OBJECTIVES:
Molecular prognostic biomarkers for gastric cancer (GC) are still limited. We aimed to identify potential messenger RNAs (mRNAs) associated with GC prognosis and further establish an mRNA signature to predict the survival of GC based on the publicly accessible databases.
METHODS:
Discovery of potential mRNAs associated with GC survival was undertaken for 441 patients with GC based on the Cancer Genome Atlas (TCGA), with information on clinical characteristics and vital status. Gene ontology functional enrichment analysis and pathway enrichment analysis were conducted to interrogate the possible biological functions. We narrowed down the list of mRNAs for validation study based on a significance level of 1.00 × 10−4, also integrating the information from the methylation analysis and constructing the protein–protein interaction network for elucidating biological processes. A total of 54 mRNAs were further studied in the validation stage, using the Gene Expression Omnibus (GEO) database (GSE84437, n = 433). The validated mRNAs were used to construct a risk score model predicting the prognosis of GC.
RESULTS:
A total of 13 mRNAs were significantly associated with survival of GC, after the validation stage, including DCLK1, FLRT2, MCC, PRICKLE1, RIMS1, SLC25A15, SLCO2A1, CDO1, GHR, CD109, SELP, UPK1B, and CD36. Except CD36, DCLK1, and SLCO2A1, other mRNAs are newly reported to be associated with GC survival. The 13 mRNA-based risk score had good performance on distinguishing GC prognosis, with a higher score indicating worse survival in both TCGA and GEO datasets.
CONCLUSIONS:
We established a 13-mRNA signature to potentially predict the prognosis of patients with GC, which might be useful in clinical practice for informing patient stratification.
INTRODUCTION
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of cancer death worldwide (1). It is imperative to identify patients with GC at a high risk for poor prognosis, so that timely and appropriate treatment strategy can be applied. Although a considerable proportion of patients are diagnosed at an advanced stage, patient outcomes can vary significantly even for patients with similar clinical features. The host heterogeneity in somatic or germline changes may have led to different prognosis.
Several studies have examined both genetic and epigenetic alterations as potential prognostic factors for GC. Alternative patterns of messenger RNA (mRNA) expression involved in cycle regulation (2), cell adhesion (3), angiogenesis (4), and tumor carcinogenesis (5) have been reported to play an important role in forecasting survival outcome of GC. One study examined the gene expression profile of 65 patients with GC based on DNA microarray data and reported 6 mRNAs associated with GC prognosis (6). Another study reported 3 mRNAs associated with GC survival based on DNA microarray data of 21 patients with GC (7). A recent study reported 25 mRNAs associated with GC prognosis using DNA microarray data of 78 patients with GC (8). These studies were limited by either small sample size or lacking suitable validation datasets, restricting the possible application of the reported mRNAs associated with GC prognosis in clinical practices. Therefore, it is imperative to identify novel mRNA signatures robustly associated with GC survival.
The booming development in high-throughput transcriptome sequencing and microarray technologies have provided opportunities to establish mRNA signatures associated with GC survival. In the present study, we took advantage of the publicly available databases to establish novel mRNA signatures, which may predict GC prognosis. Discovery of potential mRNAs was conducted among 441 patients with GC based on the Cancer Genome Atlas (TCGA), with information on clinical characteristics and vital status. Gene ontology functional enrichment analysis and pathway enrichment analysis were conducted to interrogate the possible biological functions. The independent external microarray data from the Gene Expression Omnibus (GEO) was used to validate these candidate mRNAs. A risk score model was then constructed using the validated mRNAs to testify the potential predictive value of integrated mRNA signature for GC prognosis.
METHODS
Screening mRNAs associated with GC survival in the discovery stage
All data in the TCGA-STAD (stomach adenocarcinoma) training set, including clinical information, mRNA expression data, and DNA methylation profile, were downloaded from TCGA data portal (9). We filtered the patients with missing survival information and obtained the data for 441 patients with GC who had intact follow-up clinical information. Other clinical factors, including age, gender, race, TNM (tumor, lymph nodes and metastasis) stage, family history of GC, Helicobacter pylori infection, targeted molecular therapy, and radiation therapy, were also obtained. Clinical stage involved in our study was determined according to the newly released 8th AJCC staging system (10). mRNA expression data (RNA-Seq) was extracted from Illumina RNA-Seq HiSeq platform. We performed quality control by removing mRNAs that have more than half of values as zero. As many as 19,754 mRNAs remained with quantile normalization in the R package DESeq (11).
Cox regression models were used to calculate the hazard ratios and 95% confidence intervals for the associations between each examined mRNA and GC death. Proportional assumptions for Cox proportional hazard model were tested and no violations of the assumptions were found. The primary analyses were conducted adjusting for age and gender. In a secondary model, we additionally adjusted for race, family history of GC, H. pylori infection, targeted molecular therapy, radiation therapy, and clinical stage in the Cox regression model, in addition to age and gender.
Gene ontology functional enrichment analysis
Gene ontology functional enrichment analysis based on co-occurrences with sets of genes associated with GC deaths could rapidly unravel potential cellular component, biological process, and molecular functions. To explore the encoding gene ontology for the mRNAs identified with association P values <5 × 10−3 in the discovery stage (see Table S1, Supplemental Digital Content 1, http://links.lww.com/CTG/A1), we described gene function, and relationship between these concepts based on an online website DAVID 6.8 (Database for Annotation, Visualization and Integrated Discovery, https://david.ncifcrf.gov/) (12).
Pathway enrichment analysis
Pathway enrichment analysis can help gain mechanistic insights into gene lists by identifying biological pathways that are enriched in gene lists more than simply expected by chance. To explore distinct pathways that might influence GC survival outcome, an online website KOBAS (http://kobas.cbi.pku.edu.cn/) was utilized to perform pathway enrichment analysis (13). P < 0.05 was set as the cut-off value.
Integrating prognostic mRNAs with methylation profiles
For the mRNAs significantly associated with GC survival at P < 5 × 10−3 in the discovery stage (see Table S1, Supplemental Digital Content 1, http://links.lww.com/CTG/A1), we tested whether the CpG (5′-C-phosphate-G-3′) loci located in the encoding genes were associated with GC survival and also the association between methylation and mRNA expression. In the TCGA-STAD database, DNA methylation profiles were detected using the Illumina HumanMethylation450 BeadChip, which covers 96% of CpG sites defined by the University of California, Santa Cruz. Prior probe selection was performed by removing those containing missing values, those targeting the sex chromosomes, those containing single-nucleotide polymorphism (SNP), and those not annotated with any protein-coding or non–protein-coding genes. Finally, 230,495 CpG loci were retained from the original 485,577 sites. Cox regression models were used to identify CpG loci associated with GC survival at P < 0.05, adjusting for age and gender. Spearman correlation analysis was utilized to test the associations between CpG loci and the corresponding genes mRNA expression. Correlation P < 0.05 and the absolute value of correlation coefficient >0.4 were set as the cut-off criteria defining meaningful methylation-expression associations (see Table S2, Supplemental Digital Content 1, http://links.lww.com/CTG/A1).
Construction of protein–protein interaction network and subnetwork analysis
We explored the potential protein–protein interaction (PPI) for the encoding genes of the identified mRNAs with association P < 5 × 10−3 in the discovery stage (see Table S1, Supplemental Digital Content 1, http://links.lww.com/CTG/A1). We utilized the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, https://string-db.org/) (14) database to predict protein–protein association and subsequently visualized the result by Cytoscape software (version: 3.6.1, http://www.cytoscape.org/). The plugin software ClusterONE (version: 1.0, clustering with overlapping neighborhood expansion) based on the concept of the cohesiveness score and a greedy growth process was applied to further detect significant models, which may represent molecular complexes (15) (minimum size = 5, minimum density = 0.6, and edge weights = unweighted).
Selection of key candidate mRNAs for the validation
To select key candidate mRNAs for the validation stage, we further narrowed down the list of mRNAs that we obtained in TCGA based on a significance level of 1.00 × 10−4, also integrating the information from the methylation analysis and constructing the PPI network for elucidating biological processes. In detail, we also included those mRNAs whose encoding gene methylation was associated with GC survival, as well as genes in clustered subnetworks within PPI network. A total of 54 mRNAs were included in the validation stage (see Table S3, Supplemental Digital Content 1, http://links.lww.com/CTG/A1).
The validation dataset of GC, including the transcription profile based on GPL6947 platform (Illumina HumanHT-12 V3.0 expression BeadChip) and clinical information, were obtained from GSE84437 in GEO (https://www.ncbi.nlm.nih.gov/geo/), followed by background correction and quantile normalization using R package lumi (16). All mRNA expression values were log2-transformed and standardized for comparability with the discovery set as above described.
The association analyses in the validation study were also conducted using Cox regression models, adjusting for age and gender, with one-side P < 0.05 considered as statistically significant.
Construction and validation of risk score model
The risk score model consisted of mRNAs that were successfully validated in GSE84437, based on a function
Regression coefficient (β) of every mRNA derived from the multivariate Cox proportional hazard analysis in GSE84437 was used as β score of the risk score model. X represents standardized expression value of each mRNA.
We then performed the risk score model in TCGA-STAD and GSE84437, respectively, and calculated a risk score for each patient. The risk scores were categorized into tertiles. Survival curves were drawn using Kaplan–Meier method and GC death by tertiles of risk scores were compared among subgroups using log-rank tests in both TCGA-STAD and GSE84437, respectively.
RESULTS
Discovery of potential mRNAs in the discovery stage based on TCGA
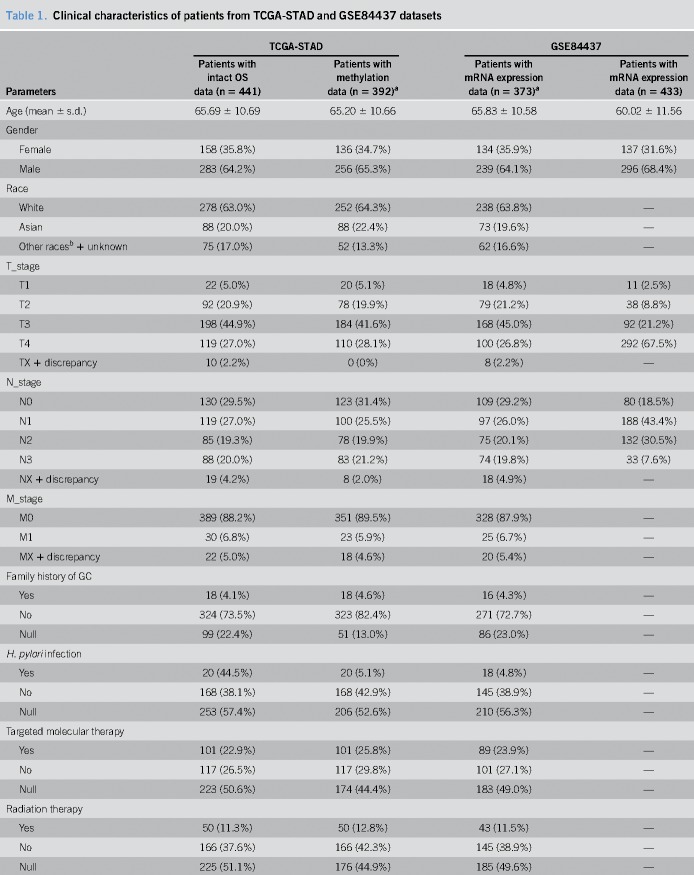
The characteristics of TCGA-STAD and GSE84437 datasets are presented in Table 1. A total of 184 mRNAs were identified at P < 5 × 10−3 associated with GC poor survival, out of 19,754 mRNAs (see Table S1, Supplemental Digital Content 1, http://links.lww.com/CTG/A1).
Table 1.
Clinical characteristics of patients from TCGA-STAD and GSE84437 datasets
Gene ontology functional enrichment analysis
To investigate the encoding gene ontology for the mRNAs identified with association P values < 5 × 10−3 in the discovery stage (see Table S1, Supplemental Digital Content 1, http://links.lww.com/CTG/A1), GO analysis was performed using online biological tool DAVID. Twelve GO terms were obtained (Figure 1a and see Table S4, Supplemental Digital Content 1, http://links.lww.com/CTG/A1). Most genes were enriched in the plasma membrane cellular component.
Figure 1.
Gene functional and pathway enrichment analysis of the 184 genes, which encode messenger RNAs significantly associated with gastric cancer death at P < 5 × 10−3 in discovery stage. P < 0.05 was considered as threshold values of significant difference in enrichment analysis. (a) Significantly enriched GO terms of the 184 genes using the online tool DAVID. (b) Significantly enriched pathways of the 184 genes using the online tool KOBAS 3.0. Four databases were utilized for analyses, including “KEGG pathway,” “Reactome,” “BioCyc,” and “PANTHER.”
Pathway enrichment analysis
A total of 4 databases, including “KEGG pathway,” “Reactome,” “BioCyc,” and “PANTHER,” were applied in pathway enrichment analysis. Top 20 pathways were shown in Figure 1b and Table S5 (Supplemental Digital Content 1, http://links.lww.com/CTG/A1). The result indicated that encoding genes of mRNAs significantly associated with GC survival in TCGA may be mainly involved in signaling transduction pathways.
Construction of PPI network and subnetwork analysis
To achieve a better understanding of the biological processes in GC survival, we adopted online database STRING (https://string-db.org/) and visualization software Cytoscape (version: 3.6.1, http://www.cytoscape.org/) to construct PPI network. Based on the criterion (minimum required interaction score = 0.4), a total of 66 genes associated with GC survival were filtered into the PPI network. It comprised 66 nodes and 58 edges (Figure 2a). Furthermore, a key subnetwork model, which could represent the overlapping protein complexes in the PPI network, was identified through the plugin software ClusterONE, containing VMF, SELP, CD36, CD44, and CD109 (Figure 2b).
Figure 2.
Protein-protein interaction network and subnetwork analysis. (a) Protein–protein interaction network constructed by the 184 genes, which encode messenger RNAs significantly associated with gastric cancer death at P < 5 × 10−3 in the discovery set. (b) The key subnetwork module extracted from the protein–protein interaction network through the plugin software ClusterONE (minimum size = 5, minimum density = 0.6, and edge weights = unweighted).
Selection of key candidate mRNAs for the validation study
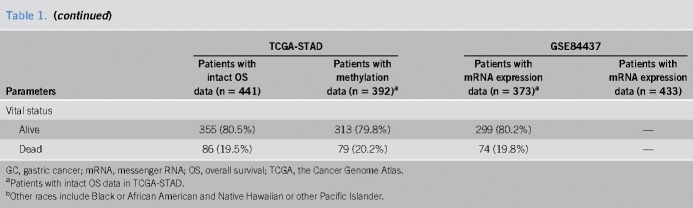
A total of 54 mRNAs (see Table S3, Supplemental Digital Content 1, http://links.lww.com/CTG/A1) were examined in the validation study based on GSE84437 datasets, with 13 of them significantly associated with GC survival at P < 0.05 in the validation. Among them, high mRNA expression of 12 genes, including RIMS1 (regulating synaptic membrane exocytosis 1), PRICKLE1 (prickle planar cell polarity protein 1), MCC (mutated in colorectal cancers), DCLK1 (doublecortin-like kinase 1), FLRT2 (fibronectin leucine rich transmembrane protein 2), SLCO2A1 (solute carrier organic anion transporter family member 2A1), CDO1 (cysteine dioxygenase type 1), GHR (growth hormone receptor), CD109 (CD109 molecule), SELP (selectin P), UPK1B (uroplakin 1B), and CD36 (CD36 molecule), were associated with poor survival, while high mRNA expression of SLC25A15 (solute carrier family 25 member 15) was associated with improved survival. The associations remained significant in the Cox regression analysis, which adjusted additionally for clinical characteristics (Table 2). Among these 13 mRNAs, SELP, CD36, and CD109 were also highlighted in the key subnetwork, which could represent the overlapping protein complexes in the PPI network, as described above (Figure 2b).
Table 2.
The HRs and P values for the association between 13 messenger RNAs and gastric cancer death in TCGA and validated in GSE84437
Construction and validation of risk score model
Based on the mRNA expression levels of these 13 genes, we established the risk score as follows:
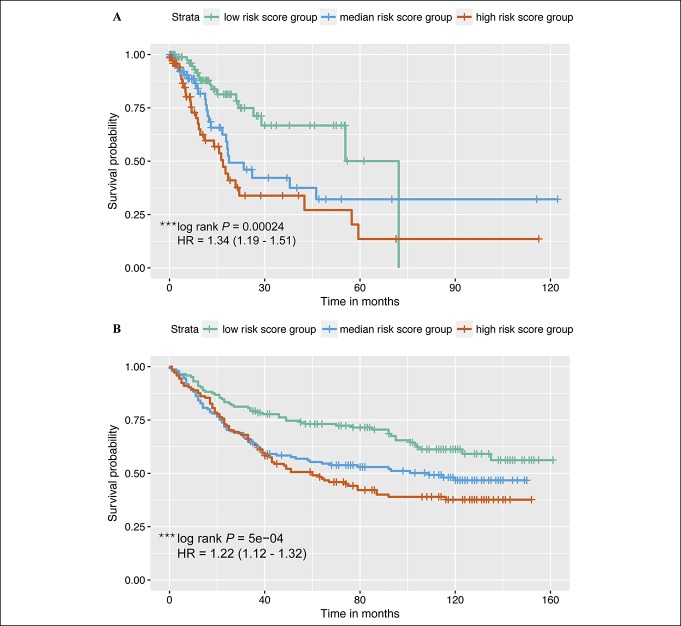
Risk score = 0.618 × DCLK1 + 0.400 × FLRT2 + 1.395 × MCC + 0.458 × PRICKLE1 + 0.881 × RIMS1 + (−0.768) × SLC25A15 + 0.404 × SLCO2A1 + 0.655 × CDO1 + 0.379 GHR + 0.969 × CD109 + 0.441 × SELP + 0.382 × UPK1B + 0.289 × CD36, with a higher score indicating worse survival potential. The risk score was independently associated with GC survival, with a hazard ratio (95% confidence interval) of 1.34 (1.19–1.51) per one score increase in TCGA-STAD and of 1.22 (1.12–1.32) per one score increase in GSE84437. Kaplan–Meier survival curves performed well on distinguishing patients with GC with differential survival status in TCGA-STAD (Figure 3). In GSE84437, the risk scores performed well in distinguishing patients by survival probability, particularly for those with the survival time of around 45 months or longer (Figure 3).
Figure 3.
Kaplan–Meier survival curves for patients in the discovery (the Cancer Genome Atlas-STAD) and validation (GSE84437) sets. The risk scores based on 13 messenger RNAs were categorized into tertiles. The green curve represents low risk score group. The blue curve represents median risk score group. The pink curve represents high risk score group. Log-rank tests were conducted to compare survival curves among subgroups in each dataset. (a) Kaplan–Meier survival curves for patients in the discovery set. (b) Kaplan–Meier survival curves for patients in the validation set.
DISCUSSION
Molecular signatures associated with GC prognosis have been sparse. In the present study, we comprehensively examined the mRNA signature associated with GC survival in the discovery stage (TCGA-STAD) based on RNA-Seq data and the validation stage (GSE84437) based on microarray data. A total of 13 mRNAs were identified significantly associated with GC survival. Among them, except for SLC25A15, 12 mRNAs were inversely associated with GC survival in both the discovery and validation dataset, including RIMS1, PRICKLE1, MCC, DCLK1, FLRT2, SLCO2A1, CDO1, GHR, CD109, SELP, UPK1B, and CD36. A 13 mRNA-based risk score model was established, which achieved good performance on predicting GC survival in both TCGA and GSE84437 datasets.
Potential function of the mRNA encoding genes was annotated based on the gene ontology functional enrichment analysis. Among the encoding genes for 13 mRNAs significantly associated with GC survival in the replication analysis, SLC25A15 was not enriched in the plasma membrane cellular component. Interestingly, also, SLC25A15 was the only mRNA inversely associated with risk of GC death in our analysis. A previous study suggested the association between plasma membrane and cancer cell migration and invasion (17). Several biomarkers based on plasma membrane proteins for GC development and progression have also been identified (18,19). The association of plasma membrane proteins with GC prognosis has been investigated as well (20,21). Thus, we assume that plasma membrane might play a key role in GC survival through regulating these identified mRNA expressions.
We also conducted pathway enrichment analysis to identify potential biological pathways associated with GC survival. Among the 13 mRNAs, we found once again that SLC25A15 was not involved in signal transduction pathway. A previous study showed that the cancer development was encoded by altering the patterns of signal transduction networks (21). Aberrant activation of several signal transduction pathways, including Hedgehog, Notch, and Wnt pathways, has already been associated with multicancer development (22). Several cancer therapeutics have been developed targeting signal transduction pathways during past few years (23). Thus, the signal transduction networks may be involved in the mechanisms underlying the associations between identified mRNAs and GC survival.
The contrasting associations with GC death that we found for SLC25A15 and other mRNAs are interesting, but the underlying mechanism is still unclear. Previous literature has not linked SLC25A15 to cancer development or prognosis. Current knowledge regarding the functional differences between SLC25A15 and other mRNAs has been very sparse. In our study, further integrating the results for the gene ontology functional enrichment analysis and pathway enrichment analysis, SLC25A15, which encodes the only mRNA inversely associated with GC death, was neither enriched in the plasma membrane cellular component nor involved in the signal transduction pathway. It is therefore reasonable to speculate that the plasma membrane cellular component and signal transduction pathway might play important roles in risk of GC death, which may partly explain the associations with GC death in the opposite directions that were found for SLC25A15 and other mRNAs. However, our study based on association analyses and function and pathway annotations cannot directly respond to the mechanisms underlying the associations. Efforts from basic laboratory are required to elucidate the gene functions, which could clarify the contrasting associations for SLC25A15 and other mRNAs with GC death.
Among the highlighted genes in our study, mRNA expression of CD36 and protein expression of DCLK1 and SLCO2A1 have been previously associated with GC survival. CD36 mRNA overexpression was associated with poor GC overall survival based on cDNA microarray data of 18 patients with GC in the discovery set and 30 patients with GC in the validation set (7). DCLK1 protein overexpression has been associated with poor GC overall survival using expression data of GC tumor specimens (n = 122) examined by immunohistochemistry (25). Our findings are consistent with these studies on the association of DCLK1 and CD36 with GC survival, though they either lacked suitable validation sets or had limited sample size. In addition, a DCLK1-based mRNA signature has also been established previously to predict GC survival (26). However, negative SLCO2A1 (also known as PGT) protein expression in GC tumor specimens (n = 96), as examined by immunohistochemistry, was associated with poor survival (27), in contrast to our findings on the inverse association between SLCO2A1 mRNA expression and GC survival. As that study was conducted based on a limited sample size, and also lacked a validation stage, the robustness of that finding may be of concern.
In previous studies, 4 mRNAs have been associated with prognosis of other cancers, but not with GC prognosis. CD109 mRNA overexpression has been associated with poor overall survival of lung adenocarcinoma (RNA-Seq) (28). SELP mRNA overexpression has been associated with the poor overall survival of gastrointestinal stromal tumors (microarray) (29). PRICKLE1 mRNA overexpression from cancer cell lines has been associated with poor metastasis-free survival of basal breast cancer (30). In addition, UPK1B mRNA overexpression has been associated with laryngeal cancer recurrence (microarray) (31). Our study is the first to report the association of these 4 mRNAs with GC survival in the same direction with the previously reported association of these mRNAs with survival of other cancers.
Methylation has been associated with the risk and (or) prognosis of GC and other cancers (32). Among the highlighted genes in our study, CDO1 methylation has been correlated with prognosis of other cancers previously (33,34), while our study is the first to report the association between CDO1 hypermethylation and mRNA overexpression and the risk of GC death (see Figure S1 and Table S3, Supplemental Digital Content 1, http://links.lww.com/CTG/A1). Several studies have identified FLRT2 methylation as potential biomarker for screening of prostate cancer (35) and breast cancer (36). However, although our study showed the association between FLRT2 mRNA overexpression and poor GC survival, we did not find the association of FLRT2 methylation with GC prognosis. In addition, our study is also the first to report an inverse association between GHR mRNA expression and methylation (see Table S2 and Figure S1, Supplemental Digital Content 1, http://links.lww.com/CTG/A1) and also the first to report GHR mRNA overexpression and hypomethylation as risky factors for GC death (see Table S3, Supplemental Digital Content 1, http://links.lww.com/CTG/A1). GHR encodes the transmembrane receptor for growth hormone, while the elevated serum level of growth hormone has been related to increased risk of GC and several other cancers (37). For MCC, RIMS1, and SLC25A15, previous evidence has been sparse regarding their associations with cancer development or prognosis.
Our study was conducted based on publicly accessible databases with a relatively big sample size of both discovery and validation datasets. A comprehensive approach was conducted by integrating prognostic mRNAs with methylation profiles and subnetwork analysis for conspicuously thorough selection of key mRNAs for validation, which yielded 13 mRNA signatures associated with GC survival. Except CD36, DCLK1, and SLCO2A1, other mRNAs are newly reported to be associated with GC survival. The 13 mRNA-based risk score model performed well in distinguishing the risk of GC prognosis, which might be useful in clinical practice regarding patient stratification in the recent future. We were able to control for the major clinical characteristics in a secondary analysis, which did not change the results materially.
We acknowledge several limitations. First, TCGA and GEO databases were derived based on different platforms for mRNA expression (RNA-Seq vs microarray). Although we deliberately performed data processing, the interpretation of the results, particularly the effect magnitudes across the 2 platforms, should be cautious. Second, our study was based on association studies and bioinformatics analysis. Further studies are warranted to clarify the mechanisms linking these mRNAs to GC poor survival. Third, our analyses were restricted by the available clinical characteristics and GC outcome variables. Gene expression may be varied by different race/ethnicity but our validation dataset (GSE84437) does not have the information on race/ethnicity. In the discovery stage, a secondary analysis additionally adjusting for race did not materially change the findings though. Further studies are warranted to examine the potential predictive value of the 13 mRNA signatures associated with other GC prognosis-related variables, such as progression-free survival, and also to examine the mRNAs associated with GC survival in certain race/ethnicity group.
In conclusion, findings based on 2 well-established cohorts suggest the 13 mRNA signatures might play an important role in predicting GC survival ahead of time. Our study may have implications for clinical practices regarding patient stratification. Exploration in the laboratory setting may contribute to the understanding of underlying molecular mechanisms and may inspire the development of novel targeted therapeutic strategies.
CONFLICTS OF INTEREST
Guarantor of the article: Wen-Qing Li and Kai-Feng Pan.
Specific author contributions: J.D.: study concept and design, data analyses, and draft of the manuscript. Z.-X.L., Y.Z., J.-L.M., T.Z., and W.-C.Y.: interpretation of data. W.-Q.L., and K.-F.P.: study concept and design, draft of the manuscript, critical revision of the manuscript for important intellectual content. All authors corrected and approved the manuscript.
Financial support: Beijing Municipal Administration of Hospitals' Ascent Plan (DFL20181102) (to K.-F.P.).
Potential competing interests: None.
Study Highlights
WHAT IS KNOWN
✓ The prognosis of patients with GC vary significantly even among those with similar clinical features.
✓ Molecular biomarkers which predict GC prognosis are still limited.
WHAT IS NEW HERE
✓ This study was conducted based on publicly accessible databases with a relatively big sample size of both discovery and validation datasets.
✓ The study yielded 13 mRNA signatures significantly associated with GC survival.
✓ Except CD36, DCLK1, and SLCO2A1, other mRNAs are newly reported to be associated with GC survival.
✓ The 13 mRNA-based risk score model performed well in distinguishing the risk of GC prognosis, which might be useful in clinical practice regarding patient stratification in the recent future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all individuals who participated in this study and donated samples.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A1
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Akama Y, Yasui W, Yokozaki H, et al. Frequent amplification of the cyclin E gene in human gastric carcinomas. Jpn J Cancer Res 1995;86:617–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graziano F, Mandolesi A, Ruzzo A, et al. Predictive and prognostic role of E-cadherin protein expression in patients with advanced gastric carcinomas treated with palliative chemotherapy. Tumour Biol 2004;25:106–10. [DOI] [PubMed] [Google Scholar]
- 4.Tanigawa N, Amaya H, Matsumura M, et al. Correlation between expression of vascular endothelial growth factor and tumor vascularity, and patient outcome in human gastric carcinoma. J Clin Oncol 1997;15:826–32. [DOI] [PubMed] [Google Scholar]
- 5.Sanz-Ortega J, Steinberg SM, Moro E, et al. Comparative study of tumor angiogenesis and immunohistochemistry for p53, c-ErbB2, c-myc and EGFr as prognostic factors in gastric cancer. Histol Histopathol 2000;15:455–62. [DOI] [PubMed] [Google Scholar]
- 6.Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res 2011;17:1850–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CN, Lin JJ, Chen JJ, et al. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J Clin Oncol 2005;23:7286–95. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Liu Y, Niu Z, et al. Prognostic value of a 25-gene assay in patients with gastric cancer after curative resection. Sci Rep 2017;7:7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Institutes of Health, National Cancer Institute. Genomic Data Commons Data Portal. Available at https://portal.gdc.cancer.gov/. Accessed October 10, 2017.
- 10.Liu JY, Peng CW, Yang XJ, et al. The prognosis role of AJCC/UICC 8th edition staging system in gastric cancer, a retrospective analysis. Am J Transl Res 2018;10:292–303. [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman BT, Huang DW, Tan Q, et al. DAVID knowledgebase: A gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 2007;8:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie C, Mao X, Huang J, et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 2012;9:471–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008;24:1547–8. [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal JK, Lauritzen SP, Scheffer L, et al. S100A11 is required for efficient plasma membrane repair and survival of invasive cancer cells. Nat Commun 2014;5:3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao W, Xu J, Wang F, et al. Plasma membrane proteomic analysis of human gastric cancer tissues: Revealing flotillin 1 as a marker for gastric cancer. BMC Cancer 2015;15:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbosh PH, McConkey DJ, Plimack ER. Targeting signaling transduction pathways in bladder cancer. Curr Oncol Rep 2015;17:58. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Wang X, Xu J, et al. Notch1 activation is a poor prognostic factor in patients with gastric cancer. Br J Cancer 2014;110:2283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoki M, Ishigami S, Uenosono Y, et al. Expression of BMP-7 in human gastric cancer and its clinical significance. Br J Cancer 2011;104:714–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolch W, Halasz M, Granovskaya M, et al. The dynamic control of signal transduction networks in cancer cells. Nat Rev Cancer 2015;15:515–27. [DOI] [PubMed] [Google Scholar]
- 23.Shimizu T, Nakagawa K. Novel signal transduction pathways: The molecular basis for targeted cancer therapies in Hedgehog/Notch/Wnt pathway. Nihon Rinsho 2015;73:1342–8. [PubMed] [Google Scholar]
- 24.Tai D, Wells K, Arcaroli J, et al. Targeting the WNT signaling pathway in cancer therapeutics. Oncologist 2015;20:1189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng QB, Yu JC, Kang WM, et al. Expression of doublecortin-like kinase 1 in human gastric cancer and its correlation with prognosis. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2013;35:639–44. [Chinese] [DOI] [PubMed] [Google Scholar]
- 26.Weygant N, Ge Y, Qu D, et al. Survival of patients with gastrointestinal cancers can be predicted by a surrogate microRNA signature for cancer stem-like cells marked by DCLK1 kinase. Cancer Res 2016;76:4090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda S, Tanigawa T, Watanabe T, et al. Reduction of prostaglandin transporter predicts poor prognosis associated with angiogenesis in gastric adenocarcinoma. J Gastroenterol Hepatol 2016;31:376–83. [DOI] [PubMed] [Google Scholar]
- 28.Shukla S, Evans JR, Malik R, et al. Development of a RNA-seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst [online] 2017;109 (.org/10.1093/jnci/djw200). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin S, Zhu W, Li J. Identification of key genes related to high-risk gastrointestinal stromal tumors using bioinformatics analysis. J Cancer Res Ther 2018;14:S243–47. [DOI] [PubMed] [Google Scholar]
- 30.Daulat AM, Bertucci F, Audebert S, et al. PRICKLE1 contributes to cancer cell dissemination through its interaction with mTORC2. Dev Cel 2016;37:311–25. [DOI] [PubMed] [Google Scholar]
- 31.Su J, Zhang Y, Su H, et al. A recurrence model for laryngeal cancer based on SVM and gene function clustering. Acta Otolaryngol 2017;137:557–62. [DOI] [PubMed] [Google Scholar]
- 32.Jones PA. Cancer: Death and methylation. Nature 2001;409:141–4. [DOI] [PubMed] [Google Scholar]
- 33.Meller S, Zipfel L, Gevensleben H, et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics 2016;11:871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deckers IA, Schouten LJ, Van Neste L, et al. Promoter methylation of CDO1 identifies clear-cell renal cell cancer patients with poor survival outcome. Clin Cancer Res 2015;21:3492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y, Davison J, Qu X, et al. Methylation profiling identified novel differentially methylated markers including OPCML and FLRT2 in prostate cancer. Epigenetics 2016;11:247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae H, Kim B, Lee H, et al. Epigenetically regulated Fibronectin leucine rich transmembrane protein 2 (FLRT2) shows tumor suppressor activity in breast cancer cells. Sci Rep 2017;7:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedek M, van der Velden LM, Strous GJ. Multimeric growth hormone receptor complexes serve as signaling platforms. J Biol Chem 2014;289:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.