA response and immunity. (A) PR1 expression levels in leaves (from L03 to L12) of 4–5-wk-old A. thaliana Col-0 plants 24 h after spray with 500 µM SA, following 500 µM ABA spray pretreatment for 24 h were determined by quantitative RT-PCR. Data represent means ± SEM (shadows) calculated from three biological replicates by using a mixed linear model. Asterisks indicate significant differences between SA and combined ABA/SA treatments (*P < 0.05 and **P < 0.01, two-tailed Student’s t tests). (B and F) Leaf numbers in 4-wk-old Col-0 (B) and 35S::miR156a (F) plants highlighting old (OL), middle (ML), and young leaves (YL). (C) RAB18 expression levels in old and young leaves of 4–5-wk-old Col-0 plants as in A. Data represent means ± SEM calculated from three biological replicates by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.05). (D) Heat map showing expression patterns of the genes that show significant expression changes 48 h after ABA spray compared with mock (q < 0.01 and |log2FC| > 1) for up-regulated (yellow; 1,291 genes) or down-regulated genes (blue; 1,712 genes) in L7 or L12 of 4–5-wk-old Col-0 plants. (E and G) Pto hrcC− (OD600 = 0.0002) was infiltrated into old, middle, and young leaves of 4–5-wk-old Col-0 (E) and 35S::miR156a (G) plants 24 h after 500 µM ABA spray or mock. Bacterial growth was measured at 2 days postinoculation. Data represent means ± SEM calculated from at least three independent experiments, each with at least four biological replicates, by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.005; **P < 0.01, two-tailed Student’s t tests).
To systematically explore the leaf age-dependency of ABA-mediated transcriptional changes, we conducted RNA sequencing (RNA-seq) experiments. Although ABA-triggered responses in old (L7) and young (L12) rosette leaves were similar overall, some gene clusters exhibited leaf age-dependent expression patterns (Fig. 1D, SI Appendix, Fig. S1 C and D, and Dataset S1). These results support our finding that expression of a subset of ABA-regulated genes is leaf age-dependent.
We next examined the physiological significance of leaf age-dependent effects of ABA on immunity. The bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pto) is known to trigger ABA accumulation through the action of type III effectors (25, 26). Therefore, we reasoned that the disarmed Pto hrcC− strain, which is unable to deliver type III effectors, can be used to detect the effect of exogenous ABA on immunity by measuring bacterial titers in leaves. We found that ABA treatment enhanced Pto hrcC− growth more strongly in old compared with young rosette leaves (Fig. 1E and SI Appendix, Fig. S1B). This is consistent with an ABA-mediated suppression of the SA response in old but not in young leaves (Fig. 1A) and with SA-mediated immunity restricting Pto hrcC− growth (27).
We speculated that the observed leaf age-dependent ABA effects might be linked to leaf developmental stage because the onset of detectable suppression of the SA response by ABA correlates approximately with the onset of a vegetative phase change from juvenile to adult leaves in Col-0 plants (28). To explore this possibility, we employed transgenic A. thaliana overexpressing the miRNA miR156a in which the expression of juvenile traits is markedly prolonged (28). However, similar to WT, we observed that ABA effects on Pto hrcC− growth were dependent on leaf age despite the juvenile trait being manifest in younger and older leaves (Fig. 1 F and G). Thus, leaf age but not vegetative phase change likely controls ABA–SA cross-talk, with marked consequences for the SA response and bacterial resistance.
ABA Suppresses Immunity via the ABA RESPONSIVE ELEMENT BINDING PROTEIN–SNAC-A Transcription Factor Cascade.
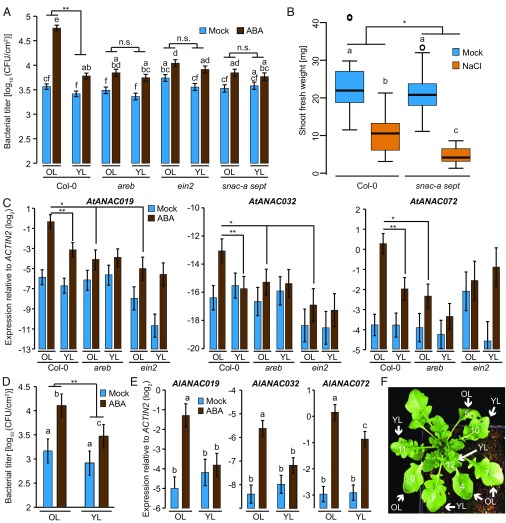
ABA RESPONSIVE ELEMENT (ABRE) BINDING PROTEIN (AREB) transcription factors (TFs) redundantly regulate a major part of ABA-mediated transcriptional changes (24) (SI Appendix, Fig. S2A). We found that ABA-triggered suppression of immunity against Pto hrcC− in old leaves was compromised in areb triple mutant plants (areb1 areb2 abf3) (24), indicating that ABA-triggered immune suppression requires AREB-mediated transcription. Next, we further dissected this transcriptional cascade. AREB TFs have been shown to bind to the promoters of SNAC-A TFs in vitro (29), and a subset of SNAC-A TFs, ANAC019, 055, and 072, whose gene expression is induced by ABA, are involved in the suppression of SA accumulation (30). These SNAC-A TFs redundantly control a subset of ABA responses such as SAG26 expression and senescence, but not RAB18 expression (23). Therefore, we tested whether SNAC-A TFs collectively regulate ABA-mediated immune suppression. Indeed, we found that ABA-mediated immune suppression in old leaves was compromised in snac-a septuple mutant (snac-a sept; anac019 anac055 anac072/rd26 anac002/ataf1 anac081/ataf2 anac102 anac032) (23) plants (Fig. 2A).
Fig. 2.
ABA triggers immune suppression in old leaves through AREB and ANAC TFs. (A) Old leaves (OL) and young leaves (YL) of 4–5-wk-old Col-0, areb1 areb2 abf3 (areb), ein2, and anac septuple mutant (snac-a sept) plants were infiltrated with Pto hrcC− (OD600 = 0.0002) 24 h after 500 µM ABA spray or mock. Bacterial growth was measured at 2 days postinoculation (dpi). Data represent means ± SEM calculated from three independent experiments, each with at least five biological replicates, by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.005). (B) Shoot fresh weight of Col-0 and snac-a sept seedlings grown on MS plates containing 100 mM NaCl or mock for 10 d. The box plots show combined data from three independent experiments, each with at least eight biological replicates. Different letters indicate significant differences (adjusted P < 0.05). (C) AtANAC019, AtANAC032, and AtANAC072 expression levels in old and young leaves of 4–5-wk-old Col-0 and areb1 areb2 abf3 (areb) plants 24 h after 500 µM ABA spray or mock were determined by quantitative RT-PCR. Data represent means ± SEM calculated from at least three biological replicates by using a mixed linear model. (D) Pto hrcC− (OD600 = 0.0002) was infiltrated into old and young leaves of 5–6-wk-old A. lyrata plants 24 h after 500 µM ABA spray or mock. Bacterial growth was measured at 2 dpi. Data represent means ± SEM calculated from three independent experiments, each with at least five biological replicates, by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.005). (E) AlANAC019, AlANAC032, and AlANAC072 expression levels in old and young leaves of 5–6-wk-old A. lyrata plants 24 h after 500 µM ABA spray or mock were determined by quantitative RT-PCR. Data represent means ± SEM calculated from at least three biological replicates by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.05). (F) Leaf numbers in 5–6-wk-old A. lyrata showing old and young leaves. (A–D) n.s., not significant (*P < 0.05 and **P < 0.01, two-tailed Student’s t tests).
Plants overexpressing ANAC002, 019, 055, or 072 show enhanced abiotic stress tolerance (3, 31). Conversely, we found that salt tolerance was impaired in snac-a sept plants (Fig. 2B and SI Appendix, Fig. S2B). Expression of ANAC019, ANAC032, and ANAC072 after ABA treatment was higher in old than in young rosette leaves in an AREB-dependent manner (Fig. 2C), suggesting that this ANAC induction is under control of the AREBs. It has been shown that SNAC-A induction by ABA also requires ETHYLENE INSENSITIVE2 (EIN2) (23), a key component of ethylene signaling (32). Accordingly, we found that ABA-triggered leaf age-dependent expression of SNAC-As and suppression of immunity against Pto hrcC− were impaired in ein2 plants (Fig. 2 A and C), whereas RAB18 induction remained intact (SI Appendix, Fig. S2A). This corroborates the conclusion that these SNAC-A TFs play an important role in ABA-mediated suppression of immunity in old leaves. Collectively, these results suggest that snac-a sept plants exhibit an altered balance between biotic and abiotic stress responses.
To explore the evolutionary conservation of the leaf age-dependent effect of ABA on immunity, we investigated Arabidopsis lyrata, a close relative of A. thaliana. We found that ABA suppresses immunity against Pto hrcC− and induces SNAC-A expression in a leaf age-dependent manner as in A. thaliana Col-0 plants (Fig. 2 D–F) (33).
PBS3 Protects Young Leaves from ABA-Triggered Immune Suppression.
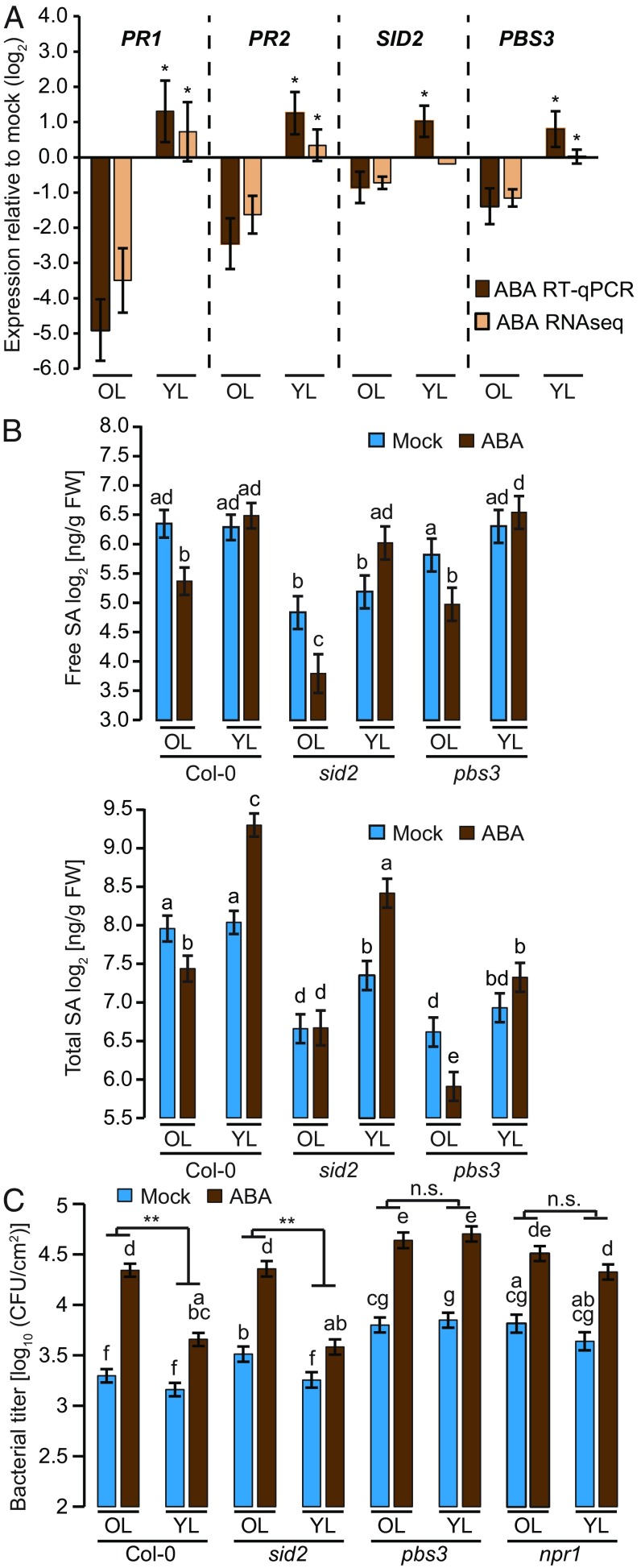
Our RNA-seq experiments described here earlier revealed that ABA mediates a strong suppression of genes enriched for the Gene Ontology term “response to other organism” specifically in old leaves (L7; cluster VI; Fig. 1D). Cluster VI included PBS3, encoding an acyl-adenylate/thioester-forming enzyme, which is known to be important for SA accumulation and signaling and for immunity against biotrophic pathogens. Cluster VI also included the SA response markers PR1 and PR2 (34), suggesting that SA signaling components are specifically suppressed in old leaves. Therefore, although it is not in cluster VI, we also included SID2 for further analysis because it encodes a key enzyme for pathogen-induced SA biosynthesis (35). ABA reduced the expression of all tested genes in old leaves but did not in young leaves (Fig. 3A). Next, we determined free and total SA levels after ABA application in Col-0, sid2, and pbs3 plants and found that ABA had contrasting effects on SA levels in old and young Col-0 leaves (Fig. 3B). Strikingly, the increased total SA, mediated by ABA treatment in young leaves of Col-0, was retained in sid2 but abolished in pbs3 plants (Fig. 3B). Given that PBS3 has been proposed to protect SA from degradation (36), we conclude that the increase in total SA elicited by ABA in young leaves may be caused by reduced SA degradation rather than increased SA biosynthesis via SID2.
Fig. 3.
PBS3 protects young leaves from ABA-triggered immune suppression. (A) The expression changes of PR1, PR2, SID2, and PBS3 in old leaves (OL) and young leaves (YL) of 4–5-wk-old Col-0 plants 48 h after 500 µM ABA spray compared with mock in RNA-seq and quantitative RT-PCR. Data represent means ± SEM of three biological replicates. Asterisks indicate significant differences in young compared with old leaves (*P < 0.05, two-tailed Student’s t tests). (B) Free and total SA amounts in old and young leaves of 4–5-wk-old Col-0, sid2, and pbs3 plants 48 h after spray with 500 µM ABA or mock. Data represent means ± SEM calculated from three biological replicates by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.05). (C) Old and young leaves of 4–5-wk-old Col-0, sid2, pbs3, and npr1 plants were infiltrated with Pto hrcC− (OD600 = 0.0002) 24 h after 500 µM ABA spray or mock. Bacterial growth was measured at 2 days postinoculation. Data represent means ± SEM calculated from three independent experiments, each with at least five biological replicates, by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.005; *P < 0.05 and **P < 0.01, two-tailed Student’s t tests). n.s., not significant.
Notably, we found that young leaves of pbs3 plants are vulnerable to ABA-mediated suppression of immunity against Pto hrcC−, whereas sid2 plants exhibited a WT-like phenotype (Fig. 3C). In addition, we found that the enhanced SA-induced PR1 expression by ABA in young leaves is compromised in pbs3 plants whereas ABA-induced RAB18 expression remains intact (SI Appendix, Fig. S3 A and B). Finally, increased free and total SA accumulation was impaired in pbs3 leaves upon treatment with flg22, a peptide epitope derived from bacterial flagellin that stimulates SA accumulation (27) (SI Appendix, Fig. S3 C and D). Thus, PBS3 is required for immunity-triggered SA accumulation. In young leaves, however, increased total SA accumulation and SA response upon ABA treatment might be important to protect young leaves from ABA-mediated immune suppression, which is impaired in pbs3 plants.
Given that NPR1, encoding the SA receptor (37, 38), is required for the majority of the SA response (37) (SI Appendix, Fig. S3B), we also included npr1 plants in our analysis. We found that, similar to pbs3, young leaves of npr1 plants are not protected against ABA-mediated immune suppression (Fig. 3C). These and previous results might suggest that NPR1 is required for PBS3 function or vice versa (39). ABA promotes NPR1 degradation, which correlated with reduced PR1 expression, whereas SA antagonizes this (6). Together, NPR1 might be protected from ABA-mediated degradation by higher SA levels in young leaves. Our finding that expression of SNRK2.8, an important regulatory interactor of NPR1 (40), is strongly suppressed by ABA in only old leaves supports this hypothesis (SI Appendix, Fig. S1 C and D).
PBS3 Regulates the Trade-Off Between Biotic and Abiotic Stress Responses.
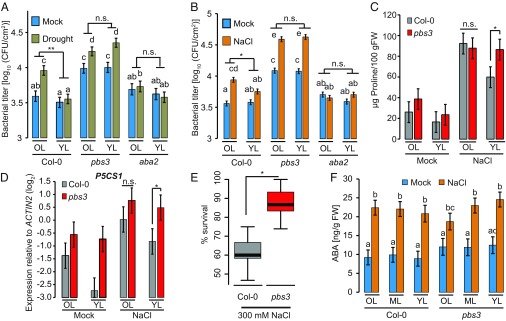
Abiotic stresses such as salinity and drought activate ABA biosynthesis (41). To test whether endogenous ABA accumulation induced by abiotic stresses triggers leaf age-dependent immune suppression, we measured Pto hrcC− growth in A. thaliana plants after salt or drought treatment. To avoid the known pleiotropic effects of SA hyperaccumulation in npr1 on plant performance (42, 43), we focused on pbs3 plants. Similar to the ABA treatment described here earlier, young leaves of Col-0 plants were protected from salt and drought stress-triggered immune suppression (Fig. 4 A and B). Abiotic stress-triggered suppression of immunity against Pto hrcC− in old leaves was not seen in aba2, which is impaired in ABA biosynthesis (44) (Fig. 4 A and B), indicating that abiotic stress-induced immune suppression is dependent on endogenous ABA and/or derived metabolites from ABA (45). In contrast to WT Col-0, young leaves of pbs3 plants were not protected from the abiotic stress-triggered immune suppression (Fig. 4 A and B). Thus, the protective role of PBS3 in young leaves is physiologically relevant.
Fig. 4.
Impact of PBS3 on leaf age-dependent stress-response trade-offs. (A and B) Old leaves (OL) and young leaves (YL) of 4–5-wk-old Col-0, pbs3, and aba2 plants were infiltrated with Pto hrcC− (OD600 = 0.0002) 2–3 wk after transfer to drought or well-watered conditions (mock) at 2-wk-old stage (A) or 2 d after water or 75 mM NaCl treatment (B). Bacterial growth was measured at 3 days postinoculation (dpi) (A) or 2 dpi (B). Data represent means ± SEM calculated from three independent experiments, each with at least five biological replicates, by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.01). (C and D) Proline (C) or P5CS1 expression levels (D) in old and young leaves of 4–5-wk-old Col-0 and pbs3 plants 6 d after 100 mM NaCl or mock treatment. Data represent means ± SEM calculated from three biological replicates by using a mixed linear model. (E) Survival rate of Col-0 and pbs3 plants after salinity stress recovery. Two-week-old plants were watered with 300 mM NaCl for 2 wk followed by recovery with water for another 1 wk. Data consist of three independent experiments, each with at least 35 plants per genotype. (F) The ABA levels in old, middle, and young leaves of 4–5-wk-old Col-0 and pbs3 plants 6 h after 100 mM NaCl or mock soil drench treatment. Data represent means ± SEM calculated from at least three biological replicates by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.05). (A–E) *P < 0.05 and **P < 0.01, two-tailed Student’s t tests; n.s., not significant.
Considering the extensive cross-talk between biotic and abiotic stress responses in both directions (2), we hypothesized that young leaves of pbs3 are hyperresponsive to abiotic stress compared with those of WT. To measure a leaf-specific abiotic stress response output, we determined the accumulation of proline, which serves as a cellular osmoprotectant (46). Upon salt stress, proline levels and expression of P5CS1, which encodes an enzyme that catalyzes the rate-limiting step in proline biosynthesis (46), were significantly higher only in young leaves of pbs3 compared with Col-0 (Fig. 4 C and D). Together with the compromised immune phenotype, it appears that, in pbs3 plants, the balance between biotic and abiotic stress responses is shifted toward abiotic stress tolerance. In agreement with this, under severe salt stress (300 mM NaCl), which can be lethal to A. thaliana plants, the survival rate of pbs3 was higher than that of Col-0 plants (Fig. 4E).
Interestingly, Col-0 and pbs3 plants accumulated comparable levels of basal and salt stress-induced ABA in leaves of different ages (Fig. 4F). We also determined the levels of other phytohormones, namely auxin (indole-3-acetic acid; IAA), jasmonic acid and its precursor 12-oxo-phytodienoic acid, SA, and the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (47) in Col-0 and pbs3 leaves of different ages upon salt stress or mock treatment. Although these hormone levels were mostly unaffected by genotype or salt stress, levels of IAA and ACC were dependent on leaf age (SI Appendix, Fig. S4), which may influence the leaf age-dependent stress cross-talk. In summary, these results suggest that, during combined stress, PBS3 lowers the abiotic stress response and enhances immunity in young leaves.
PBS3 Is Required for the Maintenance of Plant Growth and Reproduction Under Combined Stress.
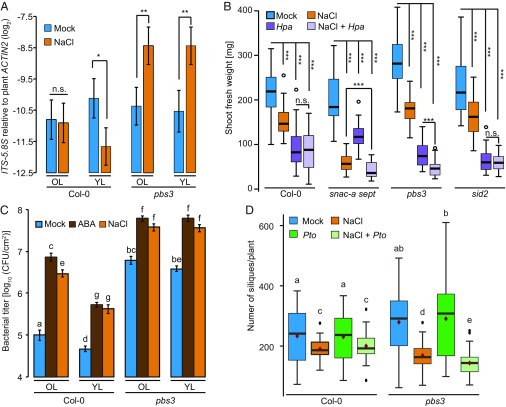
We employed two experimental systems to test whether the altered balance of leaf age-dependent stress cross-talk in pbs3 plants affects plant fitness-related traits during combined stresses. In the first system, we combined salt stress with infection with the obligate biotrophic oomycete pathogen Hyaloperonospora arabidopsidis (Hpa), whose growth is sensitive to SA-mediated immunity (48). Considering that many plant pathogens, including Hpa, require high humidity for successful infection, co-occurrence of drought stress with pathogen infection is unlikely to be common. Therefore, we used mild salt stress as an abiotic stress factor, which can co-occur with pathogen infection (49). Salt pretreatment reduced oomycete biomass in the young leaves of Col-0 WT, whereas, in pbs3 plants, salt stress promoted Hpa growth in old and young leaves (Fig. 5A). We included snac-a sept plants in the plant performance assay because, in this genotype, the balanced trade-offs of abiotic and biotic stress responses were shifted in the opposite direction compared with pbs3 plants (Figs. 2 A and B, 3C, and 4). Furthermore, we included sid2 plants because of their deficiency in SA biosynthesis but indistinguishable leaf age-dependent stress cross-talk vs. Col-0 plants (Fig. 3C). We found that single salt and Hpa stress reduced shoot fresh weight of all tested genotypes, with Hpa infection having greater negative consequences on growth of Col-0, pbs3, and sid2, except snac-a sept plants, whose growth suffered more severely from salt stress (Fig. 5B). This snac-a sept plant phenotype is consistent with our observation that snac-a sept plants exhibited lowered tolerance to mild salt stress in a germfree environment (Fig. 2B).
Fig. 5.
Leaf age-dependent variation in biotic and abiotic stress cross-talk contributes to plant fitness-related traits under combined stresses. (A) Hpa growth 8 d after inoculation in old leaves (OL) and young leaves (YL) of 4–5-wk-old Col-0 and pbs3 plants following 75 mM NaCl or water (Mock) soil drench treatment for 2 d. Data are means ± SEM calculated from at least three biological replicates by using a mixed linear model. (B) Shoot fresh weight of Col-0, snac-a sept, pbs3, and sid2 plants challenged with mock, NaCl, Hpa, or both NaCl and Hpa (Methods). The box plots show combined data from at least three independent experiments for Col-0, pbs3, and sid2 and two independent experiments for snac-a sept mutant plants, each with at least eight biological replicates. (A and B) *P < 0.05, **P < 0.01, and ***P < 0.001, two-tailed Student’s t tests; n.s., not significant. (C) Old and young leaves of 4–5-wk-old Col-0 and pbs3 plants were infiltrated with Pto cor− (OD600 = 0.0002) 1 d after water, 500 µM ABA, or 100 mM NaCl treatment. Bacterial growth was measured at 2 days postinoculation. Data represent means ± SEM calculated from three independent experiments, each with at least five biological replicates, by using a mixed linear model. Different letters indicate significant differences (adjusted P < 0.005). (D) The number of siliques in Col-0 and pbs3 plants after water (Mock), 50 mM NaCl (NaCl), Pto cor− (Pto), or both NaCl and Pto. The box plots show combined data from three independent experiments, each with at least 10 biological replicates. Statistical analysis was performed by using log-transformed silique numbers. Different letters indicate significant differences (adjusted P < 0.05).
Interestingly, the effects of combined stress on plant growth were similar to those of Hpa single stress in WT and sid2 plants, which exhibited leaf age-dependent stress cross-talk (Figs. 3C and 5B). In contrast, combined stress had more severe consequences on plant growth than the single Hpa and salt stress in pbs3 and snac-a sept plants, respectively (Fig. 5B). Thus, these results suggest that a loss in leaf age-dependent stress-response cross-talk is associated with a plant growth penalty during combined stress conditions.
In the second assay, we used salt stress and infection with Pto cor− as an additional biotic and abiotic stress combination. The Pto cor− strain lacks the phytotoxin coronatine that can suppress immune responses such as SA accumulation and MAPK activation (30, 50) but is more virulent compared with Pto hrcC− (Figs. 1E and 5C). Similar to Pto hrcC− infection, immune suppression by ABA treatment and salt stress was leaf age-dependent in Col-0 but independent of leaf age in pbs3 plants (Fig. 5C). To evaluate whether A. thaliana reproduction was affected by abiotic and biotic response cross-talk, we measured the number of siliques under single salt or Pto cor− stress or in combination in Col-0 and pbs3 plants. Under our experimental conditions, Pto cor− infection alone did not affect silique numbers of Col-0 and pbs3 plants, but moderate salt stress resulted in a reproductive penalty (Fig. 5D). In Col-0 plants, the effect of combined stress on silique numbers was similar to that of salt stress alone, whereas the negative effect of combined stress exceeded the single salt stress in pbs3 plants (Fig. 5D). Together, these findings indicate that PBS3, which is necessary for balancing leaf age-dependent biotic and abiotic stress responses based on leaf age, is required for the maintenance of growth and reproduction under combined biotic and abiotic stress.
PBS3 Coordinates Leaf Age- and Salt Stress-Dependent Phyllosphere Microbiota Assembly.
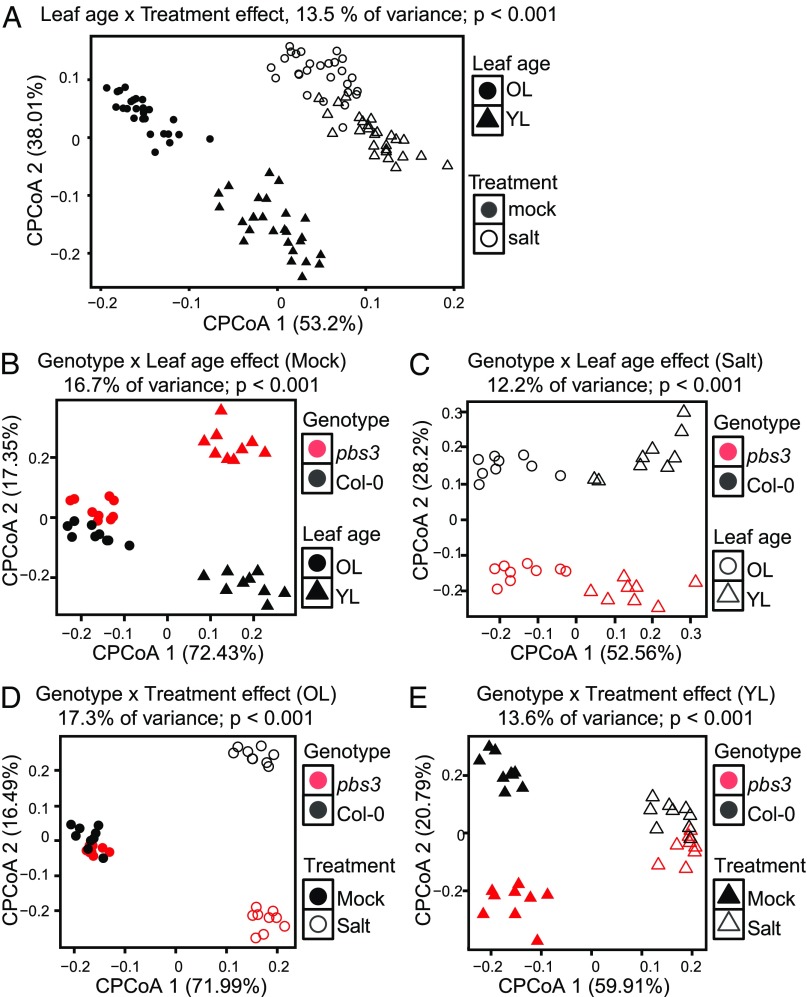
To test the impact of leaf age-dependent variation in cross-talk between ABA and SA signaling on biotic components beyond pathogens, we compared the composition of leaf-associated bacterial communities under abiotic stress or control in old and young rosette leaves. We used surface-sterilized A. thaliana seeds sown in a natural loamy soil with a characterized soil biome (7). Exposure of the plants to chronic salt stress resulted in decreases in plant fresh weight (SI Appendix, Fig. S5A). We applied bacterial 16S rRNA gene amplicon sequencing for quantitative bacterial community profiling and found upon principal coordinate analysis (PCoA) that salt treatment and leaf age were major factors in determining leaf microbiota composition (explaining 13.5% of community variation; Fig. 6A and SI Appendix, Fig. S5B). In addition, compared with Col-0, the leaves of aba2 plants were inhabited by more distinct bacterial communities under salt stress than the control condition (SI Appendix, Fig. S5 C and D), indicating that the salt stress-induced shift in the leaf-associated bacterial community is, at least in part, controlled by plant ABA signaling.
Fig. 6.
PBS3 shapes the leaf age- and salt stress-dependent assembly of leaf bacterial communities. (A–E) Canonical analysis of principle coordinates of bacterial β-diversity Bray–Curtis distances based on bacterial 16S rRNA profiling of leaf bacterial communities in WT Col-0, pbs3, and aba2 (A) or Col-0 and pbs3 plants (B–E). Plants were grown in natural Cologne soil treated with water (mock) or 75 mM NaCl (salt) for 6 wk. Constrained analysis was performed for leaf age × treatment effect (A), genotype × leaf age effect under mock (B) or salt stress (C), and genotype × treatment effect in old leaves (OL; D) or young leaves (YL; E).
Notably, young leaves of pbs3 and WT Col-0 plants hosted distinct bacterial communities in comparison with their old leaves in the control condition (16.7% of variation; Fig. 6B). Under salt stress, old and young leaves of these two genotypes all harbored distinct bacterial communities (12.2% of variation; Fig. 6C). To directly visualize the effects of salt stress on bacterial community composition in these genotypes, we also performed PCoA analysis for young and old leaves separately. This refined analysis revealed that, under salt stress, old leaves of pbs3 and Col-0 plants hosted distinct bacterial communities in comparison with the control condition (17.3% of variation; Fig. 6D). In contrast, bacterial communities in the young leaves of pbs3 and Col-0 plants were more distinct under control conditions than under salt stress (13.6% of variation; Fig. 6E). Together, these results show that pbs3 and Col-0 plants assemble distinct leaf age-dependent bacterial communities and that salt stress alters the community profiles in these two genotypes in a distinctive manner.
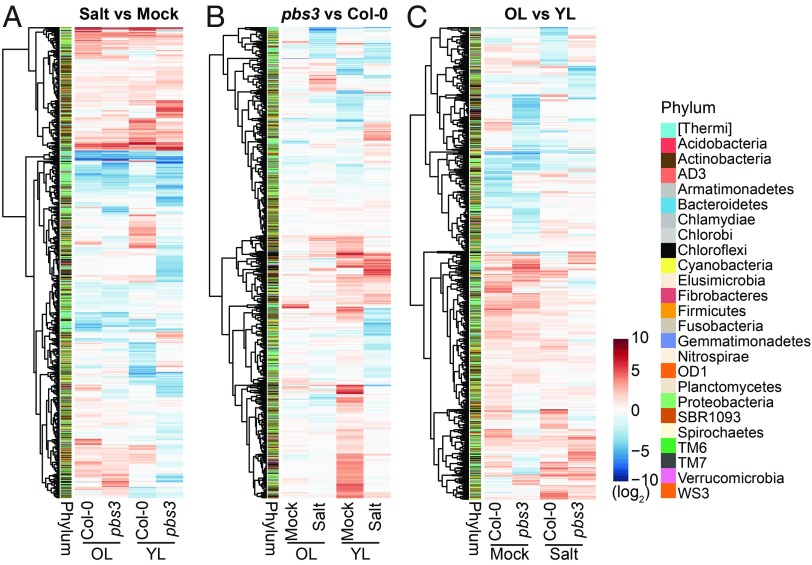
Salt stress altered the relative abundance of a broad range of bacterial operational taxonomic units (OTUs) belonging to different phylogenetic lineages rather than a specific taxonomic group in plant leaves and unplanted soil (Fig. 7A, SI Appendix, Figs. S5B and S6A, and Dataset S2). Shannon diversity analysis suggests that the different plant genotypes affected bacterial community profiles rather than overall bacterial richness (SI Appendix, Fig. S5E). Analysis at the level of OTUs revealed that PBS3 affects relative abundances of a wide range of leaf-associated bacteria including Actinobacteria, Firmicutes, and Proteobacteria (Fig. 7B and SI Appendix, Fig. S6B). Similarly, the distinctive leaf age-dependent community shifts seen in pbs3 plants influence a wide range of leaf-associated bacterial OTUs belonging to different phyla (Fig. 7C and SI Appendix, Fig. S6C). Thus, PBS3, salt stress, and leaf age broadly affected leaf-associated bacterial community assemblages. Together, these findings extend the physiological significance of variation in leaf age-dependent biotic and abiotic stress cross-talk to the leaf microbiota.
Fig. 7.
PBS3, salt stress, and leaf age determine relative bacterial abundances. (A) Heat map displaying log2 fold changes of relative abundances in bacterial OTUs under salt stress compared with mock in old leaves (OL) and young leaves (YL) of WT Col-0 and pbs3 plants. (B) Heat map displaying log2 fold changes of relative abundance for bacterial OTUs in old and young leaves of pbs3 compared with Col-0 plants under mock or salt conditions. (C) Heat map displaying log2 fold changes of relative abundance for bacterial OTUs in old leaves compared with young leaves of Col-0 and pbs3 plants under mock or salt conditions. (A–C) Plants were grown in natural Cologne soil treated with water (Mock) or 75 mM NaCl (Salt) for 6 wk. The log2 fold changes were subjected to hierarchical clustering. The phylum to which each OTU belongs is indicated by the colored bar.
Discussion
We have unveiled a genetically controlled mechanism by which A. thaliana plants balance trade-offs between conflicting biotic and abiotic stress responses by integrating these differently in young and old leaves. Moreover, we find that leaf age-dependent preference of stress responses balances trade-offs to increase plant growth and reproduction during combined stress. Thus, our findings define a plant strategy to maintain fitness in nature, where plants are often exposed to multiple stresses simultaneously (51), and demonstrate the physiological significance of stress-hormone cross-talk at the organismal level.
Young leaves serve as a better energy source compared with old leaves because cellular components such as the photosynthesis apparatus are more intact (52–55). The ODT explains why plants prioritize young leaves over old leaves for defense against insect herbivores by postulating that young leaves constitute a higher value for the whole plant, where value is correlated with the cost of having that tissue removed (22). Our study shows that young leaves exhibit higher biotic stress responses but lower abiotic stress responses compared with old leaves. Thus, our work suggests that young leaves are not simply protected from stresses because of their higher value in a manner similar to the ODT, but rather that plants actively balance the trade-off between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. pbs3 and snac-a sept plants, in which this balancing trade-off mechanism was absent, exhibit fitness penalties during combined biotic and abiotic stress conditions. In nature, actively maintaining fitness during combined stresses might be crucial for plant reproduction, and the balancing trade-off mechanism adds another dimension to our understanding of how plants cope with complex and fluctuating environments.
Our genetic analysis revealed that leaf age-dependent stress response prioritization during combined stress is controlled by PBS3 and NPR1, components of SA signaling. However, SA biosynthesis via SID2 in the isochorismate pathway was not required. Hence, this mechanism is distinct from plant age-dependent control of plant immunity as described during age-related resistance, which is fully dependent on SID2 in A. thaliana (18). Another study reported that expression of ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5), required for SA accumulation, shows leaf age-dependency in A. thaliana (56). However, EDS5 expression is higher in older compared with younger leaves. Therefore, this EDS5 expression pattern does not explain why young leaves are protected from ABA-triggered immune suppression. Thus, the leaf age-dependent trade-off between biotic and abiotic stress responses during combined stress is regulated by a mechanism distinct from the previously described age-dependent variation in stress responses. Our findings also indicate that the function of SA is not limited to plant–microbe interactions, but has wider implications for plant fitness maintenance under combined stress. Consistent with this, A. thaliana npr1 mutants exhibited reduced fitness in the field but not under controlled standard conditions (57).
Our data show that salt stress, leaf age, plant ABA biosynthesis, and PBS3 influence the structure of leaf-associated bacterial communities. Factors determining microbiota structure and contributions of plant commensals to plant health and fitness are beginning to be defined (13). Our findings demonstrate that leaf age-dependent variation in biotic and abiotic stress cross-talk is not limited to interactions with microbial pathogens, but also influences associations with resident leaf commensals. In the context of the observed differential biotic and abiotic stress response prioritization in younger and older leaves, this raises the intriguing possibility that the corresponding distinctive leaf-resident bacterial communities are adapted to contribute preferentially to biotic and abiotic stress tolerance, respectively. Irrespective of this, our work identifies a leaf age-dependent genetic intersection among immunity, the leaf-associated bacterial microbiota, and abiotic stress tolerance, which might determine plant fitness in natural environments.
Materials and Methods
Plants were grown in a chamber at 22 °C with 60% relative humidity and a 10-h light period for 4 wk before transfer to another chamber at 22 °C with 60% relative humidity and a 12-h light period before treatments. All A. thaliana plants used were in the Col-0 accession background. The details and procedures of plant materials and growth conditions, bacterial infection, Hpa infection, performance assay, quantitative PCR, RNA-seq, SA measurements, proline quantification, quantification of multiple phytohormones, bacterial 16S rRNA gene profiling, and statistical analysis, as well as the gene accession numbers used in this study, are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Yusuke Saijo for the pbs3-1 mutant; Kazuko Yamaguchi-Shinozaki and Kazuo Shinozaki for the areb1 areb2 abf3 triple and the snac-a sept mutants; Scott Poethig for the 35S::miR156a transgenic line; Anna Lisa Roth, Thorsten Thiergart, and Thomas Griebel for technical assistance with bacterial community preparation, data deposition, and SA measurements, respectively; and Neysan Donnelly and members of the laboratory of K.T. for critical reading of the manuscript. This work was supported by the Max Planck Society (K.T., R.G.-O., and P.S.-L.), the Cooperative Research Program for Agricultural Science & Technology Development Grant PJ01093907 (to P.S-L.) through Rural Development Administration, Republic of Korea, a predoctoral fellowship from the Nakajima Foundation (to T.N.), and a postdoctoral fellowship from the Alexander von Humboldt Foundation (to Y.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE114645) and in the European Nucleotide Archive, https://www.ebi.ac.uk/ena (accession no. PRJEB26793).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817233116/-/DCSupplemental.
References
- 1.Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 2.Bostock RM, Pye MF, Roubtsova TV. Predisposition in plant disease: Exploiting the nexus in abiotic and biotic stress perception and response. Annu Rev Phytopathol. 2014;52:517–549. doi: 10.1146/annurev-phyto-081211-172902. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y, et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19:1279–1290. doi: 10.1038/cr.2009.108. [DOI] [PubMed] [Google Scholar]
- 4.Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K. Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol. 2017;55:401–425. doi: 10.1146/annurev-phyto-080516-035544. [DOI] [PubMed] [Google Scholar]
- 5.Yasuda M, et al. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell. 2008;20:1678–1692. doi: 10.1105/tpc.107.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, Dommel M, Mou Z. Abscisic acid promotes proteasome-mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana. Plant J. 2016;86:20–34. doi: 10.1111/tpj.13141. [DOI] [PubMed] [Google Scholar]
- 7.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 8.Ritpitakphong U, et al. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016;210:1033–1043. doi: 10.1111/nph.13808. [DOI] [PubMed] [Google Scholar]
- 9.Vogel C, Bodenhausen N, Gruissem W, Vorholt JA. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016;212:192–207. doi: 10.1111/nph.14036. [DOI] [PubMed] [Google Scholar]
- 10.Castrillo G, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duran P, et al. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell. 2018;175:973–983.e14. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lebeis SL, et al. PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 13.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay between innate immunity and the plant microbiota. Annu Rev Phytopathol. 2017;55:565–589. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
- 14.Santos-Medellín C, Edwards J, Liechty Z, Nguyen B, Sundaresan V. Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio. 2017;8:e00764-17. doi: 10.1128/mBio.00764-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017;11:2691–2704. doi: 10.1038/ismej.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards JA, et al. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 2018;16:e2003862. doi: 10.1371/journal.pbio.2003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc Natl Acad Sci USA. 2018;115:E4284–E4293. doi: 10.1073/pnas.1717308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kus JV, Zaton K, Sarkar R, Cameron RK. Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell. 2002;14:479–490. doi: 10.1105/tpc.010481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeier J. Age-dependent variations of local and systemic defence responses in Arabidopsis leaves towards an avirulent strain of Pseudomonas syringae. Physiol Mol Plant Pathol. 2005;66:30–39. [Google Scholar]
- 20.Sperdouli I, Moustakas M. Leaf developmental stage modulates metabolite accumulation and photosynthesis contributing to acclimation of Arabidopsis thaliana to water deficit. J Plant Res. 2014;127:481–489. doi: 10.1007/s10265-014-0635-1. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo M, Oelmüller R. REDOX RESPONSIVE TRANSCRIPTION FACTOR1 is involved in age-dependent and systemic stress signaling. Plant Signal Behav. 2015;10:e1051279. doi: 10.1080/15592324.2015.1051279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCall AC, Fordyce JA. Can optimal defence theory be used to predict the distribution of plant chemical defences? J Ecol. 2010;98:985–992. [Google Scholar]
- 23.Takasaki H, et al. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015;84:1114–1123. doi: 10.1111/tpj.13067. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida T, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 25.de Torres-Zabala M, et al. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Torres Zabala M, Bennett MH, Truman WH, Grant MR. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009;59:375–386. doi: 10.1111/j.1365-313X.2009.03875.x. [DOI] [PubMed] [Google Scholar]
- 27.Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53:763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- 28.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickman R, et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013;75:26–39. doi: 10.1111/tpj.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng XY, et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11:587–596. doi: 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran L-SP, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 33.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nobuta K, et al. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007;144:1144–1156. doi: 10.1104/pp.107.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;4 14:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 36.Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF. Salicylic acid biosynthesis and metabolism. Arabidopsis Book. 2011;9:e0156. doi: 10.1199/tab.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, et al. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1:639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Ding Y, et al. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 2018;173:1454–1467.e15. doi: 10.1016/j.cell.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, et al. The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: Roles of major regulators and the phytotoxin coronatine. Mol Plant Microbe Interact. 2008;21:1408–1420. doi: 10.1094/MPMI-21-11-1408. [DOI] [PubMed] [Google Scholar]
- 40.Lee HJ, et al. Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell. 2015;27:3425–3438. doi: 10.1105/tpc.15.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finkelstein R. Abscisic acid synthesis and response. Arabidopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Q, et al. Salicylic acid-altering Arabidopsis mutants response to NO(2) exposure. Bull Environ Contam Toxicol. 2010;84:106–111. doi: 10.1007/s00128-009-9913-3. [DOI] [PubMed] [Google Scholar]
- 43.Ding Y, Shaholli D, Mou Z. A large-scale genetic screen for mutants with altered salicylic acid accumulation in Arabidopsis. Front Plant Sci. 2015;5:763. doi: 10.3389/fpls.2014.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adie BAT, et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng JK, Ye M, Li B, Noel JP. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell. 2016;166:881–893. doi: 10.1016/j.cell.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 46.Kesari R, et al. Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc Natl Acad Sci USA. 2012;109:9197–9202. doi: 10.1073/pnas.1203433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shigenaga AM, Argueso CT. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin Cell Dev Biol. 2016;56:174–189. doi: 10.1016/j.semcdb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Zeilmaker T, et al. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015;81:210–222. doi: 10.1111/tpj.12719. [DOI] [PubMed] [Google Scholar]
- 49.Bai Y, Kissoudis C, Yan Z, Visser RGF, van der Linden G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018;93:781–793. doi: 10.1111/tpj.13800. [DOI] [PubMed] [Google Scholar]
- 50.Mine A, et al. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc Natl Acad Sci USA. 2017;114:7456–7461. doi: 10.1073/pnas.1702613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandey P, Irulappan V, Bagavathiannan MV, Senthil-Kumar M. Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci. 2017;8:537. doi: 10.3389/fpls.2017.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang RH, Chang JC, Li KT, Lin TS, Chang LS. Leaf age and light intensity affect gas exchange parameters and photosynthesis within the developing canopy of field net-house-grown papaya trees. Sci Hortic. 2014;165:365–373. [Google Scholar]
- 53.Prochazkova D, Wilhelmova N. Leaf senescence and activities of the antioxidant enzymes. Biol Plant. 2007;51:401–406. [Google Scholar]
- 54.Hikosaka K. Effects of leaf age, nitrogen nutrition and photon flux density on the organization of the photosynthetic apparatus in leaves of a vine (Ipomoea tricolor Cav) grown horizontally to avoid mutual shading of leaves. Planta. 1996;198:144–150. doi: 10.1007/BF00325881. [DOI] [PubMed] [Google Scholar]
- 55.Hensel LL, Grbić V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandran D, et al. Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe. 2014;15:506–513. doi: 10.1016/j.chom.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Heidel AJ, Clarke JD, Antonovics J, Dong X. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics. 2004;168:2197–2206. doi: 10.1534/genetics.104.032193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.