Significance
Despite the usefulness of genetically engineered mouse models in cancer research, their small total blood volume and the rarity of circulating tumor cells (CTCs) preclude the use of existing liquid biopsy techniques for longitudinal CTC studies in mice. We have devised a method for collecting CTCs from an unanesthetized mouse longitudinally, spanning multiple days or weeks, to study acute perturbations (e.g., drug treatment) or potentially long-term phenotypes (e.g., tumor progression) within the same mouse. Here, we show that our optofluidic-based approach eliminates confounding biases driven by intermouse heterogeneity that can occur when CTCs are collected across different mice.
Keywords: microfluidic, GEMM, circulating tumor cells, metastasis, single-cell RNA-Seq
Abstract
Circulating tumor cells (CTCs) play a fundamental role in cancer progression. However, in mice, limited blood volume and the rarity of CTCs in the bloodstream preclude longitudinal, in-depth studies of these cells using existing liquid biopsy techniques. Here, we present an optofluidic system that continuously collects fluorescently labeled CTCs from a genetically engineered mouse model (GEMM) for several hours per day over multiple days or weeks. The system is based on a microfluidic cell sorting chip connected serially to an unanesthetized mouse via an implanted arteriovenous shunt. Pneumatically controlled microfluidic valves capture CTCs as they flow through the device, and CTC-depleted blood is returned back to the mouse via the shunt. To demonstrate the utility of our system, we profile CTCs isolated longitudinally from animals over 4 days of treatment with the BET inhibitor JQ1 using single-cell RNA sequencing (scRNA-Seq) and show that our approach eliminates potential biases driven by intermouse heterogeneity that can occur when CTCs are collected across different mice. The CTC isolation and sorting technology presented here provides a research tool to help reveal details of how CTCs evolve over time, allowing studies to credential changes in CTCs as biomarkers of drug response and facilitating future studies to understand the role of CTCs in metastasis.
Circulating tumor cells (CTCs) are an intermediate in the hematogenous spread of tumors during metastasis (1). Given their accessibility and potential prognostic and diagnostic value, CTCs have been the focus of significant clinical research efforts monitoring response to therapy and predicting risk of relapse (2–4). Over the past decade, novel microfluidic liquid biopsy-based techniques, as well as in vivo, vein catheter-based methods, have been developed to detect and collect CTCs directly from the blood of human patients (2–10). Combined with recently developed single-cell profiling methods, such as single-cell RNA sequencing (scRNA-Seq) (11–14), in-depth examination of CTCs is now possible. Such studies can provide new insights into the genomic properties of CTCs, as well as their relationship to matched primary and metastatic tumors (3, 4, 15–18).
Genetically engineered mouse models (GEMMs) of cancer, which mimic the natural multistage evolution of their human counterparts, facilitate characterization of both acute perturbations (e.g., drug treatment) and long-term phenotypic changes (e.g., tumor evolution) not possible in human subjects. However, despite the usefulness of GEMMs in cancer research, the combination of the small total murine blood volume (∼1.5 mL) and the rarity of CTCs in circulating blood (fewer than 100 cells per milliliter) (3, 19) precludes the use of existing liquid biopsy techniques for longitudinal CTC studies in mice. When repeated blood samples are required at short intervals, a maximum of 1.0% of an animal’s total blood volume can be removed every 24 h (∼16.5 μL for a 25-g mouse) (20), a miniscule volume that does not yield a sufficient sample of CTCs for analysis.
GEMMs have been developed that combine genetic perturbations (manipulation of oncogenes and tumor suppressor genes) with genetically encoded fluorescent markers, enabling the unbiased detection and isolation of CTCs from the bloodstream. In vivo flow cytometry techniques have been used to enumerate CTCs in ear capillaries or tail veins longitudinally without euthanizing animals (21–23), but these techniques do not permit isolation and downstream molecular characterization of CTCs. On the other hand, in vivo, vein catheter-based techniques allow for the direct capture and isolation of CTCs from much larger blood volumes in real time (7, 8). However, these techniques are currently limited to detecting epithelial cell adhesion molecule (EpCAM)-expressing CTCs, which may result in only a subpopulation of CTCs being detected and isolated (24).
Results
Optofluidic Platform Design and Characterization.
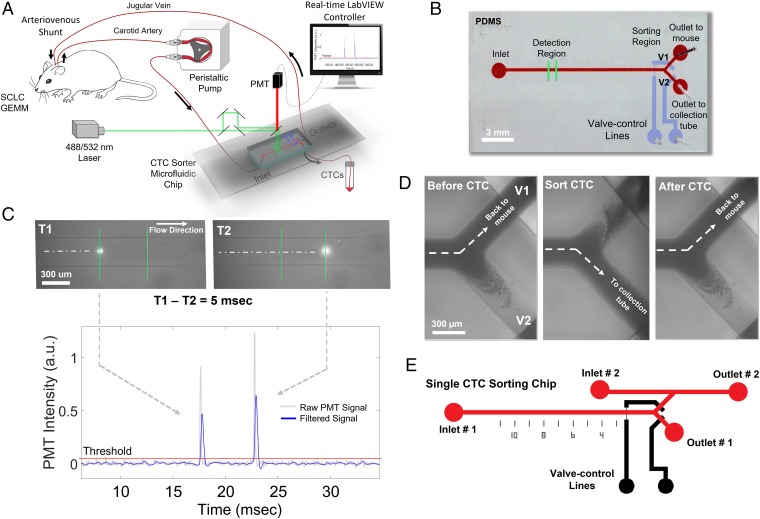
To enable longitudinal, in-depth studies of CTC biology in GEMMs and other murine cancer models, we have developed an optofluidic system capable of detecting and capturing fluorescent CTCs in living mice over several hours, days, or weeks. Key components of the system include a polydimethylsiloxane-based microfluidic CTC sorter chip, a fluorescence detector, and computer-controlled pneumatic valves (Fig. 1 A and B). A cannulated mouse with two permanent catheters easily accessible on its back allows for continuous blood withdrawal from the left carotid artery and return through the right jugular vein. Blood flows at a rate of 30 μL⋅min−1 into the CTC sorter chip. Two closely spaced laser beam lines illuminate the main flow channel of the chip. As such, each fluorescent cell that passes through the device emits two pulses of light, which are detected by a photomultiplier tube (Fig. 1C and SI Appendix, Fig. S1). The second laser line allows the controller to compute the velocity of the cells, which is essential to ensure reliable CTC capture. Similar to the ensemble-decision aliquot ranking technique for sorting CTC-containing aliquots of blood (9, 10), upon fluorescent cell detection, the controller instantly operates pneumatic valves (25) to redirect a small blood volume that includes the CTC toward a collection tube (mean ± SD = 127 ± 47 nL per sort event; Fig. 1D and SI Appendix, Supplementary Information Text and Fig. S2). Blood from the collection tube can then be further enriched for CTCs and run through a secondary single-CTC sorting chip for downstream characterization using techniques such as scRNA-Seq (Fig. 1E).
Fig. 1.
Microfluidic sorter for longitudinal CTC studies in GEMMs. (A) Peristaltic pump withdraws blood from a surgically implanted cannula in the carotid artery of a mouse at a flow rate of 30 μL⋅min−1. The blood is directed into the main flow channel of the CTC sorter chip. For tdTomato-positive cells, a green (532-nm) laser illuminates two points along the main flow channel of the CTC chip separated by a known distance. Thus, fluorescent CTCs emit two red-shifted pulses of light, which are detected by a photomultiplier tube (PMT). Based on the timing of the pulses, a LabVIEW program computes the velocity of the cells and operates computer-controlled pneumatic valves to redirect fluorescent CTCs toward a collection tube. After exiting the chip, CTC-depleted blood returns to the jugular vein of the mouse via a second surgically implanted cannula. (B) Top-view image of the CTC sorter microfluidic chip showing the inlet, outlets, and valve actuation lines (V1 and V2). (C) Illustration of the CTC detection mechanism using the two excitation laser lines. A low-pass filter is applied to the raw data for determining true peaks. (D) Outlet by which blood is returned to the mouse is briefly sealed while the opposite outlet is opened to allow for CTC isolation in real time. (E) After collection, CTCs are further enriched by a secondary CTC sorting chip designed with a parallel channel to flush CTCs into wells containing cell lysis buffer for downstream scRNA-Seq.
Validation of the Platform’s Detection Limits with Fluorescent Microbeads and Cells.
To ascertain the detection limit of our CTC sorter, we passed a sample of healthy mouse blood spiked with flow cytometry calibration beads through the system. The reference beads comprised five fluorescence intensity groups, including one with zero fluorescence. The system consistently detected the two brightest fluorescence levels (peaks 4 and 5) and approximately the brightest 30% of level 3 (peak 3) (SI Appendix, Fig. S3 A–C). This sensitivity was sufficient to detect nearly the entire population of tdTomato-expressing murine small cell lung cancer (SCLC) cells spiked in healthy mouse blood. We also tested blood isolated from autochthonous SCLC tumor-bearing mice that exhibit metastasis to distant organs in a pattern similar to metastatic spread in human patients (26). Tumors in these mice were initiated by Cre-mediated deletion of tumor suppressor genes Trp53, Rb1, and Pten in the murine lung epithelium (26). This GEMM also includes a Cre-activated tdTomato allele (27) that engenders fluorescence in all tumor cells after tumor initiation, including CTCs. The majority of CTCs from the blood of SCLC tumor-bearing mice were above the detection threshold (SI Appendix, Supplementary Information Text and Fig. S3D).
Validation of Platform’s Sorting Functionality.
After establishing that the sensitivity of the optical detector was sufficient, we characterized and optimized the sorting efficacy using blood samples from healthy mice spiked with low concentrations of tdTomato-expressing murine SCLC cells. In samples containing 100 cells per milliliter or more, over 80% of detected tdTomato-positive cells were successfully captured. For samples with only 10 cells spiked into 500 μL of healthy mouse blood, the sorted sample contained 6.0 ± 0.7 cells (mean ± SD, n = 3 repeats). Applying a slight delay in actuating the pneumatic valves until the cell has moved closer to the sorting region decreased the collected blood volume per CTC to 76 ± 28 nL (mean ± SD) without compromising the capture efficiency. At this volume, on the order of 700 neighboring white blood cells (WBCs) and over 70,000 red blood cells (RBCs) and platelets in the bloodstream are collected in addition to the target CTC on each valve actuation. These experiments demonstrate that the CTC sorter is capable of isolating fluorescent CTCs from blood even at very low concentrations (SI Appendix, Fig. S4).
Longitudinal CTC Collection from SCLC Tumor-Bearing Mice.
Next, we conducted a longitudinal study of CTCs collected from autochthonous SCLC tumor-bearing mice treated with the BET bromodomain inhibitor JQ1, which has been demonstrated to have antiproliferative effects in SCLC (28–30). CTCs were isolated from mice over a 2-h period before treatment (0 h) and at 24-h intervals following treatment initiation, continuing over 96 h (SI Appendix, Supplementary Information Text and Figs. S5 and S6A). CTCs were enriched from the samples by RBC lysis, followed by WBC depletion, using magnetic-activated cell sorting (SI Appendix, Fig. S6B) and, finally, by passing through a secondary, single-cell CTC sorting chip. Enriched CTCs were then processed using Smart-Seq2 (31) (Fig. 1E and SI Appendix, Supplementary Information Text and Fig. S6C). Cells with insufficient gene complexity for downstream analysis after scRNA-Seq were eliminated computationally, in addition to cells with high expression of immune and platelet signature genes (3, 4, 16) (SI Appendix, Supplementary Information Text). The overall yields of the process (from blood to a successful scRNA-Seq library) were 11.5% and 5.3% for samples from treated and untreated mice, respectively (median values with a range of 7.4–31% for treated samples and a range of 3.3–6.7% for untreated samples; SI Appendix, Fig. S7).
Analysis of Single-CTC Transcriptomes Across Different Mice and Within Each Mouse.
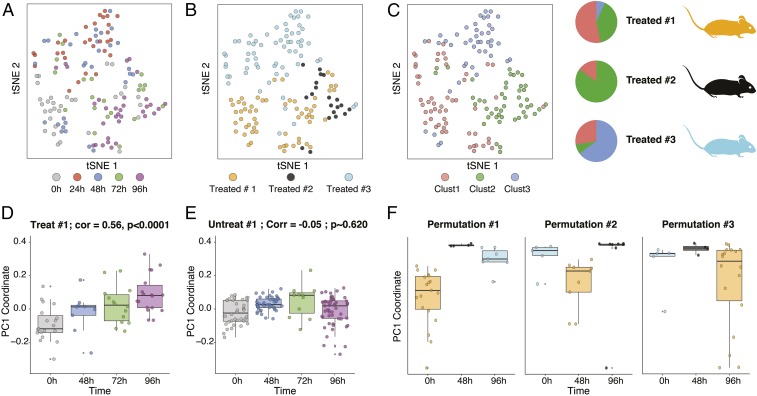
We then examined our data to determine how the information collected longitudinally from the same mouse with our system compared with the common approach of capturing CTCs across different mice using asynchronous terminal bleeds (16, 19). To analyze our longitudinal CTC data, we pooled our collected CTC transcriptomes across all mice, performed a principal component analysis (PCA) visualized by t-distributed stochastic neighbor embedding (tSNE) (12, 32, 33), and identified clusters (using k-nearest neighbors clustering) over the significant principal components (PCs) (14, 34) (Fig. 2 A–C and SI Appendix, Supplementary Information Text). This unsupervised analysis revealed that mouse of origin contributed significantly to the variation observed in our dataset, with cluster representation driven primarily by individual mice (3, 4, 6) (Fig. 2C). We next performed PCA on CTCs collected from each mouse individually. Here, we found that PC1 significantly correlated (Spearman correlation) with time since treatment (P < 0.05, Student’s t test following a Lilliefors test for normality) when independently calculated for each of the treated mice but not for either control (Fig. 2 D and E and SI Appendix, Fig. S8). This suggests that by isolating CTCs from the same animal longitudinally, we are able to eliminate potentially confounding differences between animals that could otherwise mask biologically relevant gene expression changes that occur over time.
Fig. 2.
scRNA-Seq of captured CTCs demonstrates the utility of intramouse CTC profiling. The tSNE of all CTCs collected across three JQ1-treated mice is colored by time point posttreatment (A), mouse (B), and cluster of assignment based on k-nearest neighbors clustering (C). (Top Right) Pie charts show the fractional representation of each cluster in each treated mouse. Boxplots of the first PC of CTC transcriptomes from PCAs were obtained from longitudinally following the same treated mouse [D, correlation (Corr) = 0.56] or untreated mouse (E, Corr = −0.05). Each point represents a CTC. (F) Boxplots of the first PC from three different “mock terminal bleed” permutations across three treated mice (SI Appendix, Supplementary Information Text and Fig. S9).
In comparison, the conventional approach for performing a longitudinal CTC analysis would be to begin the experiment with a cohort of mice and obtain terminal bleeds from a subset at each time point. We simulated this approach from our measurements by selecting a different treated mouse to represent each of the 0-h, 48-h, and 96-h time points (SI Appendix, Fig. S9); here, regardless of which mouse was chosen to represent which time point, we found that the mean PC1 coordinate of treated mouse 1 existed outside the interquartile range of the other mice, suggesting a consistent mouse-specific effect that dominates the first PC (Fig. 2F and SI Appendix, Fig. S10). As such, conclusions drawn from analysis of CTCs from terminal bleeds at different time points across mice would be confounded by organism-specific features from the different mice.
Supervised Analysis of Single-CTC Transcriptomes.
To more formally examine treatment-induced shifts in gene expression, we calculated differential expression across all pairs of time points within each longitudinally profiled mouse (14, 35) and, once again, simulated terminal bleed data (SI Appendix, Supplementary Information Text and Fig. S9). Our analyses showed that the majority of the differentially expressed genes within each mouse (per mouse) were unique (SI Appendix, Fig. S11). Furthermore, each per-mouse differentially expressed gene set shared little overlap (P < 0.05, hypergeometric test) with those calculated from mock terminal bleed datasets (SI Appendix, Fig. S12), regardless of the chosen mouse for the different time points. At each time point, differentially expressed genes in the mock terminal bleed data were enriched for several functional processes, such as mitochondrial function, cellular organization, and metabolism (36, 37); however, upon further inspection of the different mock terminal bleed permutations, we found that these enrichments were linked primarily to mouse rather than time point (Fig. 2F and SI Appendix, Figs. S10 and S12; e.g., housekeeping genes or ribosome), suggesting confounding mouse-to-mouse heterogeneity. This is evocative of the marked inter- and intrapatient heterogeneity observed in CTCs longitudinally collected from human patients (5–7), and suggests the importance of examining the same mouse over time.
Discussion
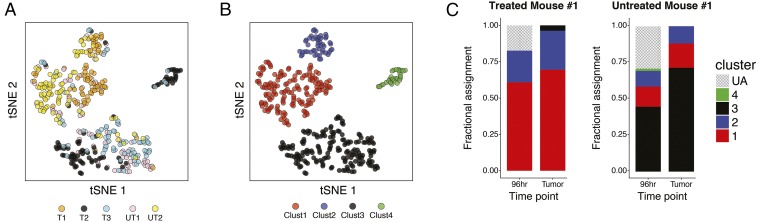
The platform outlined here represents an important advancement in the detection and continuous capture of single CTCs from the same mouse over time. Our method enables CTCs to be isolated in low blood volumes and prepares them for downstream characterization. Here, we used scRNA-Seq to show that continuous CTC capture from the same mouse eliminates biases driven by intermouse heterogeneity that can occur when CTCs are collected across different mice. Although future work will be needed to elucidate the underlying drivers of this variability, given the baseline genetic homogeneity of the animals used to generate our GEMM, one potential explanation could be underlying differences in the cellular composition of the primary tumors across different animals. scRNA-Seq results from the primary tumor samples harvested from each animal after terminal CTC collection (96 h) are consistent with this hypothesis (Fig. 3 A and B and SI Appendix, Supplementary Information Text). These data suggest that primary tumors from each mouse may contain multiple malignant gene expression states (32, 33), which appear to be shared across mice. Although some of these differences could be attributed to the presence of multiple independently evolving primary tumors within each mouse, the fact that each gene expression state is composed of cells from multiple mice suggests that these states may be a shared feature of our SCLC GEMM, although future experiments will be needed to robustly validate this finding. Intriguingly, we were able to computationally match ∼67% (62 cells) of the terminally collected CTCs (96 h, n = 92 cells from five mice) to one of these shared states (SI Appendix, Supplementary Information Text and Fig. S13). Furthermore, we observed comparable state frequencies between a mouse’s CTCs and its corresponding primary tumor sample at the terminal time point (P > 0.5, Fisher’s exact test; Fig. 3C). We note that mice with fewer than 10 CTCs, the statistical power in our comparison was limited in some cases (SI Appendix, Fig. S14). While further experimentation will be needed to corroborate this preliminary finding, our data demonstrate the potential value of having matched primary tumor samples as a reference in mouse models of cancer, and that analysis of CTCs in our murine model of SCLC may reveal similar biology to primary tumors from the same mouse, suggesting their utility as a surrogate for matched tumors under specific circumstances.
Fig. 3.
scRNA-Seq of end-point primary tumors demonstrates heterogeneity in phenotype. (A) The tSNE of primary tumor cells across treated and untreated mice, colored by mouse called from k-nearest neighbors clustering. (B) tSNE of primary tumor cells across treated and untreated mice, colored by clusters. (C) Computational cluster assignments (SI Appendix, Supplementary Information Text) for 96-h CTCs next to their matched primary for a representative treated mouse and untreated mouse plotted as bar plots (n = 18 and n = 82 cells for treated mouse 1 96-h CTCs and tumor cells, respectively; n = 52 and n = 84 cells for untreated mouse 1 96-h CTCs and tumor cells, respectively). UA, unassigned. Neither pairing is significantly different (P = 0.99 and P = 0.66 for treated mouse 1 and untreated mouse 1, respectively, by Fisher’s exact test).
Future work of this kind has the potential to shed new light on the relationship between CTCs, primary tumors, and metastases, allowing for the exploration of their utility as biomarkers and facilitating examination of how individual CTCs contribute to metastasis. Moreover, they may help elucidate the features that inform shifts observed upon perturbation, such as drug treatment. Ultimately, these data show that our platform opens the door for novel CTC experimentation, such as examining longitudinal drug responses and comparing CTCs with primary tumors (shown here), characterizing their relationship to metastases, and measuring the rate of CTC production in an acute window. With additional development, our device could enable longitudinal studies in mice to find associations between individual CTCs and clusters of CTCs, profile rare immune cells (e.g., using genetic reporters or based on tetramer staining), monitor mesenchymal cells in a variety of contexts (including wound healing and tumor formation), and measure induction rates of drugs or nanoparticles in circulating mononuclear cells.
Materials and Methods
All RNA-sequencing data generated in this study (raw data and processed data matrices) have been deposited into the Gene Expression Omnibus database hosted at the National Center for Biotechnology Information under the accession code GSE122233. Information on mouse models, cell culture, shunt surgery, optofluidic platform design and fabrication, real-time data processing and analysis, CTC and tumor cell processing and enrichment, single-cell RNA-sequencing sample preparation, and data analysis is available in SI Appendix. All animal-based procedures were approved by the Massachusetts Institute of Technology Committee on Animal Care, Division of Comparative Medicine.
Supplementary Material
Acknowledgments
We thank R. Kimmerling, S. Olcum, M. Stevens, B. A. Goods, A. S. Genshaft, C. G. K. Ziegler, N. Calistri, L. Atta, and M. B. Cole for helpful discussions. We also thank Samira Daswani for technical assistance. This work was supported, in part, by the Thomas and Sarah Kailath Fellowship (to B.H.); the A*STAR (Agency for Science, Technology and Research, Singapore) National Science Scholarship (to S.R.N.); the Lustgarten Foundation (M.G.V.H.); the Ludwig Center at MIT (S.R.M., T.J., and M.G.V.H.); Stand Up To Cancer (M.G.V.H.); the Howard Hughes Medical Institute (HHMI) Faculty Scholars Award (to M.G.V.H.); the HHMI Investigator Program (T.J.); the Searle Scholars Program (A.K.S.); the Beckman Young Investigator Program (A.K.S.); NIH Grant 1R01 CA184956 (to S.R.M. and T.J.); NIH New Innovator Award 1DP2GM119419 (to A.K.S.); the Pew-Stewart Scholars (A.K.S.), a Sloan Fellowship in Chemistry (to A.K.S.); NIH Grant 5U24AI118672 (to A.K.S.); NIH Grant 1U54CA217377 (to A.K.S. and S.R.M.); NIH Grant 1R33CA202820 (to A.K.S.); NIH Grant 2U19AI089992 (to A.K.S.); NIH Grant 1R01HL134539 (to A.K.S.); NIH Grant 2RM1HG006193 (to A.K.S.); NIH Grant 2P01AI039671 (to A.K.S.); and Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE122233).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814102116/-/DCSupplemental.
References
- 1.Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene. 2016;35:1216–1224. doi: 10.1038/onc.2015.192. [DOI] [PubMed] [Google Scholar]
- 2.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyamoto DT, et al. RNA-seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ting DT, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozkumur E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarioglu AF, et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12:685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorges TM, et al. Enumeration and molecular characterization of tumor cells in lung cancer patients using a novel in vivo device for capturing circulating tumor cells. Clin Cancer Res. 2016;22:2197–2206. doi: 10.1158/1078-0432.CCR-15-1416. [DOI] [PubMed] [Google Scholar]
- 8.Vermesh O, et al. An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo. Nat Biomed Eng. 2018;2:696–705. doi: 10.1038/s41551-018-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiro PG, et al. Sensitive and high-throughput isolation of rare cells from peripheral blood with ensemble-decision aliquot ranking. Angew Chem Int Ed Engl. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson ES, Anand RK, Chiu DT. Improved detection by ensemble-decision aliquot ranking of circulating tumor cells with low numbers of a targeted surface antigen. Anal Chem. 2015;87:9389–9395. doi: 10.1021/acs.analchem.5b02241. [DOI] [PubMed] [Google Scholar]
- 11.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 12.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohr JG, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aceto N, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vishnoi M, et al. The isolation and characterization of CTC subsets related to breast cancer dormancy. Sci Rep. 2015;5:17533. doi: 10.1038/srep17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 19.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parasuraman S, Raveendran R, Kesavan R. Blood sample collection in small laboratory animals. J Pharmacol Pharmacother. 2010;1:87–93. doi: 10.4103/0976-500X.72350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgakoudi I, et al. In vivo flow cytometry: A new method for enumerating circulating cancer cells. Cancer Res. 2004;64:5044–5047. doi: 10.1158/0008-5472.CAN-04-1058. [DOI] [PubMed] [Google Scholar]
- 22.Zettergren E, et al. Instrument for fluorescence sensing of circulating cells with diffuse light in mice in vivo. J Biomed Opt. 2012;17:037001. doi: 10.1117/1.JBO.17.3.037001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nedosekin DA, Verkhusha VV, Melerzanov AV, Zharov VP, Galanzha EI. In vivo photoswitchable flow cytometry for direct tracking of single circulating tumor cells. Chem Biol. 2014;21:792–801. doi: 10.1016/j.chembiol.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chudziak J, et al. Clinical evaluation of a novel microfluidic device for epitope-independent enrichment of circulating tumour cells in patients with small cell lung cancer. Analyst. 2016;141:669–678. doi: 10.1039/c5an02156a. [DOI] [PubMed] [Google Scholar]
- 25.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 26.McFadden DG, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenhart R, et al. Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol Cancer Ther. 2015;14:2167–2174. doi: 10.1158/1535-7163.MCT-15-0037. [DOI] [PubMed] [Google Scholar]
- 29.Kato F, et al. MYCL is a target of a BET bromodomain inhibitor, JQ1, on growth suppression efficacy in small cell lung cancer cells. Oncotarget. 2016;7:77378–77388. doi: 10.18632/oncotarget.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jahchan NS, et al. Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell Rep. 2016;16:644–656. doi: 10.1016/j.celrep.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 32.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung NC, Storey JD. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics. 2015;31:545–554. doi: 10.1093/bioinformatics/btu674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDavid A, et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.