Abstract
The 4D extended cardiac-torso (XCAT) phantom has provided a valuable tool to study the effects of anatomy and motion on medical images, especially cardiac motion. One limitation of the XCAT was that it did not have a physiological basis which to realistically simulate variations in cardiac function. In this work, we incorporate into the XCAT anatomy the four-chamber FE Living Heart Model (LHM) developed by the Living Heart Project (LHP). The LHM represents the state of the art in cardiac FE simulation because of its ability to accurately replicate the biomechanical motion of the entire heart and its variations. We create a new series of 4D phantoms capable of simulating patients with varying body sizes and shapes; cardiac positions, orientations, and dynamics. While extendable to other imaging modalities and technologies, our goal is to use the FE-enhanced XCAT models to investigate the optimal use of computed tomography (CT) for the evaluation of coronary artery disease (CAD). With the ability to simulate realistic, predictive, patient quality 4D imaging data, the enhanced XCAT models will enable optimization studies to identify the most promising systems or system parameters for further clinical validation.
Keywords: Biomedical imaging phantoms, finite element analysis, cardiac motion compensation, computed tomography, image analysis, medical diagnostic imaging, medical simulation
I. Introduction
CORONARY artery disease (CAD) is the leading cause of death for both men and women in the United States every year [1]. Since 1900, it has accounted for more deaths than any other major cause. It presents a tremendous strain on the health care system with the cost of diagnosis and treatment having been estimated at more than $70 billion annually.
Many non-invasive imaging modalities are being studied and refined to better detect, stratify, and monitor therapy for patients with CAD. However, imaging of the heart is a challenging task, primarily due to its rapid movement. There is always a degree of temporal blur (motion artifact), the impact and limitations of which on clinical utility has remained uncertain, and image noise, directly reflective of the magnitude of radiation exposure used. Additionally, patient body habitus and technical limitations may contribute to high noise and degrade spatial resolution. For imaging modalities involving ionizing radiation like computed tomography (CT), radiation dose is also an ever-present reality that needs to be minimized without compromising image quality.
Clinical trials are the best avenue for the evaluation of existing and emerging imaging technologies, but the ever-expanding number of technologies and parameters and the potential risks from radiation exposure for modalities such as CT make a trial for every application or protocol unfeasible, pragmatically, financially, and ethically. As a result, medical imaging researchers, industry, and the FDA are increasingly moving toward computer-based simulations.
In computer-based simulation, computational phantoms, models of anatomy and physiology, serve as the patients. Phantom patients can be “imaged” using algorithms that simulate the physics of different imaging devices (CT, nuclear medicine, MRI, ultrasound, etc.). From this combination, multitudes of medical imaging data can be generated entirely on the computer with complete user control over the patient anatomy and motions as well the parameters for image acquisition, reconstruction, processing and analysis. Experiments can be performed quickly and cost effectively on the computer, giving researchers the ability to answer fundamental questions that can only be practically answered by harnessing the precise controls afforded in the virtual domain. Emerging technologies can be optimized preclinically by identifying the most promising systems or system parameters for further clinical validation. The impact of imaging parameters on known anatomies can be ascertained while accurately assessing and minimizing radiation dose levels. With such technological optimization not feasible using real human subjects, computer-based simulation provides a means of quantitatively predicting and personalizing the best acquisition strategy for a given patient scanner combination.
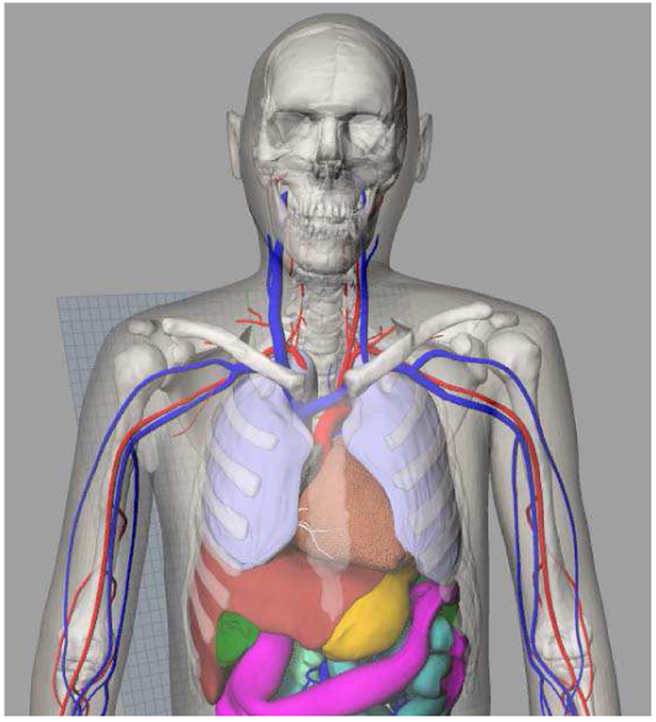
The 4D extended cardiac-torso (XCAT) phantom [2] was developed in our laboratory as a detailed computational model of the human anatomy and cardiac and respiratory motions for use in medical imaging simulation studies. Starting with adult male and female anatomies based on the Visible Human data, the XCAT has recently been expanded to include a population of models encompassing both genders and variable ages, heights, and weights [3, 4], as shown at the top of Fig. 1. With its realistic anatomical definition, the XCAT series of phantoms have been widely used in the evaluation of medical imaging devices and techniques. The XCAT has gained a particular use for cardiac applications having been applied to investigate cardiac imaging instrumentation, data acquisition techniques and image processing, and reconstruction methods [5-9].
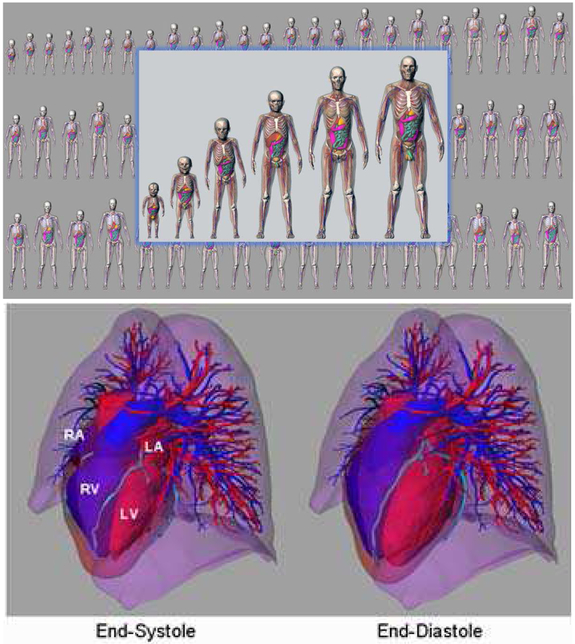
Fig. 1.
(Top) XCAT series of computational phantoms. (Bottom) Cardiac model of the XCAT phantom. Motion was based upon the analysis of tagged MRI data. Cardiac CT data was used to add details such as the coronary vessels. The right atria (RA), left atria (LA), right ventricle (RV) and left ventricle (LV) are labeled in the figure.
Despite this use, further improvements need to be made in the XCAT cardiac model. The motion of the current XCAT heart model, shown at the bottom of Fig. 1, was based upon the analysis of a single set of gated tagged MRI data of a normal male volunteer [2]. This represents one realization of the cardiac motion which was input into each new XCAT anatomy. The motion was parameterized so it could be altered to produce different variations; however, these alterations have no physiological basis. The XCAT is therefore limited in its ability to realistically simulate variations in the cardiac function.
To overcome this limitation, we have been working toward incorporating a physiologically based, finite-element (FE) mechanical model of the heart. With the ability to accurately simulate cardiac function, FE methods represent the state of the art in heart modeling. We previously developed a FE model for the XCAT left ventricle (LV) to simulate normal motion as well as altered motions due to ischemia and infarction [10, 11]. This work focused solely on the LV, however.
To expand beyond the LV to form a more complete cardiac anatomy, we incorporate into the 4D XCAT phantom the four-chamber FE Living Heart Model (LHM) [12] developed as part of the Living Heart Project (LHP) [13]. The LHP was launched by Dassault Systemes in 2014 to unite cardiovascular experts worldwide to develop a powerful simulation approach toward modeling the heart. A primary goal is to deliver validated cardiovascular models that can be used to develop safe and effective products and treatments for improved patient care. The LHP involves collaboration among 100 members including bioengineers, clinical and academic researchers, medical professionals, and regulatory authorities. The LHM model is being produced from this collaboration.
The geometry for the LHM model, shown in Fig. 2A, is based upon patient cardiac MRI data. The chambers and vessels of the heart were segmented and converted into FE meshes. The following key structures are included in the model: aortic arch, valves, coronary arteries and veins, pulmonary trunk, left and right atria and ventricles, papillary muscles and chordae tendineae, superior and inferior vena cava.
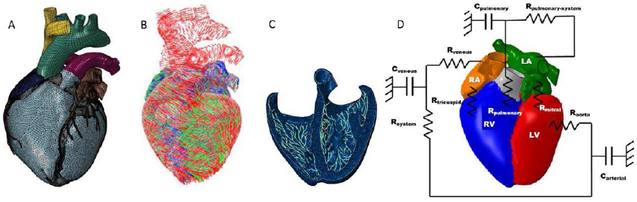
Fig. 2.
Components of the Living Heart Model including (A) the finite element mesh, (B) the fiber architecture of the muscle tissue for mechanical contraction, (C) the Purkinje network for electrical stimulation, and (D) the lumped parameter model for blood flow based on Pilla et al [14].
The LHM simulates the complete cardiac function with multiple physics. This includes the governing equations for the electro-mechanical excitation and contraction over the cardiac cycle as well as a model for blood flow and its effect on cardiac function, shown in Fig. 2 B-D. The LHM accurately simulates the motion and function of the heart as validated through a comparison of model outputs with the literature, shown in Table 1. Model parameters and material properties can all be altered within the LHM to simulate different normal and abnormal cardiac functions. Incorporated into the 4D XCAT phantom, the LHM will provide the physiological basis to simulate normal and abnormal variations in the cardiac motion within different anatomies for cardiac imaging research.
TABLE I.
COMPARISON OF LHM MODEL RESULTS
| Output | Model Result | Published Normal Ranges |
Reference |
|---|---|---|---|
| LV Ejection Fraction | 56% | >50% | [15] |
| RV Ejection Fraction | 49% | 40-60% | [16] |
| Max LV Pressure | 136.3 mmHg | 100-140 mmHg | [17] |
| Min LV Pressure | 6.8 mmHg | 3-12 mmHg | [17] |
| Max RV Pressure | 29.8 mmHg | 15-30 mmHg | [17] |
| Min RV Pressure | 0.2 mmHg | 2-8 mmHg | [17] |
| RA Pressure Range | 1.8-6.5 mmHg | 2-6 mmHg | [16] |
| LA Pressure Range | 8.7-24.5 mmHg | 4-12 mmHg | [16] |
| Maximum LV Apex-Base Shortening | 8.6 mm | 11.2 ± 3.8 mm | [18] |
| Apical Twist | 10.9° | 10.4° ± 2.6° | [19] |
II. METHODS
A. Generating a time series of heart models from the LHM
The analyses for the LHM are performed using the Abaqus finite element software (https://www.3ds.com/products-services/simulia/products/abaqus/). The LHM is analyzed in two steps. Initially the electrical conduction of the excitation wave through the entire heart is analyzed. The model contains the complete conduction system including the sino-atrial node, Purkinje fibers and so forth. Once this analysis has been completed the mechanical analysis is conducted using the activation information provided by the electrical analysis. The mechanical model contains a lumped parameter fluid load, as shown in Fig. 2. (e.g. LV pressure, arterial vascular bed, venous return pulmonary bed, etc.) which provides hemodynamic boundary conditions for the heart model. The cardiac cycle in the model is represented as three distinct phases (steps). In the first phase, the model is loaded by the diastolic filling pressure. This is followed by the systolic contraction phase which produces the systolic pressures and ejects the blood. The final phase represents cardiac relaxation.
In order to transfer the geometric information of the heart to the XCAT phantom, a custom python script was created that interrogates the completed analysis. The geometric changes within each of the three phases are output as a number of surface representations using the Polygon File format (ply) (http://paulbourke.net/dataformats/ply/). The title of each surface file identifies the geometry (e.g. ventricles, left atria etc.), the phase (e.g. systolic contraction) and the individual time increment within each of the phases. Using this script, we generated 40 time instances of the LHM over a complete cardiac cycle saving the structures at each time as a polygon mesh in the ply format.
B. Insertion of the LHM into the XCAT anatomies
The Rhinoceros modeling software (www.rhino3d.com) was used to place the LHM within the XCAT anatomies. A visual basic script was written to import the ply files from the LHM into Rhinoceros and to sort the structures into different layers according to their titles. The script was used to load frame 1 of the LHM into the same Rhinoceros file as the particular XCAT phantom. Figure 3 shows this process.
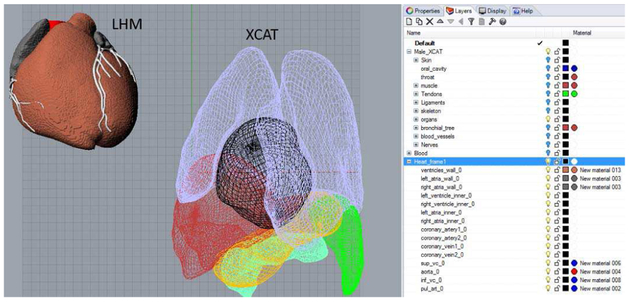
Fig. 3.
Living heart model and example XCAT anatomy as imported into the Rhinoceros modeling software. Layers are automatically created to sort the structures. Only the lungs, liver, stomach, spleen, heart, and pancreas are shown for the XCAT; the remaining structures are hidden.
Within Rhinoceros, the frame 1 LHM meshes were grouped together as a single object and manually translated, scaled, and oriented into the background anatomy using the XCAT heart surfaces and vessels as a guide, as illustrated in Fig. 4A. The rigid transformation for each XCAT was saved for later application to the surfaces for the remaining time points (Section II.C).
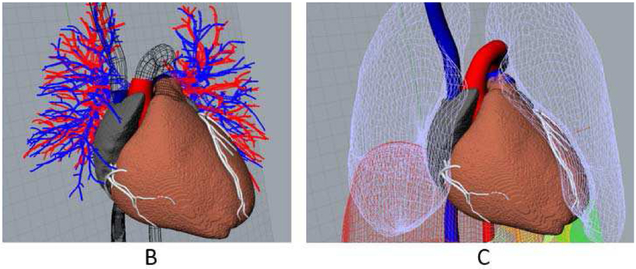
Fig. 4.
(A) The LHM is manually positioned and oriented into the XCAT using the original XCAT heart surfaces and vessels (shown in wireframe) as a guide. (B) XCAT lungs are deformed to encase the new heart and the vessel surfaces are joined with those of the LHM, plugging the heart into the background anatomy.
The XCAT lungs were deformed using the CAGEEDIT command within Rhinoceros to accommodate the new cardiac geometry. The vessel surfaces of the XCAT (aorta, pulmonary artery, pulmonary veins, superior and inferior vena cavas) were blended with those of the LHM to join the models together, as shown in Fig. 4B. Figure 5 shows an example of an enhanced LHM-XCAT phantom.
Fig. 5.
LHM combined with a male XCAT phantom.
C. Defining the heart motion within the XCAT Software
The XCAT heart model is currently defined by reading in surfaces for the structures defined at multiple points over the cardiac cycle. Time curves are then fit to the control points defining each surface creating time continuous 4D models [20, 2].
A similar format was generated for the LHM. The LHM meshes, defined for each time frame, were first transformed, applying the same rigid transformation as that calculated for the initial frame and then concatenated into a single file creating a similar set of time-changing surface files, 40 in total. The XCAT software was setup to read the new files and fit time curves to the 3D location of each vertex point of each surface mesh creating 4D models. The time curves can be sampled within the program to create any number of frames over the cardiac cycle.
D. Simulating different motion patterns
The mechanical, electrical, and blood flow parameters of the LHM can all be altered in a realistic, physiological manner to simulate different motions of the heart. To incorporate new motions within a given XCAT phantom, the FE analysis of the altered LHM would be performed using the Abaqus software as before. Using the stored transform for the particular XCAT, the procedure in II.C would be repeated to generate new surface files for the phantom.
E. Pilot study for plaque imaging
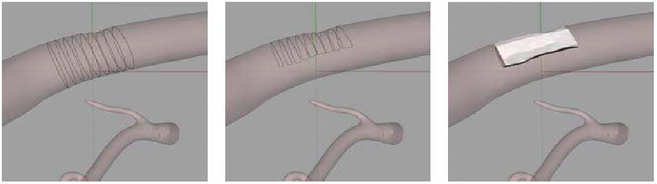
The enhanced 4D XCAT with the LHM was used in a pilot study to demonstrate the effects of cardiac motion on the imaging of cardiac plaques of known size and location. The standard adult male XCAT anatomy (50th percentile in height and weight) was used for the study. Plaques were modeled within the LHM as 3D NURBS surfaces lying on the walls of the coronary vessels. This was done using the Rhinoceros program operating on frame 1 of the LHM. Using the CONTOUR command, sections were taken through a particular vessel spaced 4 mm along its length at a desired location. The contours were cut in half to simulate a 50% blockage in the vessel. A smooth NURBS surface was then fit to the contours using the LOFT command as shown in Fig. 6. Three locations were simulated: the right coronary artery (RCA), the left anterior descending artery (LAD), and the left circumflex artery (LCX).
Fig. 6.
Modeling of a coronary plaque using Rhinoceros. Contours were taken through a vessel (left), halved to represent a 50% blockage (middle), then lofted into a NURBS surface (right).
Each plaque was set to move with the coronary artery surface in which it was placed. The frame to frame vectors for the mesh points of the given artery were used to create a 3D motion vector field. The motion field was applied to the plaque surface created for frame 1 to define it in each of the successive time points. The plaque surfaces defined at the 40 time points were added to the heart files read by the XCAT phantom.
The adult male XCAT model was then used to simulate a patient with a heart rate of 60 beats/minute. Iodine contrast was simulated within the vessels using the material definitions defined within the phantom. The attenuation value for the plaques was set to correspond to that of calcified plaques (average CT number = 1100).
CT projection data were generated from the XCAT (with and without cardiac motion) using an analytical projection algorithm developed in our laboratory [21]. Projection data were generated using a standard chest X-ray energy spectrum with a tube voltage of 120 kVp and 5 mm aluminum filtration obtained from the Catalogue of Spectral Data for Diagnostic X-rays [22]. Imaging was performed over a data acquisition window of 150 ms centered within the quiescent phase (middiastole) of the heart. During this brief period, the heart motion is reduced.
The projection data was reconstructed into CT images using a filtered backprojection algorithm. Profiles were taken along each plaque (with and without motion) to assess the effect of heart motion at the different locations within the coronary artery tree. The results were compared to those obtained with the same phantom using the original XCAT heart model.
III. RESULTS
A. LHM-XCAT
Figure 7 shows the cardiac motion of the living heart model within the XCAT anatomy, standard adult male (50th percentile in height and weight) shown as the example. The ejection fraction for the LHM model was 56% as compared to 60% for the original XCAT heart. The LV apex to base shortening and base to apex twist were 8.6 mm and 10.9 degrees respectively for the LHM (compared to 12 mm and 14 degrees for the original XCAT). All of these parameters are within the ranges found for normal heart function as can be seen in Table 1.
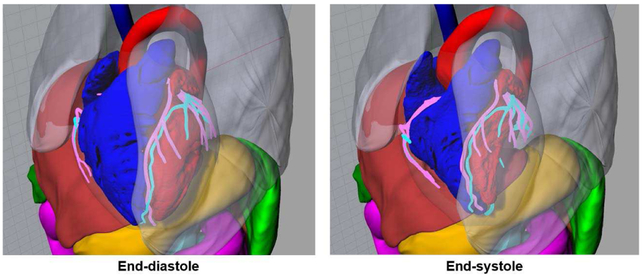
Fig. 7.
Renderings of the XCAT adult male phantom with the LHM defined at end-diastole and end-systole
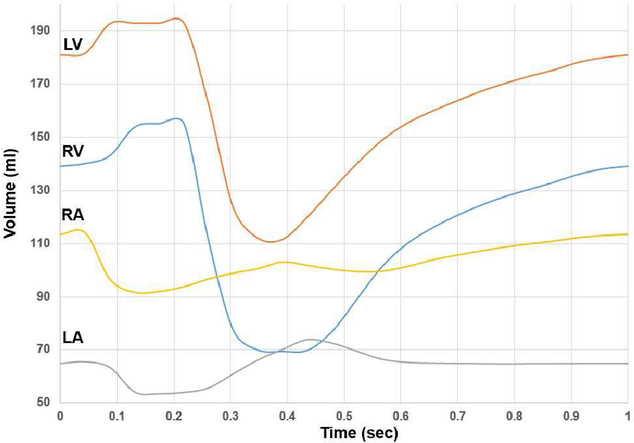
Using the time curves derived from the 40 time points, 300 time points were interpolated over one complete cardiac cycle (1 beat over the period of 1 second) for the LHM heart defined within the XCAT standard adult male. The volumes of the four chambers were calculated for each of the 300 frames and plotted in Figure 8. The figure illustrates a smooth contracting and relaxing motion following the normal pattern for the beating heart.
Fig. 8.
Volumes of the left and right atria and ventricles over the cardiac cycle (one beat over a period of one second) for the example LHM heart within the XCAT adult male.
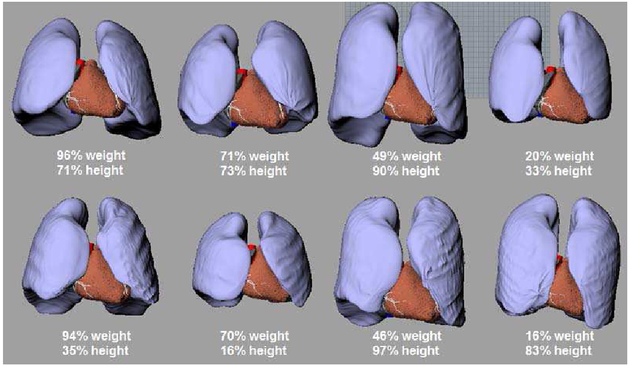
In addition to the standard male adult, the LHM was incorporated into other XCAT anatomies using the same methods. Figure 9 shows the placement of the heart within 8 anatomically variable phantoms (4 male and 4 female). Different placements, orientations, and sizes of the heart can be seen in the phantoms. The different sizes of the heart were obtained through isotropic scaling so as to not deform the FE model. As a result of this scaling, the chamber volume curves of the new hearts follow the same pattern as that in Figure 8 only with the volumes uniformly scaled up or down.
Fig. 9.
Anatomically variable LHM-XCAT phantoms. Four males (top) and four females (bottom) are shown with their weight and height percentiles indicated for each.
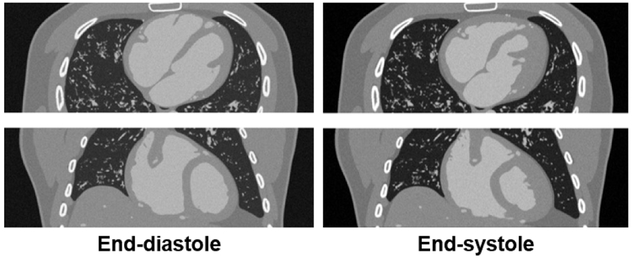
Figure 10 shows simulated CT data of an LHM-XCAT phantom defined at end-diastole and end-systole. The data demonstrates the ability of the phantoms to simulate realistic imaging data for research.
Fig. 10.
Simulated CT data of an XCAT-LHM phantom defined at end-diastole and end-systole. Transaxial (top) and coronal (bottom) slices are shown.
B. Pilot study
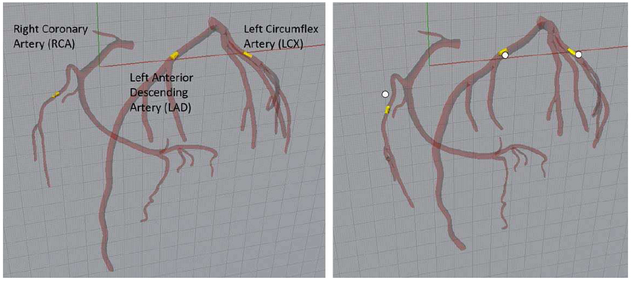
Figure 11 shows the three plaque locations selected for the simulation study. Varying amounts of cardiac motion occur at the three locations as shown in the figure. The RCA and LCX segments of the arterial tree can be seen to move the most from diastole to systole while less motion occurs at the LAD. This is similar to what has been reported by other studies [23].
Fig. 11.
Location of the three plaques simulated in the study. Plaques shown at end-diastole (left) and end-systole (right). White dots indicate the diastolic position to show the motion of each plaque.
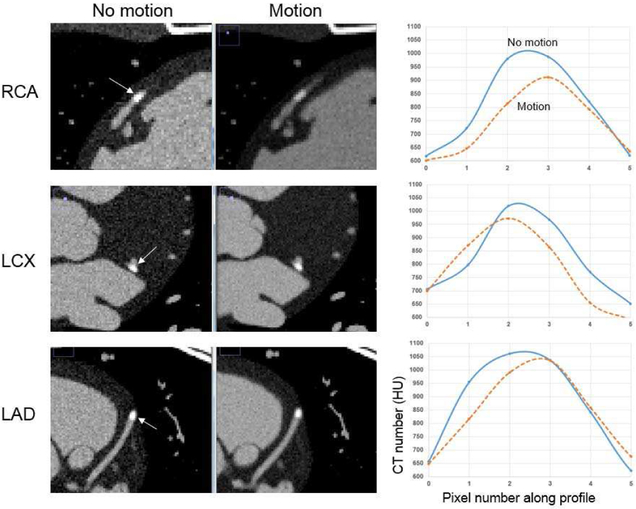
As expected, the motion of the heart, even minimal during the quiescent phase (0.6 to 0.7 sec in Fig. 8), caused a reduction in the contrast of the plaques, as seen in Fig. 12. The reduction was the most dramatic for the RCA due to its increased motion, as seen in Fig. 11. The contrast of the plaques was reduced by 7.7%, 4.6%, and 2.5% for the RCA, LCX, and LAD following the pattern of greatest to least amount of motion. Similar results were found using the original XCAT heart based on tagged MRI and 4D CT data (reduction of 8.6%, 5.5%, and 1.8% for the RCA, LCX, and LAD respectively).
Fig. 12.
(Left) Simulated CT data of the male LHM-XCAT phantom with and without cardiac motion during the quiescent phase of the heart. Zoomed in views of the coronary segments containing the plaques are shown. (Right) Profile plots through the plaques taken along the directions indicated by the arrows in the CT data.
IV. DISCUSSION
We combined our series of XCAT phantoms with the four-chamber finite element heart model from the Living Heart Project. The LHM provided the functioning heart while the XCAT library provided the surrounding anatomies. The combination of the two different computational models offered two major challenges. First, the LHM and XCAT use different formats for anatomical structures. The LHM is defined using volumetric finite element meshes while the XCAT uses NURBs and polygon mesh surfaces. To bridge this gap, a custom python script was created to convert the output of the LHM into a mesh format readable by the XCAT. second, the LHM provides a different heart anatomy than what was previously defined in each XCAT phantom. For this, we had to manually place the LHM into each individual phantom then adjust the surrounding lungs and blend the LHM vessels with those of the particular XCAT. This process took approximately 2-3 hours per phantom.
In future work, we plan to incorporate the LHM into all of our XCAT phantoms of varying sizes. For the current method, as mentioned above, the LHM was rigidly transformed to fit it within different anatomies. This captures the variation of the overall size of the heart as well as its position and orientation, but it does not capture the interior geometrical changes. To overcome this limitation, we plan to investigate the feasibility of non-rigid warping methods to fit the LHM to new anatomies. such methods can provide better fits, capturing the interior variation from heart to heart. They will also be more efficient and require less manual effort than our current technique. These methods do introduce many complexities, however, as the muscle fiber architecture and other material properties and parameters need to be determined for the deformed geometry.
The living heart model provides a realistic simulation of the cardiac motion, including the coronary arteries, similar to that of the original XCAT. Our pilot simulation study using the LHM showed similar results to those from the XCAT and to previous studies. The advantage of the LHM over the original XCAT heart, however, comes from its basis in physiology.
Based on patient data, the original XCAT cardiac model provides only one realization of the cardiac motion. Parameters can be setup to alter this motion, but such changes lack a physiological basis. The LHM, however, is defined with finite element methods that simulate the physiological and physical interactions of the tissues on multiple scales from the cell to the organ. Material properties of the tissues and other parameters can be altered in a realistic, physiologically-based manner to simulate normal and abnormal variations in the cardiac motion. Based on an analysis of several sets of gated cardiac patient CT data, we plan to alter the properties of the LHM to simulate a range of variations in the cardiac function and incorporate these into the XCAT.
For the pilot study performed in this work, we used a very simple, homogenous calcified model for the cardiac plaques. In future work, we plan to create more detailed, heterogeneous plaque models composed of varying levels of calcium and lipids, based on histological data, to incorporate into our heart models.
V. CONCLUSIONS
The series of enhanced LHM-XCAT phantoms along with detailed models for cardiac plaques will provide a valuable set of tools for objective simulation-based evaluation and optimization of cardiac imaging devices and techniques. While this work investigated an application in CT, the simulation tools are extendable to other imaging modalities and technologies. As such, the LHM-XCAT phantoms provide the necessary foundation with which to investigate different 4D imaging methods to identify the most promising systems or system parameters to better identify and characterize coronary artery disease.
ACKNOWLEDGMENT
This work was supported by research grant R01 HL131753 from the National Institutes of Health.
Contributor Information
W. Paul Segars, Carl E. Ravin Advanced Imaging Laboratories, the Department of Radiology, the Medical Physics Graduate Program, and the Department of Biomedical Engineering, Duke University, Durham, NC, 27705 USA (paul.segars@duke.edu)..
Alexander I. Veress, Department of Mechanical Engineering, University of Washington, Seattle, WA, 98195 USA.
Gregory M. Sturgeon, Carl E. Ravin Advanced Imaging Laboratories and the Department of Radiology, Duke University Medical Center, Durham, NC, 27705 USA.
Ehsan Samei, Carl E. Ravin Advanced Imaging Laboratories, the Department of Electrical and Computer Engineering, the Department of Radiology, the Department of Biomedical Engineering, the Medical Physics Graduate Program, and the Department of Physics, Duke University, Durham, NC, 27705 USA..
REFERENCES
- [1].Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. , "Heart Disease and Stroke Statistics-2013 Update A Report From the American Heart Association," Circulation, vol. 127, pp. E6–E245, January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Segars WP, Sturgeon G, Mendonca S, Grimes J, and Tsui BMW, "4D XCAT phantom for multimodality imaging research," Med Phys, vol. 37, pp. 4902–4915, September 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Segars WP, Bond J, Frush J, Hon S, Eckersley C, Williams CH, et al. , "Population of anatomically variable 4D XCAT adult phantoms for imaging research and optimization," Med Phys, vol. 40, p. 043701, April 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Segars W, Norris H, Sturgeon G, Zhang Y, Bond J, Minhas A, et al. , "The development of a population of 4D pediatric XCAT phantoms for imaging research and optimization," Medical Physics, vol. 42, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rohkohl C, Lauritsch G, Keil A, and Hornegger J, "CAVAREV–an open platform for evaluating 3D and 4D cardiac vasculature reconstruction," Phys Med Biol, vol. 55, pp. 2905–15, May 21 2010. [DOI] [PubMed] [Google Scholar]
- [6].Kim S, Chang Y, and Ra JB, "Cardiac motion correction based on partial angle reconstructed images in x-ray CT," Med Phys, vol. 42, pp. 2560–71, May 2015. [DOI] [PubMed] [Google Scholar]
- [7].Tragardh E, Ljungberg M, Edenbrandt L, Orndahl E, Johansson L, Gustafsson A, et al. , "Evaluation of inter-departmental variability of ejection fraction and cardiac volumes in myocardial perfusion scintigraphy using simulated data," EJNMMI Phys, vol. 2, p. 2, December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ghaly M, Du Y, Links JM, and Frey EC, "Collimator optimization in myocardial perfusion SPECT using the ideal observer and realistic background variability for lesion detection and joint detection and localization tasks," Phys Med Biol, vol. 61, pp. 2048–66, March 07 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Segars WP, Tsui BMW, Cai J, Yin FF, Fung GSK, and Samei E, "Application of the 4D XCAT Phantoms in Biomedical Imaging and Beyond," IEEE Trans Med Imaging, August 10 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Veress AI, Segars WP, Weiss JA, Tsui BM, and Gullberg GT, "Normal and pathological NCAT image and phantom data based on physiologically realistic left ventricle finite-element models," IEEE Trans Med Imaging, vol. 25, pp. 1604–16, December 2006. [DOI] [PubMed] [Google Scholar]
- [11].Veress AI, Segars WP, Tsui BMW, and Gullberg GT, "Incorporation of a Left Ventricle Finite Element Model Defining Infarction Into the XCAT Imaging Phantom," IEEE Transactions on Medical Imaging, vol. 30, pp. 915–927, Apr 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Baillargeon B, Rebelo N, Fox DD, Taylor RL, and Kuhl E, "The Living Heart Project: A robust and integrative simulator for human heart function," European Journal of Mechanics - A/Solids, vol. 48, pp. 38–47, 2014/11/01/ 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dassault Systemes, Living Heart Project,https://www.3ds.com/products-services/simulia/solutions/life-sciences/the-living-heart-project/.
- [14].Pilla JJ, Gorman JH, and Gorman RC, "Theoretic Impact of Infarct Compliance on Left Ventricular Function," The Annals of thoracic surgery, vol. 87, pp. 803–810, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Phibbs B, The Human Heart: A Basic Guide to Heart Disease – Second Edition: Lippincott Williams & Wilkins, 2007. [Google Scholar]
- [16].Normal Hemodynamic Parameters – Edwards Lifesciences. http://ht.edwards.com/scin/edwards/fr/sitecollectionimages/edwards/products/presep/ar04313hemodynpocketcard.pdf.
- [17].Klingensmith ME, Chen LE, Glasgow SC, Goers TA, and Melby SJ, The Washington Manual of Surgery – Fifth Edition: Lippincott Williams & Wilkins, 2005. [Google Scholar]
- [18].Rogers WJ Jr., Shapiro EP, Weiss JL, Buchalter MB, Rademakers FE, Weisfeldt ML, et al. , "Quantification of and correction for left ventricular systolic long-axis shortening by magnetic resonance tissue tagging and slice isolation," Circulation, vol. 84, pp. 721–31, August 1991. [DOI] [PubMed] [Google Scholar]
- [19].Thompson RB, Paterson I, Chow K, Cheng-Baron J, Scott JM, Esch BT, et al. , "Characterization of the relationship between systolic shear strain and early diastolic shear strain rates: insights into torsional recoil," Am J Physiol Heart Circ Physiol, vol. 299, pp. H898–907, September 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Segars WP, Lalush DS, and Tsui BMW, "A realistic spline-based dynamic heart phantom," IEEE Transactions on Nuclear Science, vol. 46, pp. 503–506, 1999. [Google Scholar]
- [21].Segars WP, Mahesh M, Beck TJ, Frey EC, and Tsui BM, "Realistic CT simulation using the 4D XCAT phantom," Med Phys, vol. 35, pp. 3800–8, August 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cranley K, Gilmore B, Fogarty G, and Desponds L, "Catalogue of Diagnostic X-ray Spectra and Other Data," Diagnostic Radiology and Magnetic Resonance Special Interest Group of the Institute of Physics and Engineering in Medicine 78, 1997. [Google Scholar]
- [23].Shechter G, Resar JR, and McVeigh ER, "Displacement and Velocity of the Coronary Arteries: Cardiac and Respiratory Motion," IEEE transactions on medical imaging, vol. 25, pp. 369–375, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]