Abstract
Purpose.
Clostridium difficile infections are the leading cause of diarrhea associated to the use of antibiotics. During infection, C. difficile initiates a sporulation cycle leading to the persistence of C. difficile spores in the host and disease dissemination. The development of vaccine and passive immunization therapies against C. difficile have focused on toxins A and B. In the present study, we used an immunoproteome-based approach to identify immunogenic proteins located on the outer layers of C. difficile spores as potential candidates for the development of immunotherapy and/or diagnostic methods against this devastating infection.
Experimental design.
To identify potential immunogenic proteins on the surface of C. difficile R20291, spore coat/exosporium extracts were separated by two-dimensional electrophoresis (2-DE) and analyzed for reactivity against C. difficile spore-specific goat sera. Finally, the selected spots were in-gel digested with chymotrypsin, peptides generated were separated by nanoUPLC followed by MS/MS using Quad-TOF-MS, corroborated by Ultimate 3000RS-nano-UHPLC coupled to Q-Exactive-Plus-Orbitrap MS.
Results.
The analysis identified 5 immnoreactive proteins: spore-coat proteins CotE, CotA and CotCB, exosporium protein CdeC, and a cytosolic methyltransferase.
Conclusion.
Our data provides a list of spore surface protein candidates as antigens for vaccine development against C. difficile infections.
Keywords: Clostridium difficile, Spores, immunoproteomics, Two-dimensional electrophoresis, CDI patients, Serologic reactivity, isoelectric focusing
Clinical relevance.
The spore forming pathogen C. difficile is the leading cause of diarrhea associated to the use of antibiotics worldwide. Current therapies to combat C. difficile infections involve the use of antibiotics that do not affect the spore form of C. difficile. Vaccines and passive immunization against C. difficile, under phase III clinical trials, focus on the neutralization of both toxins. However, given the importance of C. difficile spores in the initiation, persistence and dissemination of the disease, the identification of novel spore-protein candidates for vaccine development is crucial. Consequently, experimental evidence of the efficacy of novel protein targets on the spore surface will aid in the development of next generation vaccines.
Report.
The anaerobic bacteria Clostridium difficile is the causative agent of toxin-mediated intestinal disease[1]. C. difficile infections (CDI) have escalated as a primary healthcare challenge world-wide[2]. Disease severity may vary from a mild self-limiting diarrhea to severe pseudomembranous colitis[1]. The emergence of hypervirulent strain R20291 (typified as B1/NAP1/027) has been related to higher incidence, mortality and morbidity of the disease[1]. Mortality rates of CDI range between 1–5%, but depending of the virulence of the strain causing a specific outbreak, they may rise to 20%[2]. Despite the impact of CDI, the most challenging complications of CDI are the high rates of recurrence of the disease, which may rise to 25, 40 and 65% of the patients after a first, second and third episode of recurrent infection, respectively[2].
C. difficile spores are resilient dormant morphotypes formed during the infection and shed to the environment for transmission to new susceptible patients, and are considered the main virulence factor involved in the recurrence of the disease[3]. C. difficile spores exhibit intrinsic spore resistance properties that enable them to be resilient to all known antibiotics and attacks by the innate immune system[4–6]. Spore-targeted therapies must consider characteristics of the spore surface, which still remains poorly described. Lawley et al.[7] and Abhyankar et al.[8] performed proteomic studies in C. difficile 630 spores and determined the protein composition of the spore coat/exosporium extracts. Recently, Díaz-González et al. used three gel-free approaches to refine the proteomic composition of the outermost layer of C. difficile 630 spores[9]. These publications led to the first vaccination study using exosporium proteins of C. difficile spores, which demonstrated that CdeM and CdeC may be utilized as immunization candidates[10]. However, these studies on the composition of exosporium layer have used the laboratory C. difficile 630 strain, which has evident structural differences with epidemic strains, including R20291[11]; specifically 630 spores lack the hair-like extensions and the classical bumps observed in the electron-dense exosporium layer[11–13], suggesting that the different exosporium morphotypes might have presence/absence of exosporium structural proteins, decreasing the efficacy of their use as vaccine candidates. Further structural studies in C. difficile spore should consider epidemic strains which maintain common ultrastructure features with clinical isolated strains.
Immunoproteomics has been used to identify potential immunogenic bacterial proteins by detection with serum from vaccinated or challenged animals or patients[14]. Through the identification of immunogenic proteins, further study about the host response can help to improve vaccine strategies[14]. Examples of this approach has been applied for infection with Streptococcus pneumoniae[15,16], Burkholderia pseudomallei[17] and Neisseria meningitidis[18]. As immunoproteomics defines the subset of proteins that induce an host humoral response, most of which prove to be novel protective vaccine candidates[19], and it has been used to identify potential immunogenic antigens.
In this context, the aim of this work was to provide the first list of immunogenic proteins of the surface of C. difficile spores of an epidemically relevant strain (i.e., R20291). To prove this concept, a previously characterized anti-C. difficile spore goat serum[20] was used to immunoblot coat/exosporium spore proteins separated by 2-DE.
C. difficile strain R20291[21] was grown in BHIS broth and inoculated onto TY agar to induce sporulation as described previously[22]. Purification of C. difficile spores were made as previously described[22] until they were >99% free of vegetative cells, sporulating cells and cell debris as determined by phase contrast microscopy. Spore suspensions were stored at −80°C until use. To extract the exosporium and spore coat fragments, C. difficile spores (5×108 spores) were treated with 8 M Urea, 2 M Thiourea, 4% CHAPS and 65 mM DTT, for 90 min at 37 °C with no shaking. Treated spores were centrifuged at 15000 × g for 10 minutes and the supernatant containing the spore’ proteins were collected and stored at −80 °C until use.
To identify the immunoreactive proteins of spores of the epidemic C. difficile strain R20291, spore coat/exosporium extracts were resolved on a 2-DE Coomassie Brilliant Blue G-250 colloidal (Figure 1A) in triplicate (Fig. S1). To detect immunoreactive proteins we used a previously characterized spore-specific goat serum[20,23] as a first approach to identify novel vaccine candidates. First, as a control, we tested in a conventional one-dimensional electrophoresis the pre-immunized goat serum and found no detection of immunoreactive spore proteins (Figure S2). This goat Anti-spore serum was raised against spores of strain 630, despite the ultrastructural differences between 630 and R20291 spores, it also detects immunoreactive bands in R20291 spores; therefore, we reasoned that it was adequate as a proof of concept study.
Fig 1. Immunoproteomic analysis of the spore surface of the C. difficile strain R20291 strain.
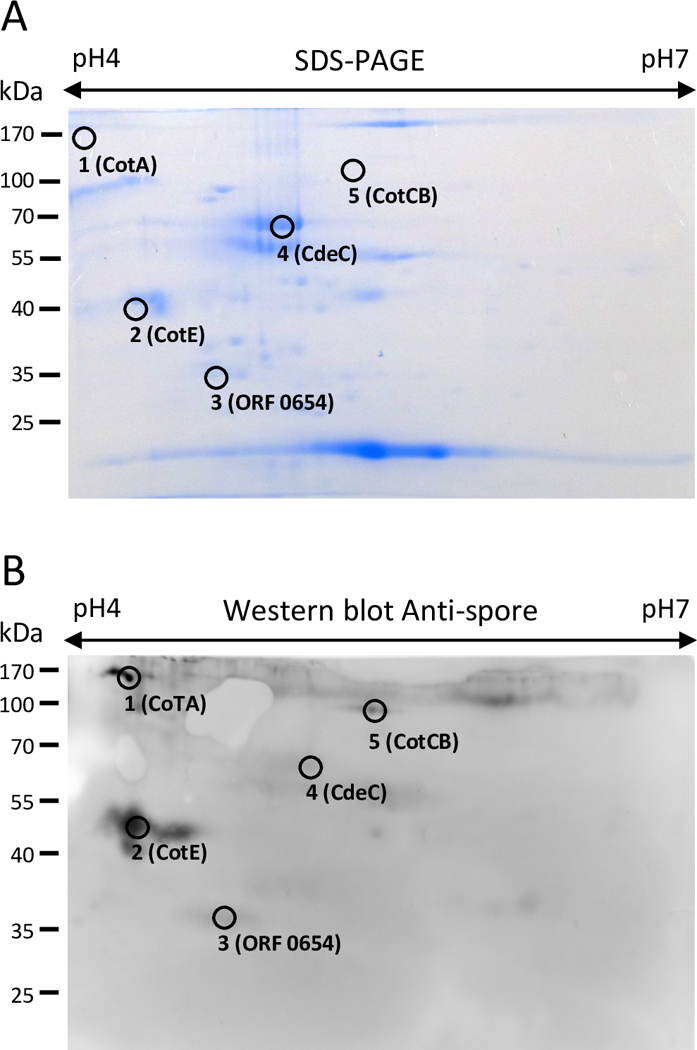
(A) 2-DE map (pI 4–7) SDS-PAGE of spore coat/exosporium extract with identified immunoreactive proteins stained with Coomassie Brilliant Blue G-250. (B) Western blot of a 2-DE gel of spore coat/exosporium extracts of R20291 epidemic strain probed with anti-spore goat serum specific for C. difficile spores and immunoreactive proteins identified. This gel is representative of three independent 2-DE gels with similar results.
Upon immunoblotting with our previously characterized goat anti-C. difficile spore serum[20], a total of 5 immunoreactive spots matched with the protein spots observed in the preparative 2-DE gel and had peptide assigned (Fig. 1A, 1B). Protein in the spots were in-gel digested with chymotrypsin, and the peptides generated were separated by nanoUPLC followed by MS/MS using a Quad-TOF MS, and corroborated by Ultimate 3000RS nano UHPLC coupled to Q Exactive Plus Orbitrap MS[24,25] (See supporting information). The immunogenic proteins detected (Table 1, Table S1 and Table S2) are mostly involved in spore coat assembly and/or exosporium assembly[11], and have been previously characterized.
TABLE 1.
Spore coat/exosporium and immunoreactive proteins of epidemic C. difficile R20291 strain identified in a 2-DE gel system
| Spot # | Gene namea | Uniprot IDa | Protein Descriptiona | Scoreb | MW (Da)c | pId | Peptidese | % coveragef |
|---|---|---|---|---|---|---|---|---|
| 1 | CDR20291_1511 | C9YLQ8 | Uncharacterized protein (CotA) | 453 | 35209 | 3.98 | 8 | 17.2 |
| 2 | CDR20291_1282 | C9YL30 | Peroxiredoxin (CotE) | 838 | 81612 | 5.25 | 40 | 33.9 |
| 3 | CDR20291_0654 | C9YJA5 | Putative carbon monoxide dehydrogenase/acetyl-CoA synthase complex, methyltransferase subunit | 288 | 29762 | 4.78 | 25 | 71.3 |
| 4 | CDR20291_0926 | C9YK25 | Uncharacterized protein (CdeC) | 330 | 46958 | 4.84 | 27 | 40 |
| 5 | CDR20291_2291 | C9YNX8 | Putative spore-coat protein (CotCB) | 249 | 21497 | 4.68 | 19 | 72.1 |
Functional annotations were retrieved from Swiss Proteina database (www.uniprot.org )
The threshold was set up by the server at the significance level P≤0.05 for random hit; individual ion scores greater than 17 were taken as significant match for identity or extensive homology, based in Mowse score (http://www.matrixscience.com )
The molecular weight of the protein identified from the database.
pI: isoelectric point of the protein identified.
The number of matching peptides to the target protein identified
The portion of the protein sequence that was observed by MS/MS analysis of the peptide mapping
CotA.
This is a spore morphogenetic protein required for the correct assembly of C. difficile spore’s overall structure, spore coat and exosporium. As observed in spores of a cotA mutant in 630 strain[26], absence of CotA leads to C. difficile spores that are sensitive to lysozyme-, ethanol- and heat-killing[26]. CotA is located primarily on the spore-coat, but is also present in the exosporium layer[9], which is consistent with CotA-removal by sonication and accessibility to CotA-specific antibodies[27]. Since this protein is also surface exposed, it is also an attractive target for C. difficile spore-specific vaccine development.
CotE.
It is a bifunctional spore coat protein with peroxiredoxin activity at its amino-terminal end and chitinase activity at its carboxy-terminal end. CotE is easily removed from the spore surface by sonication[27], suggesting an exosporium localization albeit it has also been shown to be present in the spore coat[9]. Inactivation of cotE in strain 630 leads to spores with wild-type like structure, albeit with a slight defect on heat resistance[26]. Recently, it was demonstrated that CotE interacts with intestinal mucin promoting mucin degradation, contributing to colonization and disease progression[28]. For these reasons, CotE might be an attractive target for the development of novel antibacterial therapies. On the other hand, the peroxirredoxin domain from CotE could be related to a role with inflammation[26], as it resembles a 1-Cys peroxirredoxin, which is secreted from tumoral cells and induce proinflammatory cytokines[1].
CotCB.
This spore coat protein is similar to the Bacillus subtilis CotJC manganase catalase, studies have demonstrated that it is located in the spore surface and accessible for anti-CotCB antibodies[27]. Inactivation of cotCB in the laboratory strain 630 leads to spores with a similar ultrastructure and resistant properties as wild-type spores[26]. Although it is unclear whether this protein plays a role in spore-persistence and/or infection, its immunoreactivity and accessibility to antibodies makes this another novel vaccine candidate.
CdeC.
This protein is essential for exosporium morphogenesis and the correct assembly of the spore coat of C. difficile[23]. This protein is localized in the exosporium layer, accessible to antibodies[9] and is involved in resistance to lysozyme, ethanol, and heat[23].
Cytosolic proteins of C. difficile.
It is noteworthy that several cytosolic proteins were found to be immunoreactive in the surface of epidemic R20291 spores. Among these proteins we identified a methyltransferase (CDR20291_0654). As previously suggested, it is likely that these proteins are derived from vegetative contaminants that could not be removed by the spore purification procedures[9]. Nevertheless, these proteins should be evaluated as therapeutic target for protective efficacy against C. difficile infection to elucidate whether they may or may not be considered as novel vaccine targets.
From an initial list of 184 surface proteins of C. difficile found in a recent proteomic study[9], our immunoproteomics approach reduces the number of vaccine candidates to 5. Among these spore proteins, there is evidence that vaccination against CdeC[29] and CotE[28] in a mouse model provides protection against a C. difficile infection. However, to date it has still not been demonstrated that vaccination with CotA or CotCB, as well as other cytosolic proteins found, may improve the resolution of the disease.
An important characteristic of a protein to be a good vaccine target for immunotherapy strategies is the degree of conservation in the same species (Clostridium difficile) and different species of Clostridium and with other bacteria. These criteria will avoid cross-reactivity with other microorganisms, and ensure a specific immune response against C. difficile spores. In this context, although most of the immunoreactive proteins identified in this work were specific to C. difficile, the spore coat proteins, specifically CotE, and particularly the exosporium protein CdeC, are perfect candidates for a C. difficile spore-specific recombinant protein-based vaccine.
It is remarkable that another cysteine-rich protein, CdeM, previously shown to be involved in the assembly of the exosporium layer[23] and shown to provide protective efficacy against C. difficile challenge of CdeM-immunized mice[10] was not detected by the anti-C. difficile spore goat serum. Additional exosporium proteins that were not detected in the 2-DE gels include the exosporium collagen-like BclA2, CdeA and CdeB[9] and the spore coat proteins CotD, CotF, CotG, CotB and SodA[27]. Possible explanations may include the degree of fixation of the spores used for immunization, the high degree of crosslinking of the exosporium proteins of R20291 strain, and the low detection limit of the 2-DE gel system. Further efforts to optimize this technique for C. difficile spore surface proteins might uncover additional vaccine candidates. Work to validate these immunorreactive proteins in serum of C. difficile infected patients should aid in the selection of novel vaccine candidates.
Conclusions.
The specific spore surface proteins CotA, CotE, CotCB, CdeC and one cytosolic protein were identified in this study as immunogenic proteins. Most of these antigens are unique to C. difficile, suggesting that these proteins could serve as antigen candidates for the development of immunotherapies against C. difficile infections. Further work to confirm the protective efficacy of these immunogenic proteins in animal models must be performed to fully prove their potential as vaccine alternatives.
Supplementary Material
Figure S1. Detection of proteins in C. difficile R20291 spore surface with goat anti-spore serum. Detection of R20291 spore proteins with (A) Anti-spore Goat IgG serum and (B) pre-immunized goat serum. Dilution of goat Anti-spore serum and pre-Immunized serum is 1:1000.
Fig S2. Triplicates of Immunoproteomic analysis of the spore surface of the C. difficile strain R20291 strain. (A) Spore coat/exosporium extract 2-DE map (pI 4–7) were electrophoresed on a pH 4/7 IPG Strip (Bio-Rad) and isoelectro-focused (IEF) for 3 h as described in Supplementary Information. Next, strips were placed on SDS-PAGE (1 mm thickness, 12% acrylamide) and coupled to the gel with 1% agarose containing Tris-Glycine buffer and protein spots stained with Coomassie Blue G-250. (B) Western blot of a 2-DE gel of spore coat/exosporium extracts of R20291 epidemic strain probed with anti-spore goat serum specific for C. difficile spores and immunoreactive proteins identified.
Acknowledgements
This work was supported by grants from Fondo Nacional de Ciencia y Tecnología de Chile (FONDECYT Grant 1151025 to D.P-S.) and Beca Doctorado Nacional 21151202 (M.P-G.). Research reported in this publication was supported by an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103443. The authors want to thank Mr. Doug Jennewein and Nate Brady from USD-IT Research Computing for their help in the database installation as well as server operation. The authors also thank Dr. Jaime Eyzaguirre (Universidad Andrés Bello) for support in isoelectro-focusing (IEF). M.C.R. acknowledges the support of projects PMIUAB1301 and Neuromorphics Inspired Science (grant number FA9550-16-1-0384). Data are available via ProteomeXchange with identifier PXD008592.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Conflict of interest: The authors declare that they have no conflict of interest.
References.
- [1].Rupnik M, Wilcox MH, Gerding DN, Nat. Rev. Microbiol 2009, 7, 526. [DOI] [PubMed] [Google Scholar]
- [2].Evans CT, Safdar N, Clin. Infect. Dis 2015, 60, S66. [DOI] [PubMed] [Google Scholar]
- [3].Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD, Infect. Immun 2012, 80, 2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chilton CH, Crowther GS, Ashwin H, Longshaw CM, Wilcox MH, PLoS One 2016, 11, e0161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Crowther GS, Chilton CH, Longshaw C, Todhunter SL, Ewin D, Vernon J, Karas A, Wilcox MH, J. Antimicrob. Chemother 2016, 71, 986. [DOI] [PubMed] [Google Scholar]
- [6].Paredes-Sabja D, Cofre-Araneda G, Brito-Silva C, Pizarro-Guajardo M, Sarker MR, PLoS One 2012, 7, e43635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lawley T, Croucher N, Yu L, Clare S, Sebaihia M, Goulding D, Pickard D, Parkhill J, Choudhary J, Dougan G, J. Bacteriol 2009, 191, 5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abhyankar W, Hossain AH, Djajasaputra A, Permpoonpattana P, Ter Beek A, Dekker HL, Cutting SM, Brul S, De Koning LJ, De Koster CG, J. Proteome Res 2013, 12, 4507. [DOI] [PubMed] [Google Scholar]
- [9].Díaz-González F, Milano M, Olguin-Araneda V, Pizarro-Cerda J, Castro-Córdova P, Tzeng S-C, Maier CS, Sarker MR, Paredes-Sabja D, J. Proteomics 2015, 123, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ghose C, Eugenis I, Edwards ANN, Sun X, McBride SMM, Ho DDD, Anaerobe 2016, 37, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Paredes-Sabja D, Shen A, Sorg JA, Trends Microbiol 2014, 22, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pizarro-Guajardo M, Calderón-Romero P, Paredes-sabja D, Appl. Environ. Microbiol 2016, 82, 5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pizarro-Guajardo M, Calderón-Romero P, Castro-Córdova P, Mora-Uribe P, Paredes-Sabja D, Appl. Environ. Microbiol 2016, 82, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dennehy R, McClean S, Curr. Protein Pept. Sci 2012, 13, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ling E, Feldman G, Portnoi M, Dagan R, Overweg K, Mulholland F, Chalifa-Caspi V, Wells J, Mizrachi-Nebenzahl Y, Clin. Exp. Immunol 2004, 138, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mizrachi Nebenzahl Y., Bernstein A, Portnoi M, Shagan M, Rom S, Porgador A, Dagan R, J. Infect. Dis 2007, 196, 945. [DOI] [PubMed] [Google Scholar]
- [17].Harding SV, Sarkar-Tyson M, Smither SJ, Atkins TP, Oyston PCF, Brown KA, Liu Y, Wait R, Titball RW, Vaccine 2007, 25, 2664. [DOI] [PubMed] [Google Scholar]
- [18].Mendum TA, Newcombe J, McNeilly CL, McFadden J, PLoS One 2009, 4, e5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stentzel S, Gläser R, Bröker BM, Proteomics Clin. Appl 2016, 1011. [DOI] [PubMed]
- [20].Pizarro-Guajardo M, Olguín-Araneda V, Barra-Carrasco J, Brito-Silva C, Sarker MR, Paredes-Sabja D, Anaerobe 2014, 25, 18. [DOI] [PubMed] [Google Scholar]
- [21].McEllistrem MC, Carman RJ, Gerding DN, Genheimer CW, Zheng L, Clin. Infect. Dis 2005, 40, 265. [DOI] [PubMed] [Google Scholar]
- [22].Mora-Uribe P, Miranda-Cárdenas C, Castro-Córdova P, Gil F, Calderón I, Fuentes JA, Rodas PI, Banawas S, Sarker MR, Paredes-Sabja D, Front. Cell. Infect. Microbiol 2016, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Barra-Carrasco J, Olguín-Araneda V, Plaza-Garrido Á, Miranda-Cárdenas C, Cofré-Araneda G, Pizarro-Guajardo M, Sarker MR, Paredes-Sabja D, J. Bacteriol 2013, 195, 3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun L, Zhu G, Dovichi NJ, Rapid Commun Mass Spectrom 2013, 27, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV, J. Proteome Res 2012, 11, 3487. [DOI] [PubMed] [Google Scholar]
- [26].Permpoonpattana P, Phetcharaburanin J, Mikelsone A, Dembek M, Tan S, Brisson MC, La Ragione R, Brisson AR, Fairweather N, Hong HA, Cutting SM, J. Bacteriol 2013, 195, 1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Permpoonpattana P, Tolls EH, Nadem R, Tan S, Brisson A, Cutting SM, J. Bacteriol 2011, 193, 6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hong HA, Ferreira WT, Hosseini S, Anwar S, Hitri K, Wilkinson AJ, Vahjen W, Zentek J, Soloviev M, Cutting SM, J. Infect. Dis 2017, 216, 1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ghose C, Kelly CP, Infect. Dis. Clin. North Am 2015, 29, 145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Detection of proteins in C. difficile R20291 spore surface with goat anti-spore serum. Detection of R20291 spore proteins with (A) Anti-spore Goat IgG serum and (B) pre-immunized goat serum. Dilution of goat Anti-spore serum and pre-Immunized serum is 1:1000.
Fig S2. Triplicates of Immunoproteomic analysis of the spore surface of the C. difficile strain R20291 strain. (A) Spore coat/exosporium extract 2-DE map (pI 4–7) were electrophoresed on a pH 4/7 IPG Strip (Bio-Rad) and isoelectro-focused (IEF) for 3 h as described in Supplementary Information. Next, strips were placed on SDS-PAGE (1 mm thickness, 12% acrylamide) and coupled to the gel with 1% agarose containing Tris-Glycine buffer and protein spots stained with Coomassie Blue G-250. (B) Western blot of a 2-DE gel of spore coat/exosporium extracts of R20291 epidemic strain probed with anti-spore goat serum specific for C. difficile spores and immunoreactive proteins identified.


