Significance
A mathematical model relating tumor heterogeneity at the cellular level to tumor growth at the macroscopic level is described based on a statistical mechanics framework. The model takes into account the number of accessible states available to each cell as well as their long-range coupling (population cooperation) to other cells. We show that the degree to which cell populations cooperate determines the number of independent cell states, which in turn dictates the macroscopic (volumetric) growth law. It follows that targeting cell-to-cell interactions or functional coupling among cell populations could be a way of mitigating and controlling tumor growth.
Keywords: tumor growth laws, cell coupling, statistical mechanics, tumor heterogeneity
Abstract
A tumor is made up of a heterogeneous collection of cell types, all competing on a fitness landscape mediated by microenvironmental conditions that dictate their interactions. Despite the fact that much is known about cell signaling, cellular cooperation, and the functional constraints that affect cellular behavior, the specifics of how these constraints (and the range over which they act) affect the macroscopic tumor growth laws that govern total volume, mass, and carrying capacity remain poorly understood. We develop a statistical mechanics approach that focuses on the total number of possible states each cell can occupy and show how different assumptions on correlations of these states give rise to the many different macroscopic tumor growth laws used in the literature. Although it is widely understood that molecular and cellular heterogeneity within a tumor is a driver of growth, here we emphasize that focusing on the functional coupling of states at the cellular level is what determines macroscopic growth characteristics.
A typical tumor comprises a remarkably heterogeneous agglomeration of cell types, both at the molecular level (1) and at the phenotypic/morphological level (2, 3). Why this is often the case is still somewhat open to debate (1, 4), but clearly mutational instability (5, 6), ecological niches (7, 8), and tissue microenvironmental factors (9–11) all contribute to this diversity of cell types, which, in turn, enables natural selection to act to shape the fitness landscape of the tumor (3, 12), drive tumor growth (13, 14), and select for resistant subpopulations during treatment (15, 16). A question we address in this paper is: What are the consequences of tumor heterogeneity at the cellular scale with respect to key aspects of tumor growth at the macroscale? It has long been known that cellular communication is essential for embryonic development, for example, but it has been less widely appreciated in cancer. In particular, how do cellular diversity, on the one hand, and cell–cell interactions and intercellular communication (via hormones, growth factors, neurotransmitters, and cytokines mediated by gap junction channels), on the other hand, both work in concert to determine the macroscopic growth laws of a tumor? It has been observed that the cancer problem is not merely a cell problem, it is a problem of cell interaction, not only within tissues, but with distant cells in other tissues (17). We know that communication processes among cells ultimately control many aspects of cell growth, including proliferation, differentiation, apoptosis, and a cell’s ability to adapt and respond to microenvironmental cues. We also know that disruption of cell–cell communication can lead to increased or decreased proliferation, abnormal differentiation, apoptotic alteration, and abnormal adaptive responses (18–20). In fact, it has been hypothesized that cancer essentially is a consequence of dysfunctional gap-junctional intercellular communication (21, 22).
In this paper, we present a mathematical framework that links heterogeneity/diversity at the cellular level to the many different deterministic growth laws at the volumetric level. At the cellular level, heterogeneity is modeled by allowing each of the cells to freely “occupy” one of possible independent states in a statistical mechanics formulation. A cell state could be defined either genotypically or phenotypically, keeping in mind that a cell’s phenotype can be heavily influenced by neighboring cells, microenviromental factors, or even longer-range coupling among cells. The combination of all of these various local cellular influences leads to an emergent volumetric growth law. In the early stages of progression (from a single mutated cell to a small collection of rapidly proliferating cells), the cell population is relatively unconstrained, gradually becoming more and more constrained over time due to extrinsic factors (e.g., fibroblasts, immune cells, blood vessels, and nutritional landscapes) as well as intrinsic factors (e.g., necrosis, DNA mismatch repair, chromosomal instability). The collection of all of these extrinsic and intrinsic factors is represented by a constraining function , where is the tumor cell population. This function dictates how the constraints change over time as the tumor grows [generally, is an increasing function of ]. While locally, some cells will surely have increased capacity for diversity (i.e., if they acquire chromosomal instability), or a decreased capacity for diversity (i.e., necrosis), our theory focuses on the overall macroscopic trend. This potential reduction in the number of independent states is then expressed as the ratio , which we call the “effective” number of states. The statistical mechanics formulation here uses this ratio as the fundamental microscopic (cell state) variable. We then lay out very precisely how our choice of the functional coupling denominator affects the macroscopic growth law of the tumor. Particularly relevant is the fact that while the exact cell coupling of a population may not be easily measurable, knowing the approximate macroscopic growth characteristic of a tumor (e.g., by time imaging) gives indirect insight into the functional coupling of the cell population throughout the tumor’s growth history.
1. Heterogeneity, Diversity, and Growth
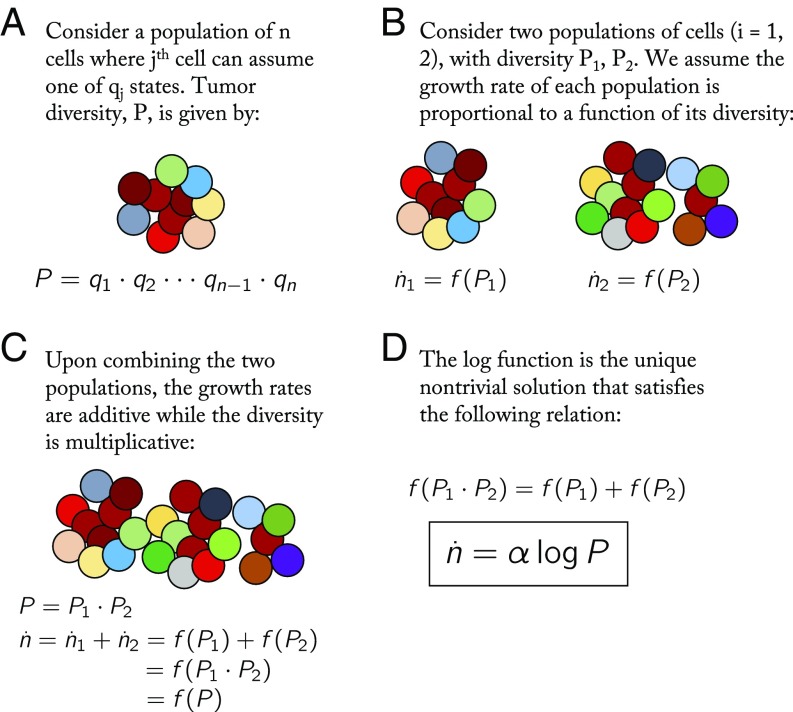
Genetic instability, a hallmark of cancer, is generally believed to be acquired early in tumorigenesis and thought to lead in a multistep fashion to other cancer hallmarks (24–26). This instability can be thought of as increasing the potential of diversity within a tumor, leading to a large number of potential genetic or morphological “states” each cell can occupy, which in turn gives rise to a combinatorial mushrooming of overall molecular/cellular configuration of the tumor. This makes a statistical mechanics approach to cancer modeling an attractive option. Kendal (27) introduced such a model based on these types of considerations, which we take as a point of departure for our work, so it is useful to first review the main features of his simple argument. Consider a population of cells where the th cell has the potential to assume one of possible states, . The number of combinations of states possible within the population is given as , a measure of diversity potential of the tumor:
| [1] |
which can be related to the growth rate of the tumor. By introducing a function, , that is a function of the number of proliferating cells, and using the requirement that for any two subpopulations and (additivity of ), we arrive at the requirement that is logarithmic (see ref. 23 for a proof of this fact):
| [2] |
where is the natural logarithm. Our basic assumption is that growth is proportional to :
| [3] |
The linkage between growth rate and measures of tumor heterogeneity is consistent with the fact that morphologic heterogeneity forms the basis of many tumor-grading classification systems (15) and is implicitly discussed in the classic paper of Nowell (3), who considered the commonly observed increased tumor aggressiveness during the natural history of solid tumor growth. It is most recently thoroughly discussed in refs. 4 and 28, and a general discussion of some of the consequences of heterogeneity can be found in ref. 13. The basic assumptions of the model are highlighted in Fig. 1. Our point of departure from Kendal’s approach (27) is to represent the number of possible states, more generally, as the ratio:
| [4] |
to allow for functional coupling as the cell population changes. Here, represents the number of uncoupled (free) states available to each cell, whereas [4] allows for the possibility that functional coupling reduces the number of effective states () the cell can occupy.
Fig. 1.
A schematic showing the key assumptions and conclusions of the model. (A) A population of n cells: colors represent current genetic or phenotypic state assumed. Each th cell can assume states. Total tumor diversity, , is the product of each cell’s diversity potential. (B) Consider two such populations of and cells growing independently. Diversity potential is given by and , respectively. (C) Considering the same two populations in combination, the growth rates must be additive, while the diversity potential is multiplicative, as shown. (D) The log function satisfies this functional requirement given in C (see ref. 23 for proofs).
To compare with Kendal’s original formulation (27), if we begin with the simplest completely uncoupled case, where we let , and (typically ) for each , the combination of states is calculated from [1] as:
| [5] |
With this, we have the growth equation:
| [6] |
which yields exponential growth:
| [7] |
with growth rate proportional to the logarithm of the number of states available to each cell. We assume that , and that tumor volume , in general, is proportional to . Therefore, with no coupling among the cells to reduce the effective number of achievable states (degrees of freedom), tumor growth is effectively exponential. This is often the case in the very earliest stages of tumor growth before the cells have coordinated, or “synced,” via any long-range communication mechanisms (discussed in section 6). To make further contact with existing literature, note that the standard exponential growth equation is simply written:
| [8] |
with , where is the doubling time of the cell population. Comparing [6] and [8], we see that:
| [9] |
or that . As the number of available states increases, the doubling time decreases (i.e., faster growth), and the two are log-related.
By contrast, suppose that the number of states each cell can achieve is reduced as the total cell population increases, due to functional coupling of the cell population. Instead of each cell acting independently as previously assumed, we now let :
| [10] |
As increases (i.e., as the tumor grows), the number of states each cell can achieve decreases linearly with . With this assumption, we have diversity as:
| [11] |
This gives rise to the growth equation:
| [12] |
whose solution is the Gompertzian function:
| [13] |
one of the most widely applied macroscopic tumor growth laws (29–33). The functional coupling of the cell population effectively reduces the number of achievable states, which will increase the doubling time (i.e., slower growth) of the tumor. These two known examples show very clearly how our choice of the functional coupling, , dictates the emergent volumetric growth equation. This can be made much more general by linking essentially all widely used tumor growth laws with specific choices of the function coupling as shown next.
2. Functional Cellular Constraints and Volumetric Growth
As the tumor grows, cell signaling and microenvironmental factors act to couple many functional aspects of the individual cells (much the way the uncoupled degrees of freedom in a mechanical system can sync, thereby reducing the effective degrees of freedom of the system as a whole). The two previous examples, (leading to exponential growth) and (leading to Gompertzian growth), are special cases. Generally speaking, as the tumor grows, the functional coupling should increase (as in the Gompertzian case); hence, should be an increasing function of . With no loss of generality, we can write as the exponential of another function (to clean up the final equation):
| [14] |
giving rise to the following relation:
| [15] |
At this point, the functional form of is unspecified, but the denominator serves to restrict the total number of “free states” a single cell can occupy. Using [1], the growth Eq. 2 then becomes:
| [16] |
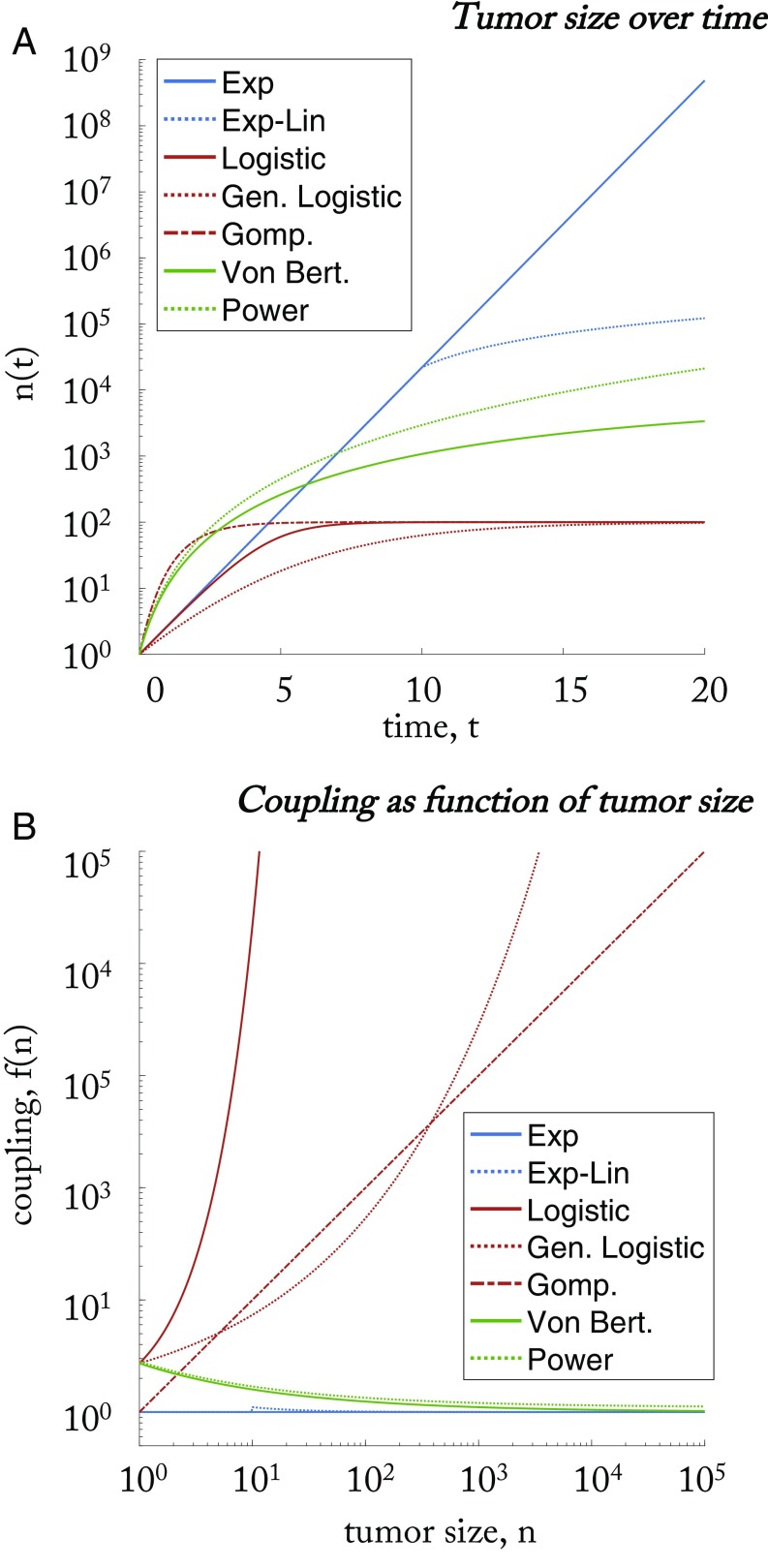
where , . The functional form determines which of the many common macroscopic growth models are in force (exponential, exponential-linear, logistic, generalized logistic, Gompertz, Von Bertanffly, and power law). An overview of results are shown in Table 1. An example commentary on the use and history of common growth models in describing tumor dynamics can be found in the report by Benzekry et al. (29).
Table 1.
Overview of growth model parameters
| Category | Model | |||
| Exponential | Exp. | 0 | 0 | |
| Exp.-linear | ||||
| Sigmoidal | Logistic | |||
| Gen. logistic | ||||
| Gomp. | ||||
| Power | Von Bert. | |||
| Power law | 0 |
Exp., exponential; Gen., generalized; Gomp., Gompertz; Von Bert., Von Bertalanffy.
Broadly, the macroscopic growth models described below can be binned into three categories: (i) exponential, (ii) sigmoidal, and (iii) power law models. Exponential models are characterized by a long period of constant proliferation cell cycle time, while sigmoidal models have eventual slowed growth until an eventual plateau. A power law model also gives rise to a similarly slowed growth, but without a specified plateau. Each of these categories is associated with a certain functional coupling: approximately constant (exponential), increasing (sigmoidal), or decreasing (power). Explicit solutions for each of the growth models are detailed below, along with a discussion on the implications of the functional form of coupling (i.e., the denominator of Eq. 15).
3. Exponential Models
A simple model of tumor growth assumes a constant cell cycle time for all proliferating tumor cells, , which leads to exponential growth (Eqs. 17 and 18).
| [17] |
| [18] |
An extension of the exponential model assumes an initial exponential phase is followed by linear tumor growth (Eq. 19), first introduced here (34):
| [19] |
The exponential-linear model is solved explicitly below.
| [20] |
For the first phase of growth (), the coefficient is equivalent to , where is the constant or exponentially distributed mean cell cycle time of the proliferative fraction within the tumor. Under the assumption of a continuously differentiable solution to Eq. 20, is uniquely determined as: , where is the initial tumor volume (29, 34).
The exponential models are shown in Fig. 2A in blue (). As explained above, a tumor grows from a single cell, and the total number of free states, m, a single cell can occupy is restricted by the exponential of a function form of (see Eq. 15). The functional coupling, , determines how the coupling changes with an increasing tumor size, . Exponential models are characterized by weak, constant coupling [ (exponential) or (exponential-linear, for large )].
Fig. 2.
Representative simulations of classical macroscopic models of tumor growth, shown here for . (A) Tumor size, , over time. Exponential models are shown in blue (). Sigmoidal models are shown in red (). Power law models are shown in green (). (B) The coupling function, , between cell states for identical parameters as A. There are three general classes of macroscopic growth models: exponential models (blue) characterized by (approximately) constant coupling; sigmoidal models (red) characterized by coupling that increases with tumor size; and power law models (green) characterized by coupling that decreases with tumor size. Exp, exponential; Exp-Lin, exponential-linear; Gen. Logistic, generalized logistic; Gomp., Gompertz; Power, power law; Von Bert., Von Bertalanffy.
4. Sigmoidal Growth Models
The logistic (Eq. 21), generalized logistic (Eq. 23), and Gompertz (Eq. 26) equations are all in a general class of equations that quantify tumor growth in a sigmoidal shape, where growth is slowed with increasing tumor size (29–31, 35).
Logistic growth (Eq. 21) is characterized by a linear decrease of relative growth rate and is often interpreted as a competition between proliferating tumor cells for space or nutrients. Logistic growth models have been used by many to describe tumor dynamics (e.g., refs. 31 and 36).
| [21] |
| [22] |
where is the carrying capacity of the tumor. The logistic model can also be written as a generalized logistic equation, below:
| [23] |
which is more conveniently written:
| [24] |
The explicit solution to the generalized logistic equation (Eq. 25) gives rise to the logistic model (Eq. 21) when (29).
| [25] |
The generalized logistic model also converges to Gompertz model (see Eq. 26, below) when (29). First introduced to describe human population growth (32), the Gompertz equation has also been used extensively in modeling tumor growth (30, 33, 37). Gompertzian growth is exponential growth with decaying growth rate:
| [26] |
This is alternatively written:
| [27] |
where . It is explicitly solved,
| [28] |
The sigmoidal models are shown in Fig. 2A in red (). The sigmoidal models are characterized by increased coupling (again, see denominator of Eq. 15) as the tumor size increases. This is shown in red in Fig. 2B and given by , which is a decreasing function of in Table 1. The functional coupling, , for all sigmoidal functions is always an increasing function of for values of .
5. Power Law Models
Another class of growth models, the power law model, has been used to derive general laws of tumor dynamics from a relationship between growth and metabolism (38, 39). Metabolic rates within the tumor often scale with a power of the total tumor size (29, 38). The Von Bertalanffy equation is written in Eq. 29, where the power law model is a special case, derived by neglecting the loss term (). Note: some often identify this model as the specific case , termed “second type growth” (29). Both the Von Bertalanffy model and the power law special case have been used to fit tumor spheroid data for murine models (40, 41) and breast cancer mammography screening data (42):
| [29] |
This model can be solved and written explicitly:
| [30] |
The power law models are shown in Fig. 2A in green (). These models are characterized by decreased coupling as the tumor size increases. This is shown in green in Fig. 2B and given by , which is a decreasing function of in Table 1. Generally, the Von Bertalanffy model is associated with second type growth, with , and the functional coupling, , for all power functions is always decreasing for values of . We now discuss the implications of the functional coupling for each category of model.
6. Underlying Biological Mechanisms
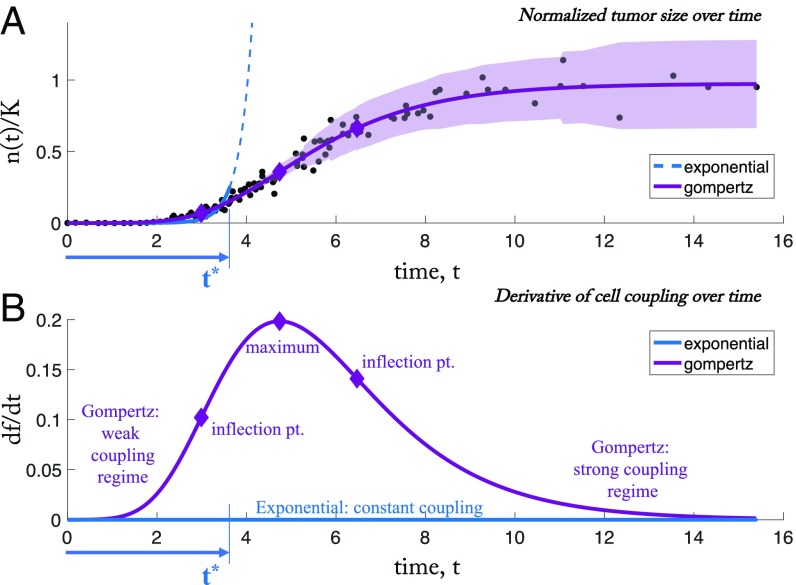
It is useful to focus on the changes in cell–cell coupling throughout the entire life history of the tumor and to discuss the biological mechanisms that give rise to these changes. To do so, we reexamine classic data reproduced from ref. 30 showing tumor growth data from 19 samples of 12 different tumors of the rat, mouse, and rabbit superimposed and normalized appropriately so that the full range of growth can be tracked from a single cell to the full carrying capacity of a tumor. We replot the data in Fig. 3A with best fits from two of the most common classical models: exponential and Gompertz. The cellular constraints (Eq. 14) implied by model fits of data from the full range of tumor initiation (a single cell) to tumor saturation (carrying capacity) give insight into how the constraints shape the tumor growth through its time history. To view how changes in the constraints affect the growth law of the tumor over time, the derivative of the function () is shown in Fig. 3. In the early stages of growth, the data are fit reasonably well by an exponential model, until time (measured by a divergence in the mean-squared error). This breakdown occurs roughly at the first inflection point of the curve plotted in Fig. 3B. As the tumor grows further, the Gompertz function predicts an increase in the effect of the constraints (), reaching a maximum at the inflection point in Fig. 3A, corresponding to the maximum point in Fig. 3B. Nonetheless, the cell–cell coupling is continually increasing over time until saturation.
Fig. 3.
Exponential and Gompertz fit of aggregate tumor data. (A) Data reproduced from ref. 30 showing growth data for 19 samples of 12 different tumors of the rat, mouse, and rabbit superimposed by scaling according to the inflection point of the growth curve (more details are available in ref. 30). The units are in days (x axis) and decimal fraction of the asymptotic tumor size, K (y axis). Residual error is shown for each data point in relation to Gompertz fit (light purple shading). An exponential curve is shown (dashed blue line) fit from time 0 to , where the mean-squared error () of that fit diverges from the of the Gompertzian fit: − 0.001 (fit parameters: ; ; ). The exponential model is a good fit only during short time intervals, which corresponds to a constant cell–cell coupling. (B) The cell–cell coupling (Eq. 14) implied by model fits of data from the full range of tumor initiation (a single cell) to tumor saturation (carrying capacity) give insight into how cell–cell coupling shapes the tumor growth. In early times, both Gompertz and exponential models give good fits in A. As the tumor growth slows, the Gompertz model predicts an increase in cell–cell coupling (), reaching a maximum rate of change at the tumor’s inflection point (inflection pt.) in A.
There are two distinct biological mechanisms that are at play during this history that are most likely responsible for these changes in coupling behavior among the tumor cells. In the early stages, the short-range communication between cells is mediated by cell gap junctions (43), which are known to be quite permeable to a wide size range of molecules and can provide a direct path for the flow of these molecules between cell interiors. A review of how this exchange of molecular information between nearby cells can control cell division is discussed in ref. 43. A failure in junctional communication is thus implicated as a key biological mechanism responsible for a transition in volumetric growth of the tumor near the time marked in our Fig. 3. As the tumor matures (past time ), longer-range intercellular communication begins to affect volumetric growth. It is widely hypothesized, with mounting evidence, that this type of longer-range intercellular communication is controlled not only by soluble factors, such as cytokines, chemokines, growth factors, and neurotransmitters, but also via microRNAs, which are the direct mechanisms by which genetic information can be exchanged. See ref. 44 for a comprehensive recent review. In fact, because of their ubiquity and stability in the bloodstream of a wide range of cancers, circulating microRNAs are being developed as a blood-based biomarker for cancer detection (45). More generally, a broad range of circulating microparticles as the key protagonists underlying a vast long-range communication network for intercellular information exchange seems increasingly implicated in a wide range of diseases from inflammatory and autoimmune diseases, as well as atherosclerosis. See ref. 46 for a comprehensive review. As the tumor progresses to carrying capacity, the biological mechanisms of long-range coupling have had sufficient chance to globally couple the tumor cells in the regime we label the strongly coupled (Gompertz) regime.
7. Therapeutic Implications
Traditional therapeutic treatments target the cancer cells directly by surgical removal or maximal eradication (chemotherapy and radiation). The linkage between cellular coupling and tumor growth leads naturally to the idea of therapeutic methods to try to disrupt or enhance the functional coupling of the cellular states to control tumor growth characteristics. This idea has been touched upon with the suggestion of multitargeted therapeutic approaches disrupting or coopting ecological interactions of tumor–host or tumor–tumor cell interactions. These approaches have been termed “ecological therapies” (20). The model developed here provides a framework for determining how those interactions guide volumetric growth, which may lead to models that optimize timing of new therapies targeting cell–cell interactions, by targeting the mediators of those interactions.
In general terms, the growth law that a tumor follows has definitive therapeutic implications. Cancers that follow exponential growth laws can be assumed to have constant cell–cell interactions (or lack thereof, in the case of blood cancers), and targeted therapies may have more promise in these scenarios, while the therapies aimed at limiting cell–cell interactions described previously will likely have little effect. Likewise, growth models with decreased coupling over time (green curves in Fig. 2B) will benefit from ecological therapies early in tumor development but not as tumor sizes increase to clinical relevant sizes. Otherwise, growth laws associated with increased coupling (red curves in Fig. 2B) show promise with respect to therapies targeting interactions.
Some have described the therapeutic implications of tumors viewed as ecosystems (47), characterized by competing subpopulations, their spatial and temporal assortment, and their interactions with their physical and chemical microenvironments (20). Much research has been done in the area of collateral sensitivity, to determine if there is pharmacological interaction (additivity or synergy) of multiple drugs in combination or sequence (see refs. 48–50), showing that drugs may have complex downstream effects on cell–cell interactions. Communication or feedback between tumor cells may provide negative (competition, predation, amensalism, parasitism) or positive (commensalism, synergism, mutualism) (19, 20) growth. Organisms compete for limited resources and cooperate for mutual advantage with interactions fluctuating with resource consumption or cell turnover. Additionally, tumor cell interactions may also be dependent on benefits derived from noncancer cells (endothelial cells, cancer-associated fibroblasts, and tumor-associated macrophages) (19, 20). Targeting these noncancer cells, from which the cancer cells are receiving benefit, should also provide therapeutic benefits during the process by which a tumor transitions from a closed system (primary tumor) to an open system of circulating cells to distant colonies (metastatic cancers). But a clear understanding of the relationship between functional coupling among cells and volumetric tumor growth is a necessary step in the direction of exploiting these connections.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heppner GH, Miller BE. Tumor heterogeneity: Biological implications and therapeutic consequences. Cancer Metastasis Rev. 1983;2:5–23. doi: 10.1007/BF00046903. [DOI] [PubMed] [Google Scholar]
- 3.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 4.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: Implications for targeted therapeutics. Br J Cancer. 2013;108:479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 6.Aguilera A, Gómez-González B. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 7.Turner JS. Homeostasis and the physiological dimension of niche construction theory in ecology and evolution. Evol Ecol. 2016;30:203–219. [Google Scholar]
- 8.Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 9.Liotta LA, Kohn EC. The microenvironment of the tumour–host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 10.Friedl P, Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Anderson ARA, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 12.Huang S. Genetic and non-genetic instability in tumor progression: Link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev. 2013;32:423–448. doi: 10.1007/s10555-013-9435-7. [DOI] [PubMed] [Google Scholar]
- 13.Marusyk A, Polyak K. Tumor heterogeneity: Causes and consequences. Biochim Biophys Acta (BBA)-Rev Cancer. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West J, Hasnain Z, Macklin P, Newton PK. An evolutionary model of tumor cell kinetics and the emergence of molecular heterogeneity driving gompertzian growth. SIAM Rev. 2016;58:716–736. doi: 10.1137/15M1044825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanton C. Intratumor heterogeneity: Evolution through space and time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: Seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 17.Potter VR. Biochemistry of cancer. In: Holland J, Freieds E, editors. Cancer Medicine. Lea and Febiger; Philadelphia: 1973. pp. 178–190. [Google Scholar]
- 18.Trosko JE, Ruch RJ. Cell-cell communication in carcinogenesis. Front Biosci. 1998;3:d208–236. doi: 10.2741/a275. [DOI] [PubMed] [Google Scholar]
- 19.Tabassum DP, Polyak K. Tumorigenesis: It takes a village. Nat Rev Cancer. 2015;15:473–483. doi: 10.1038/nrc3971. [DOI] [PubMed] [Google Scholar]
- 20.Pienta KJ, McGregor N, Axelrod R, Axelrod DE. Ecological therapy for cancer: Defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Transl Oncol. 2008;1:158–164. doi: 10.1593/tlo.08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trosko JE, Chang CC, Madhukar BV, Klaunig JE. Chemical, oncogene and growth factor inhibition of gap junctional intercellular communication: An integrative hypothesis of carcinogenesis. Pathobiology. 1990;58:265–278. doi: 10.1159/000163596. [DOI] [PubMed] [Google Scholar]
- 22.Trosko JE, Madhukar BV, Chang CC. Endogenous and exogenous modulation of gap junctional intercellular communication: Toxicological and pharmacological implications. Life Sci. 1993;53:1–19. doi: 10.1016/0024-3205(93)90606-4. [DOI] [PubMed] [Google Scholar]
- 23.D’Angelo JP, West DB. 2017. Mathematical Thinking: Problem Solving and Proofs, Pearson Modern Classics Series (Pearson, London), 2nd Ed, Chap 17.
- 24.Venkatesan S, Swanton C. Tumor evolutionary principles: How intratumor heterogeneity influences cancer treatment and outcome. Am Soc Clin Oncol Educ Book. 2016;35:e141–e149. doi: 10.1200/EDBK_158930. [DOI] [PubMed] [Google Scholar]
- 25.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—An evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 26.Douglas H, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Kendal WS. Gompertzian growth as a consequences of tumor heterogeneity. Math Biosci. 1985;73:103–107. [Google Scholar]
- 28.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nat Rev. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 29.Benzekry S, et al. Classical mathematical models for description and prediction of experimental tumor growth. PLoS Comput Biol. 2014;10:e1003800. doi: 10.1371/journal.pcbi.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laird AK. Dynamics of tumour growth: Comparison of growth rates and extrapolation of growth curve to one cell. Br J Cancer. 1965;19:278–291. doi: 10.1038/bjc.1965.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spratt JA, Fournier DV, Spratt JS, Weber EE. Decelerating growth and human breast cancer. Cancer. 1993;71:2013–2019. doi: 10.1002/1097-0142(19930315)71:6<2013::aid-cncr2820710615>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 32.Gompertz B. On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Philos Trans R Soc Lond. 1825;115:513–583. doi: 10.1098/rstb.2014.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norton L. A gompertzian model of human breast cancer growth. Cancer Res. 1988;48:7067–7071. [PubMed] [Google Scholar]
- 34.Simeoni M, et al. Predictive pharmacokinetic-pharmacodynamic modeling of tumor growth kinetics in xenograft models after administration of anticancer agents. Cancer Res. 2004;64:1094–1101. doi: 10.1158/0008-5472.can-03-2524. [DOI] [PubMed] [Google Scholar]
- 35.Steel GG, Lamerton LF. The growth rate of human tumours. Br J Cancer. 1966;20:74–86. doi: 10.1038/bjc.1966.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaidya VG, Alexandro FJ. Evaluation of some mathematical models for tumor growth. Int J Bio-Med Comput. 1982;13:19–35. doi: 10.1016/0020-7101(82)90048-4. [DOI] [PubMed] [Google Scholar]
- 37.Casey AE. The experimental alteration of malignancy with an homologous mammalian tumor material: I. Results with intratesticular inoculation. Am J Cancer. 1934;21:760–775. [Google Scholar]
- 38.Von Bertalanffy L. Quantitative laws in metabolism and growth. Q Rev Biol. 1957;32:217–231. doi: 10.1086/401873. [DOI] [PubMed] [Google Scholar]
- 39.West GB, Brown JH, Enquist BJ. A general model for ontogenetic growth. Nature. 2001;413:628–631. doi: 10.1038/35098076. [DOI] [PubMed] [Google Scholar]
- 40.Marušić M, Bajzer Ž, Freyer JP, Vuk-Pavlović S. Analysis of growth of multicellular tumour spheroids by mathematical models. Cell Proliferation. 1994;27:73–94. doi: 10.1111/j.1365-2184.1994.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 41.Marušić M, Vuk-Pavlovic S, Freyer JP. Tumor growth in vivo and as multicellular spheroids compared by mathematical models. Bull Math Biol. 1994;56:617–631. doi: 10.1007/BF02460714. [DOI] [PubMed] [Google Scholar]
- 42.Hart D, Shochat E, Agur Z. The growth law of primary breast cancer as inferred from mammography screening trials data. Br J Cancer. 1998;78:382–387. doi: 10.1038/bjc.1998.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loewenstein WR. Junctional intercellular communication and the control of growth. Biochim Biophys Acta. 1979;560:1–65. doi: 10.1016/0304-419x(79)90002-7. [DOI] [PubMed] [Google Scholar]
- 44.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: Diverse structures for exchange of genetic information. Nat Perspect. 2012;13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mause SF, Weber C. Microparticles: Protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010;107:1047–1057. doi: 10.1161/CIRCRESAHA.110.226456. [DOI] [PubMed] [Google Scholar]
- 47.Willis AJ. The ecosystem: An evolving concept viewed historically. Funct Ecol. 1997;11:268–271. [Google Scholar]
- 48.Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell. 2017;171:1678–1691. doi: 10.1016/j.cell.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doroshow JH, Simon RM. On the design of combination cancer therapy. Cell. 2017;171:1476–1478. doi: 10.1016/j.cell.2017.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basanta D, Gatenby RA, Anderson ARA. Exploiting evolution to treat drug resistance: Combination therapy and the double bind. Mol Pharm. 2012;9:914–921. doi: 10.1021/mp200458e. [DOI] [PMC free article] [PubMed] [Google Scholar]