Abstract
Objective
To explore genetic and lifestyle risk factors of MRI-defined brain infarcts (BI) in large population-based cohorts.
Methods
We performed meta-analyses of genome-wide association studies (GWAS) and examined associations of vascular risk factors and their genetic risk scores (GRS) with MRI-defined BI and a subset of BI, namely, small subcortical BI (SSBI), in 18 population-based cohorts (n = 20,949) from 5 ethnicities (3,726 with BI, 2,021 with SSBI). Top loci were followed up in 7 population-based cohorts (n = 6,862; 1,483 with BI, 630 with SBBI), and we tested associations with related phenotypes including ischemic stroke and pathologically defined BI.
Results
The mean prevalence was 17.7% for BI and 10.5% for SSBI, steeply rising after age 65. Two loci showed genome-wide significant association with BI: FBN2, p = 1.77 × 10−8; and LINC00539/ZDHHC20, p = 5.82 × 10−9. Both have been associated with blood pressure (BP)–related phenotypes, but did not replicate in the smaller follow-up sample or show associations with related phenotypes. Age- and sex-adjusted associations with BI and SSBI were observed for BP traits (p value for BI, p[BI] = 9.38 × 10−25; p[SSBI] = 5.23 × 10−14 for hypertension), smoking (p[BI] = 4.4 × 10−10; p[SSBI] = 1.2 × 10−4), diabetes (p[BI] = 1.7 × 10−8; p[SSBI] = 2.8 × 10−3), previous cardiovascular disease (p[BI] = 1.0 × 10−18; p[SSBI] = 2.3 × 10−7), stroke (p[BI] = 3.9 × 10−69; p[SSBI] = 3.2 × 10−24), and MRI-defined white matter hyperintensity burden (p[BI] = 1.43 × 10−157; p[SSBI] = 3.16 × 10−106), but not with body mass index or cholesterol. GRS of BP traits were associated with BI and SSBI (p ≤ 0.0022), without indication of directional pleiotropy.
Conclusion
In this multiethnic GWAS meta-analysis, including over 20,000 population-based participants, we identified genetic risk loci for BI requiring validation once additional large datasets become available. High BP, including genetically determined, was the most significant modifiable, causal risk factor for BI.
Introduction
Brain infarcts (BI) detected on MRI are commonly seen in older persons, being described in 8%–28% of participants in population-based cohort studies.1 Most MRI-defined BI are covert, not being associated with overt, clinical stroke symptoms.2,3 Nonetheless, they cannot be considered silent or benign, as they are often associated with subtle neurologic symptoms and with increased risk of future stroke, cognitive decline, and in some studies dementia.4,5 Most MRI-defined BI are small subcortical BI (SSBI), believed to be primarily caused by small vessel disease (SVD).6
Mechanisms and predictors of BI and SSBI remain incompletely understood. No genetic risk variants for BI and SSBI have been consistently identified to date,7–16 and findings with vascular risk factors have been inconsistent.1 Partly reflecting this uncertainty, recommendations to direct clinicians on how to best manage covert MRI-defined BI are lacking.
To enhance understanding of risk factors for BI and SSBI, we first conducted a large meta-analysis of genome-wide association studies (GWAS) from 18 population-based studies, comprising 20,949 participants from 5 ethnic groups, using the 1000 Genomes reference panel (1000G), more than doubling the size of a prior GWAS.16 Second, we examined the association of vascular risk factors with BI and SSBI in this large sample, using both vascular risk factor measurements and their genetic risk scores (GRS).
Methods
Study design and samples
The meta-analyses included 18 prospective population-based cohorts participating in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium (table e-1 and additional Methods e-1, doi.org/10.5061/dryad.hk07677). Although the cohorts contributing participants are longitudinal, this study is cross-sectional, based on the analysis of BI and SSBI at one timepoint in the subset of cohort participants with brain MRI. These cohorts comprised 5 ethnic groups and ancestries: European (n = 17,956), African (n = 1,834), Hispanic (n = 737), Malay (n = 215), and Chinese (n = 207). Some cohorts contributed to data for more than one ethnic group, resulting in a total of 23 datasets (tables e-1 to e-3, doi.org/10.5061/dryad.hk07677). Out of a total of 20,949 participants, 3,726 had MRI-defined BI. We did not exclude participants with a history of overt, clinically defined stroke prior to the MRI, except in 4 cohorts where patients with history of stroke were excluded by design. Three datasets did not contribute to the SSBI analysis either due to small numbers or absence of BI subtyping. Out of a total of 19,073 participants in the remaining 20 datasets, 3,533 had BI, of whom 2,021 (57.2%) had SSBI.
Variable definitions
Detailed MRI scanning protocols, as well as BI and SSBI definitions, for each study are described in table e-4 (doi.org/10.5061/dryad.hk07677). All protocols comprised at least T1, T2, and proton density or fluid-attenuated inversion recovery (FLAIR) sequences. On MRI, BI were defined as an area of abnormal signal intensity lacking mass effect with a size ≥3–4 mm; in the white matter, they were required to be hypointense on T1-weighted images, approaching the hypointensity of CSF, to distinguish them from diffuse white matter lesions; and they were distinguished from dilated perivascular spaces based on their irregular shape, presence of a hyperintense rim in FLAIR, and absence of a typical vascular shape following the orientation of perforating vessels.17 SSBI corresponded to BI with a size <15–20 mm, located in the basal ganglia, the white matter, or the brainstem. Participants with large BI or BI located in the cerebral cortex or cerebellum were excluded from analyses of SSBI. We also measured burden of white matter hyperintensities (WMH), a quantitative MRI marker of SVD, corresponding to signal abnormalities of variable size in the white matter, appearing as hyperintensity on T2-weighted or FLAIR images, but without cavitation. Details of WMH measurements have been described previously.18
Vascular risk factors
Vascular risk factor levels measured closest to brain MRI acquisition were used. Hypertension was defined as systolic blood pressure (SBP) ≥140 mm Hg or diastolic blood pressure (DBP) ≥90 mm Hg or use of one or more blood pressure (BP)–lowering medications. We defined pulse pressure (PP) as the difference between SBP and DBP and mean arterial pressure (MAP) as DBP + 1/3 × PP. Diabetes was defined as a previous diagnosis of diabetes, a fasting plasma glucose >7.0 mmol/L, or antidiabetic drug use. Fasting serum total cholesterol, high-density lipoprotein (HDL) cholesterol, and triglycerides were measured using enzymatic methods. Low-density lipoprotein (LDL) cholesterol was calculated using the Friedewald formula. Body mass index (BMI) was defined as the ratio of weight (kg) to the square of height (m). Active smoking was defined according to study-specific criteria. History of overt, clinically defined stroke and other cardiovascular events was based on ongoing surveillance prior to brain MRI acquisition in most studies since participant recruitment had started prior to the initial brain MRI. In studies that had brain MRI scanning at the initial visit, the history and examination at this visit were used to identify prior overt, clinically defined stroke. History of cardiovascular events included history of angina, myocardial infarction, cardiac bypass surgery, angioplasty, or peripheral vascular disease.
Genotypes
All participating discovery cohorts had genome-wide genotypes imputed on the 1000G (phase 1, version 3).19 Genome-wide genotyping platforms, quality control measures, and imputation parameters used in each study are presented in tables e-5–e-7 (doi.org/10.5061/dryad.hk07677).
Genome-wide association analyses with BI and small subcortical BI
For genome-wide association analyses with BI and SSBI, each study performed logistic regression under an additive genetic model after adjusting for age, sex, principal components of population stratification, and additional study-specific covariates, such as study site or family structure, as needed (additional Methods e-2, doi.org/10.5061/dryad.hk07677, for centralized quality control description). Our primary multiethnic GWAS meta-analysis was performed using MANTRA, based on a Bayesian framework.20 In secondary analyses, we also ran the multiethnic GWAS meta-analysis with 2 alternative methods (additional Methods e-2, doi.org/10.5061/dryad.hk07677): (1) using fixed effects inverse variance weighting with METAL21,22 and (2) using the random effects meta-analysis model implemented in METASOFT.23 During meta-analysis, genomic control correction was applied to the individual studies and ethnic-specific results to remove any residual inflation of association statistics. We did not observe any systematic inflation of association statistics (figure e-1, doi.org/10.5061/dryad.hk07677). Statistical measures from MANTRA, the primary meta-analysis method, were used to define genome-wide significance (Log10 of Bayesian factor [L10BF] > 6)24 and to choose single nucleotide polymorphisms (SNPs) for follow-up (L10BF > 4.5) in either the BI or SSBI meta-analysis. Details of functional annotation of top loci are provided in additional Methods e-3 (doi.org/10.5061/dryad.hk07677).
Follow-up and extension
For follow-up and extension studies, genotypes imputed to the 1000G reference panel were available in most instances for in silico look-up of the selected risk variants. Three follow-up studies performed de novo genotyping of the top 6 loci (additional Methods e-1, doi.org/10.5061/dryad.hk07677). The lead variant (with lowest p value) was genotyped at each suggestive or genome-wide significant locus, and if not feasible, another variant in strong linkage disequilibrium (LD, r2 > 0.8) was genotyped. A p value <0.0083, correcting for 6 loci, was considered significant evidence for replication.
The follow-up sample, in which we sought to confirm associations observed in the discovery analysis, included 6,862 participants, of whom 1,483 had BI and 630 had SSBI, from 6 community-based studies of European origin and one of Japanese origin (table e-1, doi.org/10.5061/dryad.hk07677).
As an extension, to test whether genetic variants associated with MRI-defined BI or SSBI in the discovery analysis are also associated with correlated phenotypes, we first explored their association with ischemic stroke (IS) overall and the small vessel disease subtype (IS-SVD) when available in 4 collaborative studies (table e-1, doi.org/10.5061/dryad.hk07677). Second, we explored whether genetic variants associated with MRI-defined BI and SSBI were associated with neuropathologically defined BI based on 2,940 brain autopsies in participants without dementia from the Alzheimer's Disease Genetics Consortium (ADGC). Participants with large infarcts or lacunes (n = 857, 29%) were compared to participants without any infarcts or having only microscopic infarcts (n = 2,083).25
We calculated power of the follow-up and extension studies using Quanto V1.2.3 (biostats.usc.edu/software; table e-8 and figure e-2, doi.org/10.5061/dryad.hk07677).
Association of vascular risk factors with BI and SSBI
Individual studies performed logistic regression to test for association of vascular risk factor measurements with presence or absence of at least one BI or SSBI. Analyses were performed with and without adjustments for age and sex. Analyses with BP or lipid traits as the main independent variable were additionally adjusted for treatment with disease-specific medications, and association analyses with fasting plasma glucose were limited to participants without type 2 diabetes. Except for WMH burden, the regression coefficients and standard errors for risk factors in the individual studies belonging to one ethnic group were combined using fixed-effects inverse variance-weighted meta-analysis and subsequently the betas and standard errors obtained in each ethnic group were combined using fixed-effects inverse variance-weighted meta-analysis, in the absence of heterogeneity (p < 1 × 10−6), to derive the multiethnic meta-analysis estimates. For WMH burden, the statistics were combined using the Z score–based sample size weighted meta-analysis as WMH burden was measured on different scales in participating studies.18
We then explored whether genetic variants previously shown in published GWAS to be associated with specific vascular risk factors were, in aggregate, also associated with BI and SSBI. This approach was selected to assess to what extent genetically determined vascular risk factor levels are associated with BI and SSBI and to provide evidence for a causal relation between a given vascular risk factor and risk of BI or SSBI, provided that Mendelian randomization assumptions are fulfilled.26 We combined known genetic risk variants for each individual vascular risk factor into a weighted GRS, using effect estimates from the largest published GWAS of that risk factor as weights. We then tested for association of these GRS with BI and SSBI using the inverse-variance weighting (IVW) method. Construction of the GRS, selection of variants for the GRS analysis, as well as effect estimates used as weights are detailed in additional Methods e-4 and tables e-9–e-12 (doi.org/10.5061/dryad.hk07677). For significant GRS associations with BI or SSBI, we further conducted sensitivity analyses using the MR-Egger method implemented as an R package (TwoSampleMR),27 which unlike the IVW method estimates the intercept term as part of the analysis. An intercept term significantly differing from zero suggests the presence of directional (unbalanced) pleiotropy, meaning that the pleiotropic effects of genetic variants are not balanced about the null.27 We used a conservative significance threshold of p < 0.05 for the intercept.
After Bonferroni correction for 12 independent vascular phenotypes tested for association with BI and SSBI, p < 0.0042 was considered significant for associations with vascular risk factor measurements or GRS. The number of independent vascular phenotypes, taking into account correlation between the phenotypes considered, was estimated based on individual level data from the 3C-Dijon study using the online tool matSpDlite (neurogenetics.qimrberghofer.edu.au/matSpDlite/).
Standard protocol approvals, registrations, and patient consents
Institutional review boards approved all of these studies, and all participants provided informed consent.
Data availability
Summary statistics of the top SNPs are available from Dryad for both BI and SSBI. Other data that support the findings of this study are available from the corresponding authors upon reasonable request.
Results
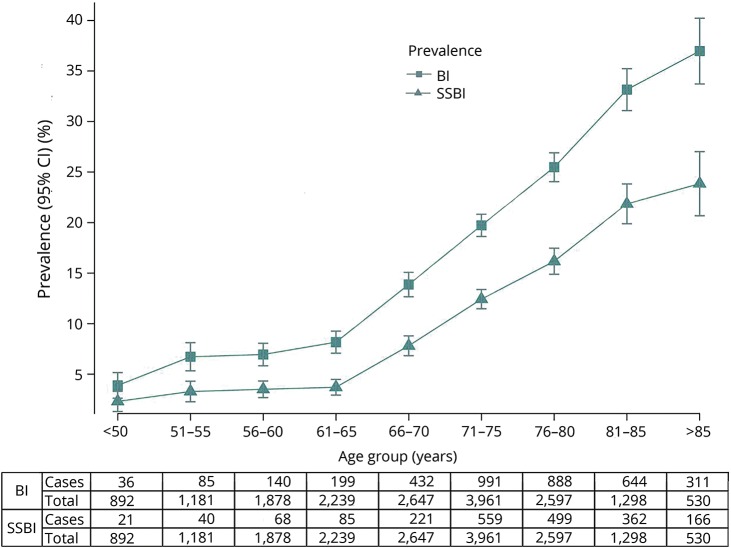
In this large population-based dataset comprising 18 cohort studies, the frequency of MRI-defined BI ranged from 4% to 38% in participating cohorts (table e-1, doi.org/10.5061/dryad.hk07677). A description of demographic characteristics in all participants with BI (n = 3,726), with SSBI (n = 2,021), and without BI (n = 17,223) is provided in tables e-2 and e-3 (doi.org/10.5061/dryad.hk07677) for individual studies. Participants with BI and SSBI were on average 6 years older and more often men compared to those without BI. In age-stratified analyses, the prevalence of BI and SSBI increased with age, most prominently beyond age 65, after which a 25.8% (range 13.9%–37.0%) increment in BI prevalence was observed compared to participants younger than 65 years (figure 1). Overall, the prevalence of BI ranged from less than 5% before age 50 to over 30% beyond age 80, with similar findings when we analyzed men and women separately (figures e-3 and e-4, doi.org/10.5061/dryad.hk07677). Only 11% of those with BI and 9% of those with SSBI had a history of stroke (12.5% and 9.8% when removing cohorts that excluded participants with history of stroke by design); hence, the vast majority of MRI-defined BI were covert.
Figure 1. Prevalence of MRI-defined brain infarcts (BI) and small subcortical brain infarcts (SSBI) by different age groups.
CI = confidence interval.
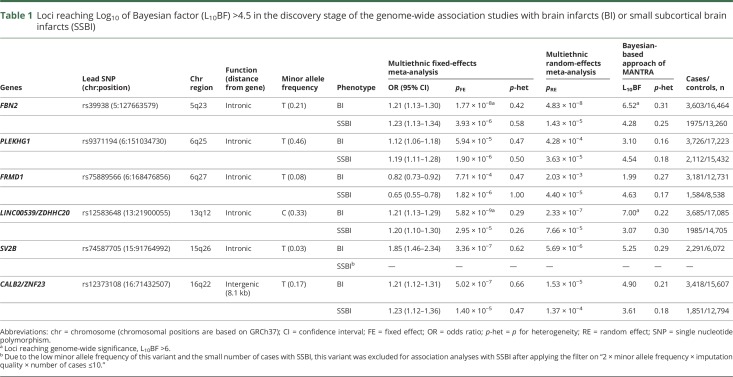
Genome-wide association plots for GWAS of BI and SSBI are displayed in figures e-5 and e-6 (doi.org/10.5061/dryad.hk07677). Two loci were associated with risk of BI at genome-wide significant level (L10BF > 6): rs39938 in FBN2 (chr5q23) and rs12583648 in LINC00539 and near ZDHHC20 (chr13q12). In addition, 2 SNPs were associated with BI at a suggestive level of significance (L10BF > 4.5): rs12373108 near CALB2/ZNF23 (chr16q22) and rs74587705 in SV2B (chr15q26) (table 1). No genome-wide significant association was observed for SSBI, but 2 loci reached the threshold for suggestive association (L10BF > 4.5): rs9371194 in PLEKHG1 (chr6q25) and rs75889566 in FRMD1 (chr6q27, table 1). These 6 loci were taken forward for the follow-up stage (table 2). For all SNPs reaching Log10BF > 4.5 in the discovery stage, association statistics are shown in table e-13 and figure e-7 (doi.org/10.5061/dryad.hk07677).
Table 1.
Loci reaching Log10 of Bayesian factor (L10BF) >4.5 in the discovery stage of the genome-wide association studies with brain infarcts (BI) or small subcortical brain infarcts (SSBI)
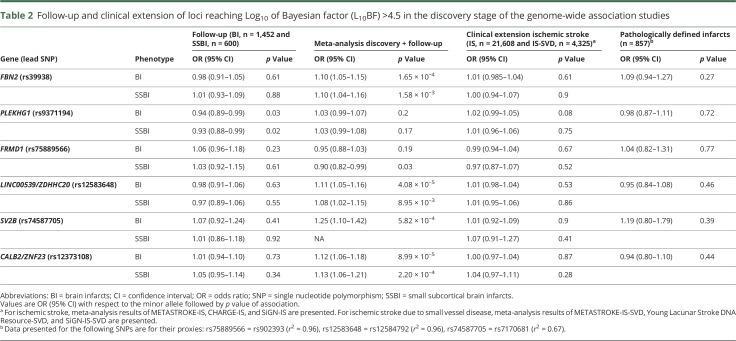
Table 2.
Follow-up and clinical extension of loci reaching Log10 of Bayesian factor (L10BF) >4.5 in the discovery stage of the genome-wide association studies
In the substantially smaller population-based follow-up studies, we could not replicate the 2 genome-wide significant or the 4 suggestive loci associated with BI or SSBI (table 2). Of the 6 loci that we followed up, we had limited power for 2 of the loci for BI (52%) and 4 of the loci for SSBI (50%–58%) (table e-8, doi.org/10.5061/dryad.hk07677). Power estimates in the follow-up study are even lower when accounting for the winner's curse phenomenon, which leads to inflated effect estimates in the discovery cohort.28 One suggestive locus for SSBI (PLEKHG1) showed nominal association with BI and SSBI in the follow-up studies (pBI = 0.03 and pSSBI = 0.02), but in the opposite direction (table 2).
Likewise, none of the genome-wide significant or suggestive loci for BI and SSBI showed association with IS (overall or IS-SVD) or pathologically defined BI in the extension studies after correcting for multiple testing (table 2 and table e-14, doi.org/10.5061/dryad.hk07677). Whereas the sample size for overall IS and IS-SVD was relatively large, it was limited for pathologically defined BI, and power was insufficient for 4 of the loci (25%–70%) (table e-8, doi.org/10.5061/dryad.hk07677).
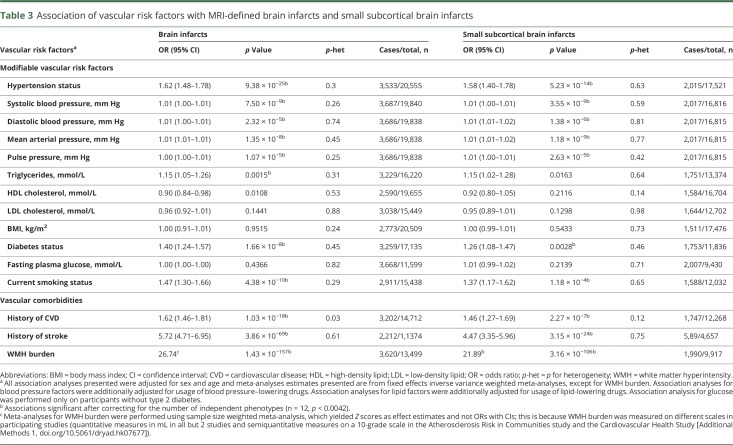
Associations of vascular risk factors with risk of BI or SSBI adjusted for age and sex are presented in table 3 (for unadjusted results, see table e-15, doi.org/10.5061/dryad.hk07677). Both BI and SSBI were significantly associated with all BP indices, the lowest p value being observed for SBP and MAP. Smoking and diabetes were also associated with both BI and SSBI. Triglycerides were significantly associated with BI only. We did not observe significant associations with levels of HDL cholesterol, LDL cholesterol, BMI, or fasting plasma glucose in nondiabetic participants. Both BI and SSBI were associated with history of cardiovascular disease and history of stroke. The most significant association by far was observed with WMH burden on brain MRI, both for BI and SSBI. As hypertension is an important risk factor for WMH as well, we additionally adjusted the regression model for hypertension to rule out a confounding effect by this variable; however, the association became even more significant (p = 5.71 × 10−172 for BI and p = 4.47 × 10−114 for SSBI) (table e-16, doi.org/10.5061/dryad.hk07677). No significant heterogeneity was seen for these associations across participating studies.
Table 3.
Association of vascular risk factors with MRI-defined brain infarcts and small subcortical brain infarcts
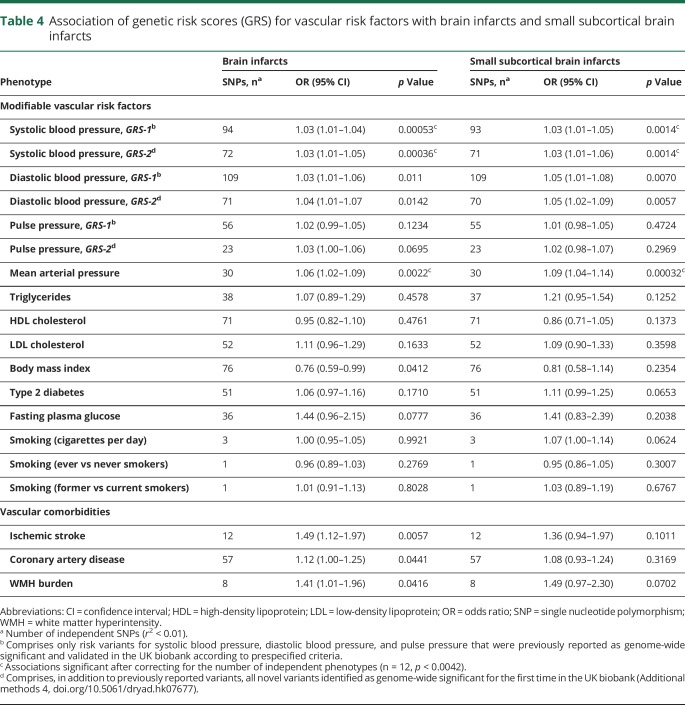
When exploring the relation of weighted genetic risk scores for vascular risk factors with BI and SSBI, we found that GRS for SBP and MAP were significantly associated with increased risk of BI and SSBI after correction for multiple testing (table 4). In sensitivity analyses using MR-Egger regression, evidence for directional pleiotropy was lacking for these associations between SBP or MAP GRS and BI or SSBI (p intercept >0.36). GRS for DBP, BMI, coronary artery disease, WMH burden, and IS were nominally associated with BI (p < 0.05, table 4), but these associations did not survive correction for multiple testing.
Table 4.
Association of genetic risk scores (GRS) for vascular risk factors with brain infarcts and small subcortical brain infarcts
Discussion
This multiethnic meta-analysis comprising over 20,000 community participants provides noteworthy insight into risk factors for MRI-defined brain infarcts. The described BI distributions across different age groups and by sex may also serve as a reference for comparison with BI and SSBI frequency in other settings. Of note, about 90% of BI were covert, not being associated with a history of stroke. In this multiethnic GWAS of BI and SSBI, we identified 2 genome-wide significant risk loci for BI, FBN2 on chr5q23 and LINC00539/ZDHHC20 on chr13q12, although these could not be replicated in a smaller follow-up sample. We further describe the association of MRI-defined BI with vascular risk factors, combining the vast majority of population-based cohort studies with BI and SSBI measurements available. We find high BP, both phenotypically expressed high BP and genetically determined risk for high BP, to be the most significant modifiable risk factor for BI. No association with cholesterol levels or BMI was found.
To identify novel genetic risk loci for MRI-defined BI and SSBI, we have more than doubled the sample size compared to the previously published GWAS of MRI-defined BI,16 used imputed genotypes based on the 1000G reference panel to increase the marker coverage, and included samples from 5 ethnicities for a broader representation of individuals from different origins. Moreover, we studied both BI and SSBI, while only BI were analyzed in the previously published GWAS meta-analysis.16 Our inability to replicate the genome-wide significant and suggestive findings could reflect false-positive results but may also be explained by insufficient power in the follow-up stage (table e-8, doi.org/10.5061/dryad.hk07677). Further studies on larger samples with MRI-defined BI are required to confirm or refute these findings. Moreover, while we could not provide evidence for an association of genome-wide significant and suggestive risk loci for BI and SSBI with IS, IS-SVD, or pathologically defined BI, this inability could reflect differences in the biology underlying these phenotypes, as well as limited power in the extension studies.
The 2 loci that crossed the genome-wide significance threshold, while requiring confirmation in larger independent samples, do harbor plausible biological candidates. Fibrillin2 (FBN2) encodes a protein that is part of the connective tissue microfibrils and elastic fiber assembly of the cell.29 Rare and common variants in FBN2 have been associated with age-related macular degeneration.30 Recent studies have also implicated common variants in FBN2 to be associated with SBP,31 although the variants differ (rs6595838-SBP and rs39938-BI, r2 = 0.017). The LINC00539/ZDHHC20 locus was a suggestive hit in a GWAS of adverse metabolic response to hydrochlorothiazide, a drug commonly used to treat hypertension.32 The lead SNP in the region could also influence the expression of the long noncoding RNA LINC00539 (table e-17, doi.org/10.5061/dryad.hk07677).
Our findings provide definitive evidence for a major and predominant association of increasing BP levels with increased risk of BI and SSBI.1,33 Beside significant associations with hypertension, a continuous association was observed for increasing levels of all BP measurements (SBP, DBP, PP, MAP), consistent with elevated BP being the major modifiable risk factor for BI, as is the case for overt, clinically defined IS.34–36 The importance and causal nature of the relation between high BP and risk of BI and SSBI is further supported by the significant association of BP genetic risk scores, for SBP and MAP, with increased risk of BI, especially SSBI, with no indication of directional pleiotropy using the MR-Egger approach.27
Previous publications on the association of BI and SSBI with vascular risk factors other than elevated BP were inconsistent.1,33,37 Our study provides evidence for a significant association of current smoking and diabetes with risk of BI and SSBI, while no association with BMI and cholesterol could be demonstrated, despite the very large sample size. These findings are consistent with epidemiologic data on IS.35 Interestingly, in contrast with cholesterol levels, a significant association of increasing triglyceride levels with BI risk was observed, although for SSBI the association did not withstand correction for multiple testing. Inconsistent results have been reported regarding association of triglycerides with overt, clinically defined IS,38,39 but the present results are in line with evidence of an association in older community-dwelling persons between high triglyceride levels and WMH burden, another MRI marker of SVD.40
As previously described, we show a significant association of WMH burden with BI and SSBI, reaching p < 10−100 in this study. Surprisingly, shared genetic variation among the top loci for WMH burden and BI was limited. While this observation could be due to lack of power, it could also suggest that WMH and BI share more environmental than genetic risk factors. A more comprehensive search for shared genetic variation between WMH burden and BI or SSBI at the genome-wide level using the LD score regression method41 could not be performed in the present study due to low variance in the BI GWAS, also hampering the calculation of BI heritability using the same method. Of note, based on estimates from previously published family-based studies, heritability for SSBI was described to be low at 29%, in contrast with a moderate to high heritability for WMH burden at 49%–80%.42–44 Hypertension is a major risk factor for WMH as well, and a BP GRS was also significantly associated with WMH burden in a prior study.18 However, the association of WMH burden with BI and SSBI was still significant after adjusting for hypertension status (table 3), or for SBP levels and BP-lowering treatment (table e-16, doi.org/10.5061/dryad.hk07677), suggesting that BP is not the only mediator of this association.
An important strength of the present study is that we have gathered nearly all large population-based studies with MRI-based identification of BI, genome-wide genotypes, and detailed vascular risk factor and comorbidity assessment, totaling over 20,000 participants covering 5 ethnic groups. Despite the unprecedented sample size, we were underpowered for the discovery of novel, robust genetic risk loci and even more so for the follow-up of genome-wide significant findings. Our ability to discover robust genetic risk variants may also have been hampered by the heterogeneity in BI and SSBI etiology, even though SVD is likely the predominant mechanism,45 and by some heterogeneity in the way BI and SSBI have been measured in participating studies. Finally, although the majority of participants had covert BI, 10% had a history of overt, clinically defined stroke, but including both covert and overt BI also enables a better representation of the spectrum of participants with MRI-defined BI in the general population. Whereas history of stroke was more common in participants with BI than those without, we do not believe that this inclusion has driven the associations we observed, given both the small number of participants with a stroke history and the significance level of the observed associations. Moreover, in this population-based setting, determining whether an MRI-defined BI could be attributed to the history of clinically defined stroke was not always possible.
In clinical practice, MRI-defined BI are commonly seen on brain MRI scans performed for various reasons in older persons. They have been shown to be powerful predictors of incident stroke and incident dementia.1,4,46 Hence BI represent an important marker for detection of high-risk individuals and initiation of preventive interventions. However, no randomized trials and no recommendations are currently available for the management of covert MRI-defined BI. The observational evidence is overwhelming for a strong causal relation between high BP and risk of BI and SSBI. A randomized trial will be needed to decide if persons with MRI-defined BI will benefit from more intensive BP-lowering strategies than is recommended currently for primary prevention.
This multiethnic, population-based study on 20,949 participants sheds important new light on susceptibility factors of MRI-defined brain infarcts, a marker of covert vascular brain injury commonly observed in older persons.
Author affiliations
From the Bordeaux Population Health Research Center (G.C., M.S., S.K., S. Schilling, C.T., S.D.), INSERM U1219, Groupe d'Imagerie Neurofonctionnelle CNRS/CEAU5293 (B.M.), and University of Bordeaux (G.C., C.T., M.S., S.K., S. Schilling, S.D., B.M.), Department of Neurology, Bordeaux University Hospital (S.D.), Bordeaux, France; Departments of Epidemiology (H.H.H.A., M.W.V., A.H., M.A.I.), Radiology & Nuclear Medicine (H.H.H.A., M.W.V., A.v.d.L., M.A.I.), Internal Medicine (A.G.U.), and Neurology (M.A.I.), Erasmus MC, Rotterdam, the Netherlands; Department of Neurology (C.L.S., V.C., H.J.A., J.R.R., P.M., S. Seshadri), Boston University School of Medicine; Department of Biostatistics (A.L.D., S. Seshadri), Boston University School of Public Health; The National Heart, Lung, and Blood Institute's Framingham Heart Study (C.L.S., V.C., A.L.D., H.J.A., J.R.R., P.M., S. Seshadri), MA; Cardiovascular Health Research Unit, Department of Medicine (J.C.B., B.M.P.), and Departments of Epidemiology (B.M.P., W.T.L.), Health Services (B.M.P.), Biostatistics (K. Rice, Q.W.), Neurology (W.T.L.), and Pathology (T.J.M.), University of Washington, Seattle; Pathology (T.J.M.), Standford University, California; Institute for Community Medicine (A. Teumer), Department of Psychiatry and Psychotherapy (H.J.G.), Institute of Diagnostic Radiology and Neuroradiology (N.H.), and Department of Neurology (B.v.S.), University Medicine Greifswald, Germany; Clinical Division of Neurogeriatrics, Department of Neurology (E.H., R.S.), Institute for Medical Informatics, Statistics and Documentation (E.H.), and Gottfried Schatz Research Center (for Cell Signaling, Metabolism and Aging), Institute of Molecular Biology and Biochemistry (Y.S., H.S.), Medical University of Graz, Austria; Department of Cardiology (S.T., J.W.J.), Section of Gerontology and Geriatrics, Department of Internal Medicine (S.T.), Molecular Epidemiology (P.E.S., M.B., J.D.), Medical Statistics and Bioinformatics (H.-W.U.), and Department of Radiology (J.v.d.G.), Leiden University Medical Center, the Netherlands; Department of Pharmacology (S.H., C.C., M.K.I.) and Department of Ophthalmology, Yong Loo Lin School of Medicine (C.-Y.C.), National University of Singapore; Icelandic Heart Association (A.V.S., V.G.), Kópavogur, Iceland; Institute of Molecular Medicine (X.J., M.F.) and Human Genetics Center (M.F.), University of Texas Health Science Center at Houston; Institute for Stroke and Dementia Research (R.M., M.D.), Klinikum der Universität München, Ludwig-Maximilians-Universität, Munich, Germany; Clinical Neurosciences (M.T., L.C.A.R.-J., H.S.M.), University of Cambridge, UK; School of Life Sciences (S.B.), University of Lincoln, United Kingdom; German Center for Neurodegenerative Diseases (DZNE) (L.C.A.R.-J.), Population Health Sciences, Bonn, Germany; Department of Medical Genetics (S.L.P.), Department of Neurology, Brain Center Rudolf Magnus (M.K.I.), Department of Epidemiology, Julius Center for Health Sciences and Primary Care (M.I.G., Y.v.d.G.), Department of Genetics, Center for Molecular Medicine (P.I.W.d.B.), and Department of Cardiology, Division Heart & Lungs (F.W.A.), University Medical Center Utrecht, and Utrecht University, the Netherlands; Institut Pasteur de Lille (V.C., P.A.), Lille University, INSERM, Lille University Hospital, France; Department of Neurology (Y.-C.Z.), Peking Union Medical College Hospital, Beijing, China; John P. Hussman Institute for Human Genomics (G.W.B., A.H.B., S.H.B.), Department of Neurology (R.L.S., T.R.), Evelyn F. McKnight Brain Institute (R.L.S., T.R.), Department of Epidemiology and Public Health Sciences (R.L.S., T.R.), and Dr. John T. Macdonald Foundation Department of Human Genetics (R.L.S., S.H.B., A.H.B.), Miller School of Medicine, University of Miami, FL; Department of Pathology and Laboratory Medicine (G.D.S.), University of Pennsylvania School of Medicine, Philadelphia; Departments of Neurology & Radiology (O.K.), Landspitali National University Hospital; Faculty of Medicine (V.G.), University of Iceland, Reykjavik; Department of Neurology (D.S.K.), Mayo Clinic, Rochester, MN; Departments of Data Science (M.E.G.) and Medicine (B.G.W., T.H.M.), University of Mississippi Medical Center, Jackson; Department of Neurology (R.F.G.), Johns Hopkins University School of Medicine, Baltimore, MD; Department of Gene Diagnostics and Therapeutics (F.T., N.K.), Research Institute, National Center for Global Health and Medicine, Tokyo; The Third Department of Internal Medicine (S.Y.) and Department of Functional Pathology (T.N.), Shimane University School of Medicine, Japan; Rush University Medical Center (K.B.R., N.T.A., D.A.E.), Chicago, IL; Brigham and Women's Hospital (K. Rexrode); Center for Translational & Computational Neuroimmunology, Department of Neurology (P.L.D.J.), Columbia University Medical Center, New York, NY; Kaiser Permanente Washington Health Research Institute (B.M.P.), Seattle, WA; Institute for Translational Genomics and Population Sciences, Los Angeles Biomedical Research Institute, and Division of Genomic Outcomes, Department of Pediatrics (J.I.R.), Harbor-UCLA Medical Center, Torrance; Departments of Pediatrics, Medicine, and Human Genetics (J.I.R.), UCLA, Los Angeles, CA; Department of Neurology (O.L.L.), University of Pittsburgh, PA; Singapore Eye Research Institute (J.L.); Duke-NUS Graduate Medical School (C.-.YC., T.Y.W., M.K.I.), Singapore; Singapore Eye Research Institute (C.-.YC., T.Y.W., M.K.I.), Singapore National Eye Centre; Memory Aging & Cognition Centre (MACC), National University Health System (M.K.I.), Singapore; Genetic Epidemiology Unit, Department of Epidemiology and Biostatistics (S.J.v.d.L., N.A., C.M.v.D.), Erasmus MC University Medical Center, Rotterdam, the Netherlands; Department of Neurology (C.D.), University of California at Davis; Brain Research Imaging Centre (J.M.W., M.d.C.V.H., M.E.B., S.M.M.), Centre for Clinical Brain Sciences (J.M.W., M.d.C.V.H., M.E.B., C.L.M.S.), Edinburgh Dementia Research Centre (J.M.W., M.d.C.V.H.), Centre for Cognitive Ageing and Cognitive Epidemiology (J.M.W., M.d.C.V.H., M.L., D.L., I.J.D., M.E.B., J.M.S., S.M.M.), Alzheimer Scotland Dementia Research Centre (J.M.S.), and Institute of Genetics and Molecular Medicine (C.L.M.S.), University of Edinburgh, UK; Department of Neurosciences, Experimental Neurology and Leuven Research Institute for Neuroscience and Disease (LIND) (R.L.), KU Leuven–University of Leuven; Center for Brain & Disease Research (R.L.), VIB, Laboratory of Neurobiology; Department of Neurology (R.L.), University Hospitals Leuven, Belgium; Florey Institute of Neuroscience and Mental Health (V.T.), University of Melbourne, Australia; Centre for Healthy Brain Ageing, Psychiatry (H.B., W.W., A. Thalamuthu, N.J.A., E.C., K.A.M., P.S.S.), Dementia Centre for Research Collaboration (H.B.), and School of Medical Sciences (P.R.S., J.B.K.), University of New South Wales, Sydney; Mathematics & Statistics (N.J.A.), Murdoch University, Perth; Neuroscience Research Australia (K.A.M., P.R.S., A.Thalamuthu), Randwick; Brain and Mind Centre (J.B.K.), The University of Sydney, Camperdown; Queensland Brain Institute (M.J.W.), University of Queensland, Brisbane; National Ageing Research Institute (D.A.), Melbourne; Academic Unit for Psychiatry of Old Age (D.A.), University of Melbourne, Australia; Department of Cerebrovascular Diseases (G.B.B.), Fondazione IRCCS Istituto Neurologico “Carlo Besta,” Milan, Italy; CTSU, Nuffield Department of Population Health (J.C.H., C.M.v.D.), and Nuffield Department of Clinical Neurosciences (P.M.R.), University of Oxford, UK; National Institute of Neurological Disorders and Stroke (C.B.W.), NIH, Bethesda, MD; Institute of Cardiovascular and Medical Sciences, Faculty of Medicine (D.J.S.), and Robertson Centre for Biostatistics (I.F.), University of Glasgow, UK; German Center for Neurodegenerative Diseases (DZNE) (K.W., H.J.G.), Site Rostock, Greifswald, Germany; Interfaculty Institute for Genetics and Functional Genomics (U.V.), University of Greifswald, Germany; John Hunter Hospital (C.L.), Hunter Medical Research Institute and University of Newcastle, Callaghan, Australia; Neurovascular Research Group (NEUVAS) (J.J.-C.), Neurology Department, IMIM–Hospital del Mar, Barcelona, Spain; Institute of Cardiovascular Research (P.S.), Royal Holloway University of London & St Peters and Ashford Hospital, UK; Center for Human Genetic Research and Department of Neurology (J.R.), Program in Medical and Population Genetics, Broad Institute, Massachusetts General Hospital, Harvard Medical School, Boston; University of Cincinnati College of Medicine (D.W.), OH; Department of Neurology (J.W.C., S.J.K.), University of Maryland School of Medicine and Baltimore VAMC; Department of Neurology (J.F.M.), Mayo Clinic Jacksonville, FL; Department of Neurology (A.S.), Jagiellonian University, Krakow, Poland; Department of Clinical Sciences Lund, Neurology (A.L.), Lund University; Department of Neurology and Rehabilitation Medicine (A.L.), Skåne University Hospital; Department of Clinical Sciences Malmö (O.M.), Lund University, Sweden; Neuroscience Institute (R.P.G.), Saint Francis Medical Center, School of Health and Medical Sciences, Seton Hall University, South Orange, NJ; College of Public Health (D.K.A.), University of Kentucky, Lexington; Institute of Biomedicine (C.J.), the Sahlgrenska Academy at University of Gothenburg, Sweden; Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, College of Pharmacy (J.A.J.), and Division of Cardiovascular Medicine, College of Medicine (J.A.J.), University of Florida, Gainesville; Department of Neurology (O.R.B.), University of British Columbia, Vancouver, Canada; Department of Epidemiology and Population Health (S.W.-S.), Albert Einstein College of Medicine, Bronx, NY; Stroke Center, Department of Neurology (J.-M.L.), Washington University School of Medicine, St. Louis, MO; Department of Medicine (B.D.M., P.F.M.), University of Maryland School of Medicine, Baltimore; Center for Public Health Genomics (S.S.R.), University of Virginia School of Medicine, Charlottesville; Durrer Center for Cardiovascular Research (F.W.A.), Netherlands Heart Institute, Utrecht, the Netherlands; Institute of Cardiovascular Science, Faculty of Population Health Sciences (F.W.A.), and Farr Institute of Health Informatics Research and Institute of Health Informatics (F.W.A.), University College London, UK; Peninsula Clinical School (V.S., C.M., M.C.), Frankston Hospital, Central Clinical School, and School of Clinical Sciences (T.P.), Monash Health, Monash University, Melbourne; Menzies Institute for Medical Research (V.S., R.M., M.C.), University of Tasmania, Hobart; Western Sydney University (R.T.), New South Wales, Australia; Departments of Neurology and Public Health Sciences (B.B.W.), University of Virginia, Charlottesville; Neuropsychiatric Institute (P.S.S.), Prince of Wales Hospital, Randwick, Australia; Munich Cluster for Systems Neurology (SyNergy) (M.D.), Munich, Germany; Intramural Research Program (L.J.L.), National Institute on Aging, NIH, Bethesda, MD; and University of Texas Health Sciences Center and Glenn Biggs Institute for Alzheimer's and Neurodegenerative Diseases (C.L.S., S. Seshadri), San Antonio.
Acknowledgment
The authors thank Dr. Anton J.M. de Craen (September 1966–January 2016) from the PROSPER and LLS study for his contribution to this work; the Older Australian Twins Study (OATS) participants, their supporters, and the OATS Research Team; all study participants and their relatives, general practitioners, and neurologists for their contributions; P. Veraart for help in genealogy; J. Vergeer for supervision of the laboratory work; P. Snijders for help in data collection; Dr. Anne Boland (CNG) for technical help in preparing the DNA samples for analyses; the University of Newcastle for funding; and the men and women of the Hunter region who participated in this study.
Glossary
- 1000G
1000 Genomes reference panel
- BI
brain infarcts
- BMI
body mass index
- BP
blood pressure
- DBP
diastolic blood pressure
- FLAIR
fluid-attenuated inversion recovery
- GRS
genetic risk scores
- GWAS
genome-wide association studies
- HDL
high-density lipoprotein
- IS
ischemic stroke
- IS-SVD
small vessel disease subtype of ischemic stroke
- IVW
inverse-variance weighting
- L10BF
Log10 of Bayesian factor
- LD
linkage disequilibrium
- LDL
low-density lipoprotein
- MAP
mean arterial pressure
- PP
pulse pressure
- SNP
single nucleotide polymorphism
- SBP
systolic blood pressure
- SSBI
small subcortical brain infarcts
- SVD
small vessel disease
- WMH
white matter hyperintensities
Contributor Information
Collaborators: Stroke Genetics Network (SiGN), the International Stroke Genetics Consortium (ISGC), METASTROKE, Alzheimer's Disease Genetics Consortium (ADGC), and the Neurology Working Group of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium, Peter Almgren, Christopher D. Anderson, Donna K. Arnett, John Attia, Hakan Ay, Oscar R. Benavente, Steve Bevan, Robert D. Brown, Mariana Bustamante, Yu-Ching Cheng, John W. Cole, Ioana Cotlarciuc, Carlos Cruchaga, Paul IW. de Bakker, Hossein Delavaran, Martin Dichgans, Gunnar Engström, Myriam Fornage, Raji P. Grewal, Laura Heitsch, Elizabeth Holliday, Laure Ibanez, Andreea Ilinca, Marguerite R. Irvin, Rebecca D. Jackson, Christina Jern, Jordi Jimenez-Conde, Julie A. Johnson, Katarina Jood, Brett M. Kissela, Steven J. Kittner, Dawn O. Kleindorfer, Daniel Labovitz, Cathy C. Laurie, Jin-Moo Lee, Robin Lemmens, Christopher Levi, Linxin Li, Arne G. Lindgren, Jane Maguire, Hugh S. Markus, Patrick F. McArdle, Olle Melander, James F. Meschia, Braxton D. Mitchell, Martina Müller-Nurasyid, Bo Norrving, Leema Reddy Peddareddygari, Joanna Pera, Sara L. Pulit, Kathryn Rexrode, Marta Ribasés, Jaume Roquer, Natalia S. Rost, Peter M. Rothwell, Tatjana Rundek, Ralph L. Sacco, Reinhold Schmidt, Pankaj Sharma, Agnieszka Slowik, Carolina Soriano-Tárraga, Tara Stanne, Konstantin Stauch, O C. Stine, Cathie LM. Sudlow, Vincent N.S. Thijs, Sylvia Wasssertheil-Smoller, David Weir, Stephen R. Williams, Quenna Wong, Daniel Woo, Bradford B. Worrall, Huichun Xu, Sudha Seshadri, Hyacinth I Hyacinth, Sandro Marini, Paul Nyquist, Cathryn Lewis, Bjorn Hansen, Bo Norrving, Jonathan Rosand, Alessandro Biffi, Christina Kourkoulis, Chris Anderson, Anne-Katrin Giese, Ralph Sacco, Pankaj Sharma, Jong-Won Chung, Gyeong-Moon Kim, Steven Lubitz, Romain Bourcier, Joanna Howson, Alessandra Granata, Anna Drazyk, Hugh Markus, Joanna Wardlaw, Braxton Mitchell, John Cole, Jemma Hopewell, Robin Walters, Iain Turnbull, Bradford Worrall, Josh Bis, Alex Reiner, Raj Dhar, Laura Heitsch, Jin-Moo Lee, Kameshwar Prasad, Chloé Sarnowski, Hugo Javier Aparicio, Qiong Yang, Daniel Chasman, Kathryn Rexrode, Chia-Ling Phuah, Guiyou Liu, Mitchell Elkind, Leslie Lange, Natalia Rost, Michael James, Jill Stewart, Dina Vojinovic, Vincent Thijs, Eugenio Parati, Giorgio Boncoraglio, Ramin Zand, Philippe Bijlenga, Magdy Selim, Caspar Grond-Ginsbach, Daniel Strbian, Liisa Tomppo, Hanne Sallinen, Dorothea Pfeiffer, Nuria Torres, Miguel Barboza, Melanie Laarman, Roberta Carriero, Elizabeth Holliday, Jordi Jimenez-Conde, Carolina Soriano, Dipender Gill, Stephanie Debette, Aniket Mishra, Jer-Yuarn Wu, Tai-Ming Ko, Silvia Bione, Katarina Jood, Turgut Tatlisumak, Lukas Holmegaard, Suo Yue, Anna bersano, Joanna Pera, Agnieszka Slowik, Christopher Levi, Kristina Schlicht, Robin Lemmens, Toshiharu Ninomiya, Saskia Lesnik Oberstein, Tsong-Hai Lee, Rainer Malik, Martin Dichgans, Arne Lindgren, Johan Wasselius, Mattias Drake, Olle Melander, Martin Stenman, Andreea Ilinca, Katherine Crawford, Umme Lena, Farrah Mateen, Hakan Ay, Ona Wu, Markus Schirmer, Steve Cramer, Polina Golland, Robert Brown, James Meschia, Owen A. Ross, Guillaume Pare, Mike Chong, Tatjana Rundek, Katrina Gwinn, Christopher Chen, Jim Koenig, Eva Giralt, Danish Saleheen, Frank-Erik de Leeuw, Karin Klijn, Yoichiro Kamatani, Michiaki Kubo, Yukinori Okada, Annie Pedersen, Maja Olsson, Juan José Martín, Huichun Xu, Eng King Tan, Petrea Frid, Chaeyoung Lee, David Tregouet, Thomas Leung, Richard Choy, Christina Jern, Keat Wei Loo, Gabriel Rinkel, Paulo Franca, Iscia Cendes, Caty Carrera, Israel Fernandez-Cadenas, Joan Montaner, Helen Kim, Mayowa Owolabi, Reecha Sofat, Mark Bakker, Ynte Ruigrok, Allard Hauer, Sara L. Pulit, Sander W. van der Laan, Ryan Irvin, Murali Sargurupremraj, Alessandro Pezzini, Foad Abd-Allah, David Liebeskind, Matthew Traylor, Rhea Tan, John Danesh, Loes Rutten-Jacobs, Amanda Donatti, Wagner Avelar, Joseph Broderick, Daniel Woo, Cathie Sudlow, Kristiina Rannikmae, Caitrin Wheeler McDonough, Tom van Agtmael, Matthew Walters, Martin Söderholm, Erik Lorentzen, Sandra Olsson, Tara Stanne, Martina Olsson, Rufus Akinyemi, Ioana Cotlatciuc, Patrick McArdle, Tushar Dave, Steven Kittner, John Attia, James E Faber, Iona Millwood, Elsa Valdés Márquez, Michelangelo Mancuso, Riina Vibo, Janika Korv, Jane Maguire, Myriam Fornage, Jennifer Majersik, Adam DeHavenon, Matthew Alexander, Michele Sale, Andrew Southerland, Debra Owens, Bruce Psaty, W. T. Longstreth, Jr, Stacey Quintero Wolfe, Carl Langefeld, Carlos Cruchaga, Jan Konrad, Kevin Sheth, Guido Falcone, Kathleen Donahue, Alexis N Simpkins, Tan Wei Liang Byorn, Bernard Chan, Phil Clatworthy, Jose Florez, Eric Harshfield, Atsushi Hozawa, Chung Hsu, Chaur-Jong Hu, Laure Ibanez, Masafumi Ihara, Marcos Lange, Soo Ji Lee, I-Hui Lee, Patricia Musolino, Hirofumi Nakatomi, Kwang-Yeol Park, Stephen S Rich, Chris Riley, Joohon Sung, Hideaki Suzuki, Katie Vo, Kazuo Washida, Laura Garcia Ibenez, Agnieszka Slowik, Albert Hofman, Ale Algra, Alex P Reiner, Alexander S F Doney, Andreas Gschwendtner, Andreea Ilinca, Anne-Katrin Giese, Arne Lindgren, Astrid M Vicente, Bo Norrving, Børge G Nordestgaard, Braxton D Mitchell, Bradford B Worrall, Bruce M Psaty, Cara L Carty, Cathie Sudlow, Christopher D Anderson, Christopher Levi, Claudia L Satizabal, Colin N A Palmer, Dale M Gamble, Daniel Woo, Danish Saleheen, E Bernd Ringelstein, Einar Valdimarsson, Elizabeth Holliday, Gail Davies, Ganesh Chauhan, Gerard Pasterkamp, Giorgio Boncoraglio, Gregor Kuhlenbäumer, Gudmar Thorleifsson, Guido J Falcone, Guillame Pare, Helena Schmidt, Hossein Delavaran, Hugh S Markus, Hugo J Aparicio, Ian Deary, Ioana Cotlarciuc, Israel Fernandez-Cadenas, James Meschia, Jemma C Hopewell, Jingmin Liu, Joan Montaner, Joanna Pera, John Cole, John R Attia, Jonathan Rosand, Jose M Ferro, Joshua Bis, Karen Furie, Kari Stefansson, Klaus Berger, Konstantinos Kostulas, Kristina Rannikmae, M Arfan Ikram, Marianne Benn, Martin Dichgans, Martin Farrall, Massimo Pandolfo, Matthew Traylor, Matthew Walters, Michele Sale, Mike Nalls, Myriam Fornage, Natalie R van Zuydam, Pankaj Sharma, Patricia Abrantes, Paul IW de Bakker, Peter Higgins, Peter Lichtner, Peter M Rothwell, Philippe Amouyel, Qiong Yang, Rainer Malik, Reinhold Schmidt, Robert Clarke, Robin Lemmens, Sander W van der Laan, Sara L Pulit, Sherine Abboud, Sofia A Oliveira, Solveig Gretarsdottir, Stephanie Debette, Stephen R Williams, Steve Bevan, Steven J Kittner, Sudha Seshadri, Thomas Mosley, Thomas WK Battey, Turgut Tatlisumak, Unnur Thorsteinsdottir, Vincent NS Thijs, W T Longstreth, Wei Zhao, Wei-Min Chen, Yu-Ching Cheng, Marilyn S. Albert, Roger L. Albin, Liana G. Apostolova, Steven E. Arnold, Sanjay Asthana, Craig S. Atwood, Clinton T. Baldwin, M. Michael Barmada, Lisa L. Barnes, Sandra Barral, Thomas G. Beach, James T. Becker, Gary W. Beecham, Duane Beekly, David A. Bennett, Eileen H. Bigio, Thomas D. Bird, Deborah Blacker, Bradley F. Boeve, Adam Boxer, James R. Burke, Jeffrey M. Burns, Joseph D. Buxbaum, Goldie S. Byrd, Guiqing Cai, Nigel J. Cairns, Laura B. Cantwell, Chuanhai Cao, Cynthia M. Carlsson, Regina M. Carney, Minerva M. Carrasquillo, Steven L. Carroll, Helena C. Chui, David G. Clark, David H. Cribbs, Elizabeth A. Crocco, Carlos Cruchaga, Philip L. De Jager, Charles DeCarli, F. Yesim Demirci, Malcolm Dick, Dennis W. Dickson, Ranjan Duara, Nilufer Ertekin-Taner, Denis A. Evans, Kelley M. Faber, M. Daniele Fallin, Kenneth B. Fallon, David W. Fardo, Martin R. Farlow, Lindsay A. Farrer, Steven Ferris, Tatiana M. Foroud, Matthew P. Frosch, Douglas R. Galasko, Marla Gearing, Daniel H. Geschwind, Bernardino Ghetti, John R. Gilbert, Rodney C.P. Go, Alison M. Goate, Neill R. Graff-Radford, Robert C. Green, Patrick Griffith, John H. Growdon, Jonathan L. Haines, Hakon Hakonarson, Ronald L. Hamilton, Kara L. Hamilton-Nelson, Vahram Haroutunian, Lindy E. Harrell, Lawrence S. Honig, Ryan M. Huebinger, Christine M. Hulette, Bradley T. Hyman, Gregory A. Jicha, Lee-Way Jin, Gyungah Jun, M. Ilyas Kamboh, Anna Karydas, John S.K. Kauwe, Jeffrey A. Kaye, Ronald Kim, Neil W. Kowall, Joel H. Kramer, Walter A. Kukull, Brian W. Kunkle, Frank M. LaFerla, James J. Lah, Rosalyn Lang-Walker, Eric B. Larson, James B. Leverenz, Allan I. Levey, Ge Li, Andrew P. Lieberman, Mark W. Logue, Oscar L. Lopez, Kathryn L. Lunetta, Constantine G. Lyketsos, Wendy J. Mack, Jennifer J. Manly, Daniel C. Marson, Eden R. Martin, Frank Martiniuk, Deborah C. Mash, Eliezer Masliah, Richard Mayeux, Ann C. McKee, Marsel Mesulam, Bruce L. Miller, Carol A. Miller, Joshua W. Miller, Thomas J. Montine, John C. Morris, Jill R. Murrell, Adam C. Naj, Thomas O. Obisesan, John M. Olichney, Vernon S. Pankratz, Joseph E. Parisi, Amanda Partch, Henry L. Paulson, Margaret A. Pericak-Vance, William Perry, Elaine Peskind, Ronald C. Petersen, Aimee Pierce, Wayne W. Poon, Huntington Potter, Joseph F. Quinn, Ashok Raj, Towfique Raj, Murray Raskind, Eric M. Reiman, Barry Reisberg, Christiane Reitz, John M. Ringman, Erik D. Roberson, Howard J. Rosen, Roger N. Rosenberg, Mark A. Sager, Mary Sano, Andrew J. Saykin, Gerard D. Schellenberg, Julie A. Schneider, Lon S. Schneider, William W. Seeley, Amanda G. Smith, Joshua A. Sonnen, Salvatore Spina, Robert A. Stern, Russell H. Swerdlow, Rudolph E. Tanzi, Tricia A. Thornton-Wells, John Q. Trojanowski, Juan C. Troncoso, Debby W. Tsuang, Otto Valladares, Vivianna M. Van Deerlin, Linda J. Van Eldik, Badri N. Vardarajan, Harry V. Vinters, Jean Paul Vonsattel, Li-San Wang, Sandra Weintraub, Kathleen A. Welsh-Bohmer, Jennifer Williamson, Thomas S. Wingo, Sarah Wishnek, Randall L. Woltjer, Clinton B. Wright, Steven G. Younkin, Chang-En Yu, Lei Yu, Ganesh Chauhan, Audrey Y. Chu, Myriam Fornage, Joshua C. Bis, Aki S. Havulinna, Muralidharan Sargurupremraj, Albert Vernon Smith, Hieab H.H. Adams, Seung Hoan Choi, Stella Trompet, Melissa E. Garcia, Ani Manichaikul, Alexander Teumer, Stefan Gustafsson, Traci M. Bartz, Céline Bellenguez, Jean Sebastien Vidal, Xueqiu Jian, Olafur Kjartansson, Kerri L. Wiggins, Claudia L. Satizabal, Flora Xue, Samuli Ripatti, Yongmei Liu, Joris Deelen, Marcel den Hoed, Susan R. Heckbert, Kenneth Rice, Nicholas L. Smith, Quenna Wong, Hugo J. Aparicio, Julie E. Buring, Paul M Ridker, Claudine Berr, Jean-François Dartigues, Anders Hamsten, Patrik K. Magnusson, Nancy L. Pedersen, Lars Lannfelt, Lars Lind, Cecilia M. Lindgren, Andrew P. Morris, Albert Hofman, Peter J. Koudstaal, Marileen LP. Portegies, André G. Uitterlinden, Anton JM de Craen, Ian Ford, J. Wouter Jukema, David J Stott, Norrina B. Allen, Michele M. Sale, Andrew D Johnson, David A. Bennett, Philip L. De Jager, Charles C. White, Hans Jörgen Grabe, Marcello Ricardo Paulista Markus, Oscar L Lopez, Jerome I. Rotter, Michael A. Nalls, Rebecca F. Gottesman, Michael E. Griswold, David S. Knopman, B. Gwen Windham, Alexa Beiser, Erkki Vartiainen, Curtis R. French, Tobias Kurth, Bruce M. Psaty, Tamara B. Harris, Stephen S Rich, Anita L. deStefano, Carsten Oliver Schmidt, Veikko Salomaa, Thomas H. Mosley, Erik Ingelsson, Cornelia M. van Duijn, Christophe Tzourio, Lenore J Launer, M. Arfan Ikram, Daniel I. Chasman, W. T. Longstreth, Jr, Sudha Seshadri, Stéphanie Debette, Benjamin F.J. Verhaaren, Stéphanie Debette, Joshua C. Bis, Jennifer A. Smith, M. Kamran Ikram, Hieab H. Adams, Ashley H. Beecham, Kumar B. Rajan, Lorna M. Lopez, Sandra Barral, Mark A. van Buchem, Jeroen van der Grond, Albert V. Smith, Katrin Hegenscheid, Neelum T. Aggarwal, Mariza de Andrade, Elizabeth J. Atkinson, Marian Beekman, Alexa S. Beiser, Susan H. Blanton, Eric Boerwinkle, Adam M. Brickman, R. Nick Bryan, Ganesh Chauhan, Christopher P.L.H. Chen, Vincent Chouraki, Anton J.M. de Craen, Fabrice Crivello, Ian J. Deary, Joris Deelen, Philip L. De Jager, Carole Dufouil, Mitchell S.V. Elkind, Denis A. Evans, Paul Freudenberger, Rebecca F. Gottesman, Vilmundur Guðnason, Mohamad Habes, Susan R. Heckbert, Gerardo Heiss, Saima Hilal, Edith Hofer, Albert Hofman, Carla A. Ibrahim-Verbaas, David S. Knopman, Cora E. Lewis, Jiemin Liao, David C.M. Liewald, Michelle Luciano, Aad van der Lugt, Oliver O. Martinez, Richard Mayeux, Bernard Mazoyer, Mike Nalls, Matthias Nauck, Wiro J. Niessen, Ben A. Oostra, Bruce M. Psaty, Kenneth M. Rice, Jerome I. Rotter, Bettina von Sarnowski, Helena Schmidt, Pamela J. Schreiner, Maaike Schuur, Stephen S. Sidney, Sigurdur Sigurdsson, P. Eline Slagboom, David J.M. Stott, John C. van Swieten, Alexander Teumer, Anna Maria Töglhofer, Matthew Traylor, Stella Trompet, Stephen T. Turner, Christophe Tzourio, Hae-Won Uh, André G. Uitterlinden, Meike W. Vernooij, Jing J. Wang, Tien Y. Wong, Joanna M. Wardlaw, B. Gwen Windham, Katharina Wittfeld, Christiane Wolf, Clinton B. Wright, Qiong Yang, Wei Zhao, Alex Zijdenbos, J. Wouter Jukema, Ralph L. Sacco, Sharon L.R. Kardia, Philippe Amouyel, Thomas H. Mosley, W. T. Longstreth, Jr, Charles C. DeCarli, Cornelia M. van Duijn, Reinhold Schmidt, Lenore J. Launer, Hans J. Grabe, Sudha S. Seshadri, M. Arfan Ikram, Myriam Fornage, Joshua C. Bis, Maryam Kavousi, Nora Franceschini, Aaron Isaacs, Gonçalo R Abecasis, Ulf Schminke, Wendy Post, Albert V. Smith, L. Adrienne Cupples, Hugh S Markus, Reinhold Schmidt, Jennifer E. Huffman, Terho Lehtimäki, Jens Baumert, Thomas Münzel, Susan R. Heckbert, Abbas Dehghan, Kari North, Ben Oostra, Steve Bevan, Eva-Maria Stoegerer, Caroline Hayward, Olli Raitakari, Christa Meisinger, Arne Schillert, Serena Sanna, Henry Völzke, Yu-Ching Cheng, Bolli Thorsson, Caroline S. Fox, Kenneth Rice, Fernando Rivadeneira, Vijay Nambi, Eran Halperin, Katja E. Petrovic, Leena Peltonen, H. Erich Wichmann, Renate B. Schnabel, Marcus Dörr, Afshin Parsa, Thor Aspelund, Serkalem Demissie, Sekar Kathiresan, Muredach P. Reilly, Kent Taylor, Andre Uitterlinden, David J. Couper, Matthias Sitzer, Mika Kähönen, Thomas Illig, Philipp S. Wild, Marco Orru, Jan Lüdemann, Alan R. Shuldiner, Gudny Eiriksdottir, Charles C. White, Jerome I. Rotter, Albert Hofman, Jochen Seissler, Tanja Zeller, Gianluca Usala, Florian Ernst, Lenore J. Launer, Ralph B. D'Agostino, Sr, Daniel H. O'Leary, Christie Ballantyne, Joachim Thiery, Andreas Ziegler, Edward G. Lakatta, Ravi Kumar Chilukoti, Tamara B. Harris, Philip A. Wolf, Bruce M. Psaty, Joseph F Polak, Xia Li, Wolfgang Rathmann, Manuela Uda, Eric Boerwinkle, Norman Klopp, Helena Schmidt, James F Wilson, Jorma Viikari, Wolfgang Koenig, Stefan Blankenberg, Anne B. Newman, Jacqueline Witteman, Gerardo Heiss, Cornelia van Duijn, Angelo Scuteri, Georg Homuth, Braxton D. Mitchell, Vilmundur Gudnason, and Christopher J. O’Donnell
Author contributions
Drs. Chauhan, Adams, Satizabal, Bis, Teumer, and Sargurupremraj contributed equally to this work. Drs. Ikram, Fornage, Launer, Seshadri, Longstreth, and Debette jointly supervised this work. Study design/conception: Drs. Schofield, Tzourio, Schellenberg, Kato, Psaty, Slagboom, Stott, Ford, Jukema, Fornage, Seshadri, Longstreth, and Debette. Statistical analysis: Drs. Schofield, Chauhan, Sargurupremraj, Satizabal, Teumer, Hofer, Trompet, Hilal, Smith, Malik, Saba, Takeuchi, Rice, van der Lee, Luciano, Deelen, Uh, Beecham, Thalamuthu, Armstrong, Grabe, and McWhirter. Sample/phenotype contribution: Drs. Phan, Schofield, Kwok, Hilal, Malik, Traylor, Tzourio, Schellenberg, Knopman, Griswold, Windham, Mosley, Schmidt, Schmidt, Yamaguchi, Nabika, Psaty, Lopez, Chen, Cheng, Amin, Aparicio, DeCarli, del Carmen Valdés Hernández, Deary, Starr, Bastin, Maniega, Beekman, Lemmens, Brodaty, Wright, Boncoraglio, Hopewell, Blanton, Wright, Sacco, Wen, Chong, van der Grond, Stott, Ford, Jukema, van der Lugt, Wittfeld, Grabe, Hosten, von Sarnowski, Völker, Jimenez-Conde, Sharma, Rosand, Cole, Thijs, Lindgren, Rundek, Rexrode, Arnett, Johnson, Benavente, Wasssertheil-Smoller, Lee, Mitchell, McArdle, Moran, Callisaya, Mather, Sachdev, van Duijn, Dichgans, Kittner, Markus, Launer, Longstreth, and Debette. Drafting the article: Drs. Chauhan, Seshadri, Longstreth, and Debette. Critical revision of the article: Drs. Chauhan, Satizabal, Teumer, Hofer, Trompet, Malik, Traylor, Tzourio, Windham, Mosley, Schmidt, Yamaguchi, Kato, Psaty, Rice, Lopez, Chen, Aparicio, DeCarli, del Carmen Valdés Hernández, Luciano, Deary, Bastin, Maniega, Slagboom, Beekman, Deelen, Uh, Lemmens, Wright, Boncoraglio, Wen, Thalamuthu, Armstrong, van der Grond, Stott, Ford, Jukema, van der Lugt, Wittfeld, Grabe, Hosten, von Sarnowski, Jimenez-Conde, Rosand, Thijs, Lindgren, Rundek, Lee, Mitchell, Rich, Geerlings, McWhirter, Callisaya, Mather, Sachdev, Dichgans, Kittner, Markus, Launer, Seshadri, Longstreth, Debette, Adams, Bis, Jian, Pulit, Amouyel, Mazoyer, Zhu, Sargurupremraj, Kaffashian, Beecham, Montine, Kjartansson, Gudnason, Gottesman, Rajan, Aggarwal, De Jager, Evans, Rotter, Liao, Wong, Ikram, Chouraki, DeStefano, Romero, Maillard, Wardlaw, Ames, Vernooij, Hofman, Uitterlinden, Levi, Sudlow, Woo, Meschia, Slowik, Melander, Grewal, Rothwell, Jern, van der Graaf, de Bakker, Asselbergs, Thomson, Rutten-Jacobs, Bevan, Worrall, Ikram, Smith, and Schilling. Funding: Drs. Schofield, Trompet, Tzourio, Schellenberg, Mosley, Schmidt, Schmidt, Psaty, Cheng, Deary, Starr, Bastin, Slagboom, Wright, Wen, Stott, Ford, Jukema, Völker, Rosand, Lindgren, Johnson, Srikanth, Mather, Sachdev, Markus, Fornage, Seshadri, Longstreth, and Debette. Supervision: Drs. Kwok, Schellenberg, Psaty, Amin, Slagboom, Wen, Jukema, Grabe, Mather, Sachdev, van Duijn, Fornage, Seshadri, Longstreth, and Debette. Final approval of the version to be published: Drs. Satizabal, Teumer, Hofer, Trompet, Hilal, Malik, Traylor, Tzourio, Schellenberg, Griswold, Windham, Mosley, Schmidt, Schmidt, Takeuchi, Nabika, Kato, Psaty, Rice, Lopez, Chen, Cheng, van der Lee, Amin, Aparicio, DeCarli, del Carmen Valdés Hernández, Luciano, Deary, Starr, Bastin, Maniega, Slagboom, Beekman, Deelen, Uh, Lemmens, Brodaty, Wright, Boncoraglio, Hopewell, Beecham, Wen, Thalamuthu, Armstrong, Chong, van der Grond, Stott, Ford, Jukema, van der Lugt, Wittfeld, Grabe, Hosten, von Sarnowski, Völker, Jimenez-Conde, Rosand, Cole, Thijs, Lindgren, Rundek, Rexrode, Johnson, Wasssertheil-Smoller, Wong, Mitchell, Rich, McArdle, Geerlings, McWhirter, Moran, Callisaya, Mather, Sachdev, van Duijn, Dichgans, Markus, Fornage, Launer, Seshadri, Longstreth, Debette, Kittner, Adams, Bis, Jian, Pulit, Amouyel, Mazoyer, Zhu, Sargurupremraj, Kaffashian, Beecham, Montine, Kjartansson, Gudnason, Gottesman, Rajan, Aggarwal, De Jager, Evans, Rotter, Liao, Wong, Ikram, Chouraki, DeStefano, Romero, Maillard, Wardlaw, Ames, Vernooij, Hofman, Uitterlinden, Levi, Sudlow, Woo, Meschia, Slowik, Melander, Grewal, Rothwell, Jern, der Graaf, de Bakker, Asselbergs, Thomson, Rutten-Jacobs, Bevan, Worrall, Ikram, and Smith.
Study funding
CHAP: R01-AG-11101, R01-AG-030146, NIRP-14-302587. SMART: This study was supported by a grant from the Netherlands Organization for Scientific Research–Medical Sciences (project no. 904-65–095). LBC: The authors thank the LBC1936 participants and the members of the LBC1936 research team who collected and collated the phenotypic and genotypic data. The LBC1936 is supported by Age UK (Disconnected Mind Programme grant). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross-council Lifelong Health and Wellbeing Initiative (MR/K026992/1). The brain imaging was performed in the Brain Research Imaging Centre (https://www.ed.ac.uk/clinical-sciences/edinburgh-imaging), a center in the SINAPSE Collaboration (sinapse.ac.uk) supported by the Scottish Funding Council and Chief Scientist Office. Funding from the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the UK Medical Research Council is acknowledged. Genotyping was supported by a grant from the BBSRC (ref. BB/F019394/1). PROSPER: The PROSPER study was supported by an investigator-initiated grant obtained from Bristol-Myers Squibb. Prof. Dr. J.W. Jukema is an Established Clinical Investigator of the Netherlands Heart Foundation (grant 2001 D 032). Support for genotyping was provided by the seventh framework program of the European commission (grant 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging grant 050-060-810). SCES and SiMES: National Medical Research Council Singapore Centre Grant NMRC/CG/013/2013. C.-Y.C. is supported by the National Medical Research Council, Singapore (CSA/033/2012), Singapore Translational Research Award (STaR) 2013. Dr. Kamran Ikram received additional funding from the Singapore Ministry of Health's National Medical Research Council (NMRC/CSA/038/2013). SHIP: SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs, as well as the Social Ministry of the Federal State of Mecklenburg–West Pomerania, and the network “Greifswald Approach to Individualized Medicine (GANI_MED)” funded by the Federal Ministry of Education and Research (grant 03IS2061A). Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthineers, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. Whole-body MRI was supported by a joint grant from Siemens Healthineers, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the Caché Campus program of the InterSystems GmbH. OATS (Older Australian Twins Study): OATS was supported by an Australian National Health and Medical Research Council (NHRMC)/Australian Research Council (ARC) Strategic Award (ID401162) and by a NHMRC grant (ID1045325). OATS was facilitated via access to the Australian Twin Registry, which is supported by the NHMRC Enabling Grant 310667. The OATS genotyping was partly supported by a Commonwealth Scientific and Industrial Research Organisation Flagship Collaboration Fund Grant. NOMAS: The Northern Manhattan Study is funded by the NIH grant “Stroke Incidence and Risk Factors in a Tri-Ethnic Region” (NINDS R01NS 29993). TASCOG: NHMRC and Heart Foundation. AGES: The study was funded by the National Institute on Aging (NIA) (N01-AG-12100), Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament), with contributions from the Intramural Research Programs at the NIA, the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute of Neurological Disorders and Stroke (NINDS) (Z01 HL004607-08 CE). ERF: The ERF study as a part of European Special Populations Research Network (EUROSPAN) was supported by European Commission FP6 STRP grant no. 018947 (LSHG-CT-2006-01947) and also received funding from the European Community's Seventh Framework Programme (FP7/2007–2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme “Quality of Life and Management of the Living Resources” of 5th Framework Programme (no. QLG2-CT-2002-01254). High-throughput analysis of the ERF data was supported by a joint grant from Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). Exome sequencing analysis in ERF was supported by the ZonMw grant (project 91111025). Najaf Amin is supported by the Netherlands Brain Foundation (project no. F2013[1]-28). ARIC: The Atherosclerosis Risk in Communities study was performed as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HSN268201100006C, HSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL70825, R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and NIH contract HHSN268200625226C. Infrastructure was partly supported by grant no. UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. This project was also supported by NIH R01 grant NS087541 to M.F. FHS: This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (contracts no. N01-HC-25195 and no. HHSN268201500001I), and its contract with Affymetrix, Inc. for genotyping services (contract no. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This study was also supported by grants from the NIA (R01s AG033040, AG033193, AG054076, AG049607, AG008122, and U01-AG049505) and the NINDS (R01-NS017950, UH2 NS100605). Dr. DeCarli is supported by the Alzheimer's Disease Center (P30 AG 010129). ASPS: The research reported in this article was funded by the Austrian Science Fund (FWF) grant nos. P20545-P05, P13180, and P20545-B05, by the Austrian National Bank Anniversary Fund, P15435, and the Austrian Ministry of Science under the aegis of the EU Joint Programme–Neurodegenerative Disease Research (JPND) (jpnd.eu). LLS: The Leiden Longevity Study has received funding from the European Union's Seventh Framework Programme (FP7/2007–2011) under grant agreement no. 259679. This study was supported by a grant from the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology, and the Netherlands Consortium for Healthy Ageing (grant 050-060-810), all in the framework of the Netherlands Genomics Initiative, Netherlands Organization for Scientific Research (NWO), UnileverColworth, and by BBMRI-NL, a Research Infrastructure financed by the Dutch government (NWO 184.021.007). CHS: This CHS research was supported by contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, N01HC15103, and HHSN268200960009C and grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, R01HL085251, and R01HL130114 from the NHLBI with additional contribution from NINDS. Additional support was provided through R01AG023629 from the NIA. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Rotterdam Study: The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research (NWO) Investments (no. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/NWO project no. 050-060-810. The Rotterdam Study is funded by Erasmus MC Medical Center and Erasmus MC University, Rotterdam, Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. M.A.I. is supported by an NWO Veni grant (916.13.054). The 3-City Study: The 3-City Study is conducted under a partnership agreement among the Institut National de la Santé et de la Recherche Médicale (INSERM), the University of Bordeaux, and Sanofi-Aventis. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study is also supported by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Mutuelle Générale de l’Education Nationale (MGEN), Institut de la Longévité, Conseils Régionaux of Aquitaine and Bourgogne, Fondation de France, and Ministry of Research–INSERM Programme “Cohortes et collections de données biologiques.” C.T. and S.D. have received investigator-initiated research funding from the French National Research Agency (ANR) and from the Fondation Leducq. S.D. is supported by a starting grant from the European Research Council (SEGWAY), a grant from the Joint Programme of Neurodegenerative Disease research (BRIDGET), from the European Union's Horizon 2020 research and innovation programme under grant agreements No 643417 & No 640643, and by the Initiative of Excellence of Bordeaux University. Part of the computations were performed at the Bordeaux Bioinformatics Center (CBiB), University of Bordeaux. This work was supported by the National Foundation for Alzheimer's Disease and Related Disorders, the Institut Pasteur de Lille, the Labex DISTALZ, and the Centre National de Génotypage. ADGC: The Alzheimer Disease Genetics Consortium is supported by NIH. NIH-NIA supported this work through the following grants: ADGC, U01 AG032984, RC2 AG036528; NACC, U01 AG016976; NCRAD, U24 AG021886; NIA LOAD, U24 AG026395, U24 AG026390; Banner Sun Health Research Institute, P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01 AG025259, R01AG33193; Columbia University, P50 AG008702, R37 AG015473; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, UO1 AG06781, UO1 HG004610; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, MO1RR00096, UL1 RR029893, 5R01AG012101, 5R01AG022374, 5R01AG013616, 1RC2AG036502, 1R01AG035137; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG30146; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582, UL1RR02777; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573, P50, P50 AG016575, P50 AG016576, P50 AG016577; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383, AG05144; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991. The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by NINDS grant NS39764, NIMH MH60451, and by GlaxoSmithKline. Genotyping of the TGEN2 cohort was supported by Kronos Science. The TGen series was also funded by NIA grant AG041232, the Banner Alzheimer's Foundation, The Johnnie B. Byrd Sr. Alzheimer's Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council [MRC], local NHS trusts, and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council), South West Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England [HEFCE], Alzheimer's Research Trust [ART], BRACE, as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), The Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, Brain Net Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, and Universitat de Barcelona). ADNI: Funding for ADNI is through the Northern California Institute for Research and Education by grants from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., Alzheimer's Association, Alzheimer's Drug Discovery Foundation, the Dana Foundation, and the National Institute of Biomedical Imaging and Bioengineering and NIA grants U01 AG024904, RC2 AG036535, and K01 AG030514. Support was also provided by the Alzheimer's Association (LAF, IIRG-08-89720; MAP-V, IIRG-05-14147) and the US Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. SiGN: Stroke Genetic Network (SiGN) was supported in part by award nos. U01NS069208 and R01NS100178 from NINDS. Genetics of Early-Onset Stroke (GEOS) Study was supported by the NIH Genes, Environment and Health Initiative (GEI) grant U01 HG004436, as part of the GENEVA consortium under GEI, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488); and the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the NIH to Johns Hopkins University (contract no. HHSN268200782096C). Assistance with data cleaning was provided by the GENEVA Coordinating Center (U01 HG 004446; PI Bruce S. Weir). Study recruitment and assembly of datasets were supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and Prevention, and by grants from NINDS and the NIH Office of Research on Women's Health (R01 NS45012, U01 NS069208-01). METASTROKE: ASGC: Australian population control data were derived from the Hunter Community Study. This research was funded by grants from the Australian National and Medical Health Research Council (NHMRC Project Grant ID: 569257), the Australian National Heart Foundation (NHF Project Grant ID: G 04S 1623), the University of Newcastle, the Gladys M Brawn Fellowship scheme, and the Vincent Fairfax Family Foundation in Australia. E.G.H. was supported by a Fellowship from the NHF and National Stroke Foundation of Australia (ID: 100071). J.M. was supported by an Australian Postgraduate Award. BRAINS: Bio-Repository of DNA in Stroke (BRAINS) is partly funded by a Senior Fellowship from the Department of Health (UK) to P.S., the Henry Smith Charity, and the UK-India Education Research Institutive (UKIERI) from the British Council. GEOS: Genetics of Early Onset Stroke (GEOS) Study, Baltimore, was supported by GEI Grant U01 HG004436, as part of the GENEVA consortium under GEI, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488), and the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract no. HHSN268200782096C). Assistance with data cleaning was provided by the GENEVA Coordinating Center (U01 HG 004446; PI Bruce S. Weir). Study recruitment and assembly of datasets were supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and Prevention, and by grants from NINDS and the NIH Office of Research on Women's Health (R01 NS45012, U01 NS069208-01). HPS: Heart Protection Study (HPS) (ISRCTN48489393) was supported by the UK MRC, British Heart Foundation, Merck and Co. (manufacturers of simvastatin), and Roche Vitamins Ltd. (manufacturers of vitamins). Genotyping was supported by a grant to Oxford University and CNG from Merck and Co. J.C.H. acknowledges support from the British Heart Foundation (FS/14/55/30806). ISGS: Ischemic Stroke Genetics Study (ISGS)/Siblings With Ischemic Stroke Study (SWISS) was supported in part by the Intramural Research Program of the NIA, NIH project Z01 AG-000954-06. ISGS/SWISS used samples and clinical data from the NIH-NINDS Human Genetics Resource Center DNA and Cell Line Repository (ccr.coriell.org/ninds), human subjects protocol nos. 2003-081 and 2004-147. ISGS/SWISS used stroke-free participants from the Baltimore Longitudinal Study of Aging (BLSA) as controls. The inclusion of BLSA samples was supported in part by the Intramural Research Program of the NIA, NIH project Z01 AG-000015-50, human subjects protocol no. 2003-078. The ISGS study was funded by NIH-NINDS Grant R01 NS-42733 (J.F.M.). The SWISS study was funded by NIH-NINDS Grant R01 NS-39987 (J.F.M.). This study used the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (biowulf.nih.gov). MGH-GASROS: MGH Genes Affecting Stroke Risk and Outcome Study (MGH-GASROS) was supported by NINDS (U01 NS069208), the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research 0775010N, the NIH and NHLBI's STAMPEED genomics research program (R01 HL087676), and a grant from the National Center for Research Resources. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research resources. Milan: Milano–Besta Stroke Register Collection and genotyping of the Milan cases within CEDIR were supported by the Italian Ministry of Health (grant nos.: RC 2007/LR6, RC 2008/LR6; RC 2009/LR8; RC 2010/LR8; GR-2011-02347041), FP6 LSHM-CT-2007-037273 for the PROCARDIS control samples. WTCCC2: Wellcome Trust Case-Control Consortium 2 (WTCCC2) was principally funded by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z and WT084724MA). The Stroke Association provided additional support for collection of some of the St George's, London cases. The Oxford cases were collected as part of the Oxford Vascular Study, which is funded by the MRC, Stroke Association, Dunhill Medical Trust, National Institute of Health Research (NIHR), and the NIHR Biomedical Research Centre, Oxford. The Edinburgh Stroke Study was supported by the Wellcome Trust (clinician scientist award to C.L.M.S.) and the Binks Trust. Sample processing occurred in the Genetics Core Laboratory of the Wellcome Trust Clinical Research Facility, Western General Hospital, Edinburgh. Much of the neuroimaging occurred in the Scottish Funding Council Brain Imaging Research Centre (https://www.ed.ac.uk/clinical-sciences/edinburgh-imaging), Division of Clinical Neurosciences, University of Edinburgh, a core area of the Wellcome Trust Clinical Research Facility, and part of the SINAPSE (Scottish Imaging Network: A Platform for Scientific Excellence) collaboration (sinapse.ac.uk), funded by the Scottish Funding Council and the Chief Scientist Office. Collection of the Munich cases and data analysis was supported by the Vascular Dementia Research Foundation. This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreements no. 666881, SVDs@target (to M.D.) and no. 667375, CoSTREAM (to M.D.); the DFG as part of the Munich Cluster for Systems Neurology (EXC 1010 SyNergy) and the CRC 1123 (B3) (to M.D.); the Corona Foundation (to M.D.); the Fondation Leducq (Transatlantic Network of Excellence on the Pathogenesis of Small Vessel Disease of the Brain) (to M.D.); the e:Med program (e:AtheroSysMed) (to M.D.) and the FP7/2007-2103 European Union project CVgenes@target (grant agreement no. Health-F2-2013-601456) (to M.D.). M.F. and A.H. acknowledge support from the BHF Centre of Research Excellence in Oxford and the Wellcome Trust core award (090532/Z/09/Z). VISP: The GWAS component of the Vitamin Intervention for Stroke Prevention (VISP) study was supported by the US National Human Genome Research Institute (NHGRI), grant U01 HG005160 (PI Michèle Sale and Bradford Worrall), as part of the Genomics and Randomized Trials Network (GARNET). Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the NIH to Johns Hopkins University. Assistance with data cleaning was provided by the GARNET Coordinating Center (U01 HG005157; PI Bruce S. Weir). Study recruitment and collection of datasets for the VISP clinical trial were supported by an investigator-initiated research grant (R01 NS34447; PI James Toole) from the US Public Health Service, NINDS, Bethesda, MD. Control data obtained through the database of genotypes and phenotypes (dbGAP) maintained and supported by the United States National Center for Biotechnology Information, US National Library of Medicine. WHI: Funding support for WHI-GARNET was provided through the NHGRI GARNET (grant no. U01 HG005152). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GARNET Coordinating Center (U01 HG005157). Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the GEI (U01 HG004424). R.L. is a senior clinical investigator of FWO Flanders. F.W.A. is supported by a Dekker scholarship-Junior Staff Member 2014T001–Netherlands Heart Foundation and UCL Hospitals NIHR Biomedical Research Centre.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Vermeer SE, Longstreth WT Jr, Koudstaal PJ. Silent brain infarcts: a systematic review. Lancet Neurol 2007;6:611–619. [DOI] [PubMed] [Google Scholar]
- 2.Longstreth WT Jr. Brain vascular disease: overt and covert. Stroke 2005;36:2062–2063. [DOI] [PubMed] [Google Scholar]
- 3.Saini M, Ikram K, Hilal S, Qiu A, Venketasubramanian N, Chen C. Silent stroke: not listened to rather than silent. Stroke 2012;43:3102–3104. [DOI] [PubMed] [Google Scholar]
- 4.Debette S, Beiser A, DeCarli C, et al. Association of MRI markers of vascular brain injury with incident stroke, mild cognitive impairment, dementia, and mortality: the Framingham Offspring Study. Stroke 2010;41:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernick C, Kuller L, Dulberg C, et al. Silent MRI infarcts and the risk of future stroke: the Cardiovascular Health Study. Neurology 2001;57:1222–1229. [DOI] [PubMed] [Google Scholar]
- 6.Longstreth WT Jr, Dulberg C, Manolio TA, et al. Incidence, manifestations, and predictors of brain infarcts defined by serial cranial magnetic resonance imaging in the elderly: the Cardiovascular Health Study. Stroke 2002;33:2376–2382. [DOI] [PubMed] [Google Scholar]
- 7.Schilling S, DeStefano AL, Sachdev PS, et al. APOE genotype and MRI markers of cerebrovascular disease: systematic review and meta-analysis. Neurology 2013;81:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison AC, Ballantyne CM, Bray M, Chambless LE, Sharrett AR, Boerwinkle E. LPL polymorphism predicts stroke risk in men. Genet Epidemiol 2002;22:233–242. [DOI] [PubMed] [Google Scholar]
- 9.van Rijn MJ, Bos MJ, Yazdanpanah M, et al. Alpha-adducin polymorphism, atherosclerosis, and cardiovascular and cerebrovascular risk. Stroke 2006;37:2930–2934. [DOI] [PubMed] [Google Scholar]
- 10.Song J, Kim OJ, Kim HS, et al. Endothelial nitric oxide synthase gene polymorphisms and the risk of silent brain infarction. Int J Mol Med 2010;25:819–823. [DOI] [PubMed] [Google Scholar]
- 11.Jenny NS, Tracy RP, Ogg MS, et al. In the elderly, interleukin-6 plasma levels and the -174G>C polymorphism are associated with the development of cardiovascular disease. Arterioscler Thromb Vasc Biol 2002;22:2066–2071. [DOI] [PubMed] [Google Scholar]
- 12.Fornage M, Chiang YA, O'Meara ES, et al. Biomarkers of inflammation and MRI-defined small vessel disease of the brain: the Cardiovascular Health Study. Stroke 2008;39:1952–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serizawa M, Nabika T, Ochiai Y, et al. Association between PRKCH gene polymorphisms and subcortical silent brain infarction. Atherosclerosis 2008;199:340–345. [DOI] [PubMed] [Google Scholar]
- 14.van Oijen M, Cheung EY, Geluk CE, et al. Haplotypes of the fibrinogen gene and cerebral small vessel disease: the Rotterdam scan study. J Neurol Neurosurg Psychiatry 2008;79:799–803. [DOI] [PubMed] [Google Scholar]
- 15.Jeon YJ, Kim OJ, Kim SY, et al. Association of the miR-146a, miR-149, miR-196a2, and miR-499 polymorphisms with ischemic stroke and silent brain infarction risk. Arterioscler Thromb Vasc Biol 2013;33:420–430. [DOI] [PubMed] [Google Scholar]
- 16.Debette S, Bis JC, Fornage M, et al. Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke 2010;41:210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu YC, Dufouil C, Tzourio C, Chabriat H. Silent brain infarcts: a review of MRI diagnostic criteria. Stroke 2011;42:1140–1145. [DOI] [PubMed] [Google Scholar]
- 18.Verhaaren BF, Debette S, Bis JC, et al. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet 2015;8:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol 2011;35:809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Chua HX, Chen P, et al. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum Mol Genet 2013;22:2303–2311. [DOI] [PubMed] [Google Scholar]
- 23.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franceschini N, van Rooij FJ, Prins BP, et al. Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am J Hum Genet 2012;91:744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beecham GW, Hamilton K, Naj AC, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer's disease and related dementias. PLoS Genet 2014;10:e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet 2015;16:327–350. [DOI] [PubMed] [Google Scholar]
- 27.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong H, Prentice RL. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics 2008;9:621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trask TM, Ritty TM, Broekelmann T, Tisdale C, Mecham RP. N-terminal domains of fibrillin 1 and fibrillin 2 direct the formation of homodimers: a possible first step in microfibril assembly. Biochem J 1999;340:693–701. [PMC free article] [PubMed] [Google Scholar]
- 30.Ratnapriya R, Zhan X, Fariss RN, et al. Rare and common variants in extracellular matrix gene Fibrillin 2 (FBN2) are associated with macular degeneration. Hum Mol Genet 2014;23:5827–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren HR, Evangelou E, Cabrera CP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet 2017;49:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Del-Aguila JL, Beitelshees AL, Cooper-Dehoff RM, et al. Genome-wide association analyses suggest NELL1 influences adverse metabolic response to HCTZ in African Americans. Pharmacogenomics J 2014;14:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fanning JP, Wesley AJ, Wong AA, Fraser JF. Emerging spectra of silent brain infarction. Stroke 2014;45:3461–3471. [DOI] [PubMed] [Google Scholar]
- 34.Volpe M, Rosei EA, Ambrosioni E, Leonetti G, Trimarco B, Mancia G. Reduction in estimated stroke risk associated with practice-based stroke-risk assessment and awareness in a large, representative population of hypertensive patients: results from the ForLife study in Italy. J Hypertens 2007;25:2390–2397. [DOI] [PubMed] [Google Scholar]
- 35.Hankey GJ, Study I, the ET. Stroke: fresh insights into causes, prevention, and treatment. Lancet Neurol 2011;10:2–3. [DOI] [PubMed] [Google Scholar]
- 36.Tikhonoff V, Zhang H, Richart T, Staessen JA. Blood pressure as a prognostic factor after acute stroke. Lancet Neurol 2009;8:938–948. [DOI] [PubMed] [Google Scholar]
- 37.Fanning JP, Wong AA, Fraser JF. The epidemiology of silent brain infarction: a systematic review of population-based cohorts. BMC Med 2014;12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freiberg JJ, Tybjaerg-Hansen A, Jensen JS, Nordestgaard BG. Nonfasting triglycerides and risk of ischemic stroke in the general population. JAMA 2008;300:2142–2152. [DOI] [PubMed] [Google Scholar]
- 39.Emerging Risk Factors C, Di Angelantonio E, Sarwar N, et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA 2009;302:1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schilling S, Tzourio C, Dufouil C, et al. Plasma lipids and cerebral small vessel disease. Neurology 2014;83:1844–1852. [DOI] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015;47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carmelli D, DeCarli C, Swan GE, et al. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke 1998;29:1177–1181. [DOI] [PubMed] [Google Scholar]
- 43.Atwood LD, Wolf PA, Heard-Costa NL, et al. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke 2004;35:1609–1613. [DOI] [PubMed] [Google Scholar]
- 44.Fornage M, Debette S, Bis JC, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol 2011;69:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arvanitakis Z, Capuano AW, Leurgans SE, Buchman AS, Bennett DA, Schneider JA. The relationship of cerebral vessel pathology to brain microinfarcts. Brain Pathol 2017;27:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermeer SE, Prins ND, den Heijer T, Hofman A, Koudstaal PJ, Breteler MM. Silent brain infarcts and the risk of dementia and cognitive decline. N Engl J Med 2003;348:1215–1222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Summary statistics of the top SNPs are available from Dryad for both BI and SSBI. Other data that support the findings of this study are available from the corresponding authors upon reasonable request.