Abstract
Extending previous work on quantile classifiers (q-classifiers) we propose the q*-classifier for the class imbalance problem. The classifier assigns a sample to the minority class if the minority class conditional probability exceeds 0 < q* < 1, where q* equals the unconditional probability of observing a minority class sample. The motivation for q*-classification stems from a density-based approach and leads to the useful property that the q*-classifier maximizes the sum of the true positive and true negative rates. Moreover, because the procedure can be equivalently expressed as a cost-weighted Bayes classifier, it also minimizes weighted risk. Because of this dual optimization, the q*-classifier can achieve near zero risk in imbalance problems, while simultaneously optimizing true positive and true negative rates. We use random forests to apply q*-classification. This new method which we call RFQ is shown to outperform or is competitive with existing techniques with respect to tt-mean performance and variable selection. Extensions to the multiclass imbalanced setting are also considered.
Keywords: Weighted Bayes Classifier, Response-based Sampling, Class Imbalance, Minority Class, Random Forests
1. Introduction
Random forests, introduced by Leo Breiman [1], is an increasingly popular learning algorithm that offers fast training, excellent performance, and great flexibility in its ability to handle all types of data [2, 3]. It provides its own internal generalization error estimate (i.e., out-of-bag error) as well as measures of variable importance [1, 4, 5, 6] and class probability estimates [7]. Random forests has been used for predictive tasks as varied as modeling mineral prospectivity [8], lake water level forecasting [9], identifying potentially salvageable tissue after acute ischemic stroke [10], identifying biomarkers for diagnosis of Kawasaki disease [11], classifying childhood onset schizophrenia [12], electrical load forecasting [13], and for pedestrian detection [14]. The random forests algorithm has also been generalized beyond classification and regression, most importantly to random survival forests, where each terminal node of a tree in the forest provides a survival function estimate [15, 16]. Random survival forests has been used to analyze survival problems with great success; for example, in esophageal cancer staging [17, 18].
In biomedical and other real world applications, a common problem is the occurrence of imbalanced data, defined as data featuring high-imbalance in the frequency of the observed class labels (see Section 2 for a formal definition). Some examples are disease prediction [19] and diagnosing aviation turbulence [20]. Imbalanced data has been observed to seriously hinder the classification performance of learning algorithms, including random forests and other ensemble methods because their decisions are based on classification error [21] and where there is high-imbalance in the frequency of the observed class labels a low error rate can be achieved by classifying all of the samples as members of the majority class.
Classification of class imbalanced data sets has been identified as a top problem in machine learning [22] and there is an ever increasing body of literature devoted to this extremely important problem. He and Garcia [23] and Sun et al. [24] systematically reviewed classification in the presence of class imbalance. As examples of more specialized reviews, Galar et al. [21] focused specifically on using ensembles to learn class imbalanced data and Lopez et al. [25, 26] explored imbalanced data characteristics. Finally, three very recent, useful reviews deserve mentioning: Krawczyk [27] thoroughly reviewed open research challenges in learning imbalanced data; Haixiang et al. [28] exhaustively reviewed existing papers on imbalanced data published between 2006–2016 and categorized them with respect to method and the journals in which they were published; and Das et al. [29] provided a comprehensive review of current approaches to imbalanced data and class overlap and open issues with the same in the broader context of data irregularities.
Section 2 formally defines the class imbalance problem and provides a breakdown of methods that have been used to address this problem. As discussed there, of the various methods proposed, under-sampling the majority class so that its cardinality matches that of the minority class is among the most popular. In the context of random forests, undersampling the majority class provides improved classification performance with respect to the minority class [30] and appears to be the most common approach when using random forests to learn imbalanced data due to the fact that it was implemented in Breiman’s original Fortran code [4] used by the randomForest R-package [31]. This method is called balanced random forests (BRF) and and it is an example of what has been referred to in the literature [32] as a data level method, which transform the distributions of the classes in the training data. We will show that BRF has an important connection to our approach even though our method is not an example of a data level method.
In Section 3, we propose our new approach to the class imbalance problem using a density-based argument. This results in a classifier that can be seen to be an example of a quantile classifier, or q-classifier [33], which classifies samples based on whether the conditional probability of the minority class exceeds a specified threshold 0 < q < 1. Theorem 2 shows that the specific threshold value q* of this classifier (q*-classifier) has the useful property that it maximizes the true positive and true negative rates. Moreover, because it can be equivalently expressed as a cost-weighted Bayes classifier, it is also shown to minimize weighted risk. Because of this dual optimization, unlike the traditional Bayes classifier (which is a q = 0.5 median classifier), the q*-classifier can achieve near zero risk in highly imbalanced data, while simultaneously optimizing true positive and true negative rates. Furthermore, we show surprisingly that balanced sampling as used by BRF also has this optimality property of maximizing the true positive and true negative rates (Theorem 3). Moreover, we show that the q*-classifier’s optimality continues to hold even if sampling strategies are imposed (Theorem 4). Because we choose to implement q*-classification using the full data, this means means balanced sampling, while achieving the same optimality property, comes at the cost of efficiency since it only uses a fraction of the data. We apply q*-classification with random forests, which we call RFQ, using a large comparative benchmark study (Section 4) and find it highly competitive and not only that but we are able to identify conditions under which RFQ significantly outperforms BRF (Section 5). These findings are further confirmed using synthetic data and through in-depth case study analyses. Section 6 shows RFQ also outperforms BRF with respect to variable selection. Section 7 considers the extension of RFQ to the multiclass imbalanced setting. Section 8 compares RFQ to boosting methods. Section 9 concludes with a discussion of our findings.
2. Imbalanced data setting
We now formally define the imbalanced data setting and introduce notation to be used throughout the paper. Denote the learning data by ℒ = (Xi, Yi)1≤i≤N where is the d-dimensional feature and Yi ∈ {0, 1} is the binary ordinal response. It is assumed that (Xi, Yi) are i.i.d. from a common distribution ℙ. Let (X, Y ) denote an independent generic data point with distribution ℙ.
Our goal is to build an accurate classifier for Y given X = x when the learning data is imbalanced. To help quantify what is meant by “imbalancedness”, we start by first defining the imbalance ratio (IR). Following the convention in the literature, we assume that the majority class labels are 0, and outnumber the minority class labels, 1.
Definition 1. The imbalance ratio (IR) is defined as IR = N0/N1 where N0 and N1 denote the cardinality of the majority and minority samples, respectively. A data set is imbalanced if IR ≫ 1.
It has been observed that class imbalance is not a problem in and of itself and does not necessarily lead to poor generalization in classification. If the training data is such that the classes can be separated in the feature space, then good classification will be achieved irrespective of IR. Rather the problem is that of training the classifier on too few minority examples in the presence of class overlap and small subgroups of minority class examples surrounded by majority class examples in the data space (some-times referred to as “small disjuncts”), which frequently characterize imbalanced data. This combination of characteristics, termed “concept complexity,” [34] make it difficult for a classifier to construct a good decision boundary leading to poor classification performance [35, 36, 37].
In order to quantify the complexity of imbalanced data, we adopt the approaches in [38] and [36], where they evaluate the distribution of the two classes in the local neighborhood of each minority example using k = 5 nearest neighbors. We adopt their taxonomy of types of minority examples except that we make no distinction between minority examples with 4/5 and 5/5 nearest neighbors of the majority class.
Definition 2. A minority class example is safe, borderline, or rare if 0 to 1, 2 to 3, or 4 to 5 of its 5 nearest neighbors are of the majority class, respectively.
We show in Section 5 that the percentage of minority class samples that are rare plays an important role in explaining differences between the q*-classifier and BRF.
Now we define some formal notions of imbalancedness. Following [39], we distinguish between marginally imbalanced and conditionally imbalanced data.
Definition 3. The data is marginally imbalanced if p(x) ≪1/2 for all where p(x) = ℙ{Y = 1|X = x}.
Thus, marginally imbalanced data is data for which the probability of the minority class is close to zero throughout the feature space.
Definition 4. The data is conditionally imbalanced if there exists a set with nonzero probability, ℙ{X ∈ A} > 0, such that ℙ{Y = 1|X ∈ A} ≈ 1 and p(x) ≪1/2 for x ∉ A.
In contrast to marginally imbalanced data, conditional imbalancedness occurs when the probability of the minority class is close to 1 given the features lie in a certain set, and approximately zero otherwise. In both cases, it is assumed that the minority class is rare.
2.1. Related work
As briefly mentioned in the Introduction, there is a vast literature on methods that have been used for the class imbalance problem. Methods to address the problem can be broadly grouped into data level methods, which transform the distributions of the classes in the training data, and algorithmic level methods, which adapt existing learning algorithms or develop new ones [32].
2.1.1. Data level methods
Data level methods, by far the most popular approach to imbalanced data [28], can be further subdivided into those that undersample the majority class or oversample the minority class to achieve balanced training data.
One-Sided Sampling [40] selectively subsamples the majority class, removing only majority class instances that are either redundant with other majority class instances or have minority class in-stances as their 1-NN. These “Tomek links” are removed since a small amount of attribute noise can push these examples to the incorrect side of the decision boundary.
Balanced Random Forests (BRF) [30], discussed in the Introduction, undersamples the majority class so that its cardinality matches that of the minority class in each bootstrap sample. BRF is a common approach when using random forests due to the fact it is implemented in the popular randomForest R-package [31].
Neighborhood Balanced Bagging [38] focuses bootstrap sampling toward minority examples that are difficult to learn while simultaneously decreasing probabilities of selecting examples from the majority class. The extent to which an example is considered difficult to learn is quantified by determining the number of majority examples among its k-nearest neighbors.
Synthetic Minority Over-sampling Technique (SMOTE) [41, 42] generates new artificial minority class examples by interpolating among the k-nearest neighbors that are of the minority class (i.e., artificial minority instances are introduced on the lines between each minority instance and its k-nearest minority class neighbors until the class frequencies are approximately balanced).
A number of methods that combine boosting with sampling the data at each boosting iteration have been developed; SMOTEBoost [43] combines SMOTE with boosting, RUSBoost [44] combines random undersampling with boosting, and EUSBoost [45] combines evolutionary undersampling with boosting.
2.1.2. Algorithmic level methods
As an alternative to sampling the data to balance the cardinality of the classes, learning algorithms can be modified to improve classification over the minority class.
SHRINK [46] labels all the instances in a region as minority class provided the region contains at least one minority class example. SHRINK then searches over these regions for the optimal minority class region with the greatest number of minority class samples relative to majority class samples. However, SHRINK fails in data sets where there exists more than one substantial cluster of minority class instances and it provides no advantage in data sets without significant class overlap.
Hellinger Distance Decision Trees (HDDT) [47] use Hellinger distance, a measure of distributional divergence, as the splitting criterion. The authors argue that the skew insensitivity of Hellinger distance makes it superior to standard splitting rules such as the Gini index in the presence of imbalanced data.
Near-Bayesian Support Vector Machines (NBSVM) [48] combines decision boundary shifting with unequal regularization costs for the majority and minority classes. NBSVM uses the empirical relative frequencies of the two classes as estimates of the prior probabilities to shift the decision boundary toward the Bayes optimal decision boundary. However, the performance of NBSVM is kernel-dependent (as with standard SVM) and is poor when the minority class is compact in comparison to the majority class.
Class Switching according to Nearest Enemy Distance [49] adapts a technique proposed by Breiman [50] to highly imbalanced data by switching the labels of majority class samples with a probability proportional to their Euclidean distance to the closest minority class sample.
3. An optimal quantile classifier for class imbalanced data
Our approach falls under the class of algorithmic level procedures. Following [33], we define a quantile classifier (q-classifier) as
where 0 < q < 1 is a prespecified quantile threshold. If we have q = 1/2, which [33] term a median classifier, we obtain the familiar Bayes classifier:
As noted in [33], minimizing loss subject to unequal misclassification costs is equivalent to classification based on p(x) ≥ q using thresholds q other than 1/2. This will be demonstrated presently and in so doing explain why imbalanced data is so challenging for classifiers.
Define the risk for a classifier (x) as
| (1) |
Here ℓ0, ℓ1 > 0 are fixed constants associated with the cost of making one of the two classification errors: A0 is the cost of misclassifying a majority class instance; ℓ1 is the cost for misclassifying a minority class instance. Assigning specific losses leads to the interpretation of (1) as a cost-weighted risk function. Under uniform weights ℓ0 = ℓ1 = 1, the risk (1) simplifies to classification error, which we denote as .
Under the cost-weighted risk (1), the optimal classifier is the cost-weighted Bayes rule, defined as
| (2) |
which we recognize as a quantile classifier with q = ℓ0/(ℓ0 + ℓ1). The following well known result establishes the optimality of the cost-weighted Bayes classifier [51]. For convenience we provide a proof in Appendix A.
Theorem 1. The cost-weighted Bayes rule is optimal in that its risk satisfies for any classifier Its risk equals
| (3) |
Thus, (3) is the smallest weighted risk achievable by a decision rule.
Now consider what happens in imbalanced data if performance is measured using misclassification error, ℓ0 = ℓ1 = 1. In this case, the cost-weighted Bayes classifier reduces to the (unweighted) Bayes classifier. Assuming marginal imbalance, i.e. p(x)≪ 0.5, the Bayes rule is δB(x) = 0, thereby classifying all observations as majority class labels. Under classification error we know this must be the optimal rule. In particular by (3), the Bayes error equals r(δB) = 𝔼[min{p(X), 1 − p(X)}] = 𝔼 [p(X)] ≈ 0 which is essentially perfect.
3.1. A density-based approach
We see that classification error provides a strong incentive for learning algorithms to correctly classify majority class samples at the expense of misclassifying minority class samples. This is obviously problematic and a better approach is to demand good performance from a classifier under both types of classification errors. Define the TNR (true negative) and TPR (true positive) value for a classifier as follows:
Our goal is to find a classifier that achieves both high TNR and TPR values in imbalance problems. The Bayes rule, δB, does not achieve this goal because it has a TNR value of 1 but a TPR value of 0.
Definition 5. A classifier is said to be TNR+TPR-optimal if it maximizes the sum of the rates, TNR + TPR.
To achieve the goal of TNR+TPR optimality, we introduce the following classifier derived from a density-based approach. The classifier, denoted by δD(x), assigns an instance x to the minority class if its data density for minority class labels, fX|Y (x|1), is larger than the data density for majority class labels, fX|Y (x|0):
Basing the classifier on the conditional density of the features, fX|Y, rather than the conditional density of the response, p(x), removes the effect of the prevalence of the minority class labels. This is one way to see how δD is able to handle imbalancedness. More directly we can show that δD is TNR+TPR-optimal. Here is an informal argument showing this. First notice that for a classifier to achieve TNR+TPR-optimality it should maximize the probability of the events this being equivalent to tracking the regions of the data space where the respective conditional densities are maximized. The value of TNR + TPR conditional on ℒ for the classifier equals
The right-hand side is the TNR + TPR value for δD, which shows that it is the optimal TNR+TPR-rule. A more formal proof of this fact is given shortly.
Before proceeding, we introduce a table of notation that will be particularly useful in this and subsequent Sections (see Table 1).
Table 1:
Notation used throughout the paper.
| generic classifier | |
| misclassification costs for majority and minority classes | |
| π | the marginal probability that Y = 1, π = ℙ {Y = 1} |
| = N1/N | relative frequency of minority class |
| p(x) | conditional class probability function, p(x) = ℙ {Y = 1|X = x} |
| fX (x | density for X |
| δq(x) | quantile classifier (q-classifier) |
| δB(x) | Bayes classifier |
| δWB(x) | cost-weighted Bayes classifier |
| δq* (x) | q*-classifier (quantile classifier with q = π) |
3.2. The q*-classifier
While it is convenient theoretically to describe the density-based classifier in terms of the conditional density of the data, in practice it will be difficult to implement the classifier as stated. However, we can rewrite δD(x) using the following identity:
Cancelling the common value fX (x) in the numerator and denominator, and using the notation of Table 1, we have
| (4) |
With a little bit of rearrangement, we now see that (4) is a q-classifier with q = π (notice analogously for the Bayes classifier that δB(x) = 1{∆B (x)≥1} where ∆B(x) = p(x)/(1 − p(x)), which is a q-classifier with q = 0.5). This leads to the following definition of the proposed classifier.
Definition 6. Call δq* (x) = 1{p(x)≥π} the q*-classifier (and keep in mind δq* = δD).
Although [33] introduced the extremely useful concept of a quantile classifier, they did not address how to select the optimal q. In deriving the q*-classifier, we have informally argued that q should be π. In the following result, we formally justify our selection of q by showing that the q*-classifier is able to achieve a near zero risk while jointly optimizing TNR and TPR.
Theorem 2. The q*-classifier is TNR+TPR-optimal. Furthermore, it is the cost-weighted Bayes rule (2) under misclassification costs ℓ0 = π and ℓ1 = (1 − π).
Theorem 2 shows that the q*-classifier is not only TNR+TPR-optimal, but also weighted risk optimal under misclassification costs ℓ0 = π and ℓ1 = (1 − π). In particular, by (3) of Theorem 1 we have
Notice that the right-hand side will be nearly zero for both types of imbalanced data: marginally and conditionally imbalanced. Moreover, unlike the Bayes rule, which also achieves a near zero risk, Theorem 2 shows the q*-classifier is able to do this while satisfying the requirement of a jointly optimized TNR and TPR.
Proof of Theorem 2. Maximizing TNR and TPR is equivalent to minimizing FPR = 1− TNR and FNR = 1− TPR. For any classifier , we have by definition
Minimizing the above expression does not change if we multiply by ℓ0ℓ1 throughout. Therefore, minimizing the FPR and FNR rate is equivalent to minimizing
which is the weighted risk r(, ℓ0, ℓ1) where ℓ0 = π and ℓ1 = 1 − π. By Theorem 1, this is minimized by the weighted Bayes rule (2), which equals the q*-classifier under the stated choices for ℓ0 and ℓ1. □
3.3. Response-based sampling: balancing the data
One common strategy to overcome the imbalance problem is to undersample the majority class to evenly balance the data. We can describe this process more formally by introducing auxiliary variables Si ∈ {0, 1} where values Si = 1 indicate subsampled cases. The subsampled learning data is defined as ℒS = {(Xi, Yi) : Si = 1, i = 1, …, N } where data values are selected with probability that depend only on the value of Y and not X. This is called response-based sampling. In particular,
| (5) |
Where
By (5), the probability a randomly selected Y from ℒS equals Y = 1 is
| (6) |
Likewise, . In order to have balanced labels we must have πS = 1/2, or equivalently πS = 1 − πS, which implies by (6)
| (7) |
The factor (7) calls to mind the factor in (4) that modulates the difference between δB and δD. This is not a coincidence as we now show. In what follows, we expand upon the justification for undersampling provided by [33], which can be inferred from [52]. Let be the Bayes rule constructed using ℒS (call this the subsampled Bayes rule). For a given x,
where by definition . By Bayes theorem,
Consequently,
By definition,
Noting that
we have
Applying a similar argument to and cancelling the common value fX (x) and , deduce that
| (8) |
Therefore,
Under (7), the right-hand side equals π/(1 − π). Hence, under (7). This implies that the subsampled Bayes rule is TNR+TPR-optimal under (7).
Theorem 3. Under balanced subsampling (7), the subsampled Bayes rule is TNR+TPR-optimal.
3.4. The q*-classifier is invariant to response-based sampling
In contrast, the q*-classifier is unaffected by response-based sampling and retains its TNR+TPR-optimality no matter what the target balance ratio is. Let be the q*-classifier constructed using . By definition, where . Equivalently,
Therefore, using (8),
| (9) |
where we have used the following identity which follows from (6)
In other words, (compare (9) to (4)). We can therefore conclude that δq* remains TNR+TPR-optimal. Combined with Theorem 3 we have therefore established the following.
Theorem 4. Under response-based sampling of the form (5), and therefore is TNR+TPR-optimal. Moreover, under balanced sampling (7), all three methods are equivalent:
and all three methods are TNR+TPR-optimal.
4. Application to random forests
In practice, the value of p(x) is unknown and therefore must be estimated. In this scenario, when we refer to q*-classification we mean classification using an estimated value for p(x) to classify observations using the quantile q = π. In general, we can apply q-classification based on any specified 0 < q < 1. Here we investigate the performance of q*-classification when applied with random forests. We refer to this procedure as RFQ. As a comparison procedure, we will use balanced random forests, which we continue to refer to as BRF. We also use the standard random forests algorithm as comparison and refer to this as RF.
Algorithm 1 provides a description of the RF classification algorithm. The algorithm requires the following parameters: ntree (number of trees trained in the forest), nodesize (target terminal node size), and mtry (number of random features used to split a tree node). RFQ and BRF apply Algorithm 1 exactly as RF does but with the following one line modifications:
RFQ: Line 17 of Algorithm 1 is modified as follows. In place of median (Bayes) classification, RFQ applies q*-classification,
BRF: Line 5 of Algorithm 1 is modified as follows. Rather than selecting a bootstrap sample of size N, a sample of size 2N1 is used, where the probabilities for minority and majority class instances to be selected for the bootstrap sample are πS (1) = (N0/N1)πS (0), thus satisfying the balancing condition (7). Keep in mind that BRF uses the Bayes rule for classification; thus the classification rule used in Line 17 is the same for BRF.
4.1. Performance comparisons on benchmark imbalanced data
In theory, both BRF and RFQ will possesses the TNR+TPR-property: this is true for BRF by Theorem 3 because it satisfies the balancing condition (7), while for RFQ this holds by Theorem 2 because it applies q*-classification. However this is predicated on knowledge of the true probability function p(x), which in practice must be estimated, and therefore performance in practice may be very different. In particular, an advantage of RFQ is that is uses a much larger sample size than BRF which should increase its efficiency in estimating p(x).
To see how the two methods performed in practice we tested them using a diverse collection of 143 benchmark imbalanced data sets (see Figure 1 for summary statistics of the data sets; Supplementary Materials Appendix C provides background information on the data). Analyses were performed in R [53] using the R-package randomForestSRC [54]. Forests of size ntree = 1000 were used for each training data set. Default settings for random forests were used: trees were grown to purity (nodesize = 1), and random feature selection was set at mtry = d/3. Tree node splits (Lines 6–12 of Algorithm 1) were implemented using Gini splitting. The value q* = π required for RFQ was estimated using the empirical relative frequency of the minority class labels, = N1/(N0 + N1). In addition to BRF and RF, we also considered standard random forests under Hellinger distance splitting [47], and BRF with Hellinger splitting.
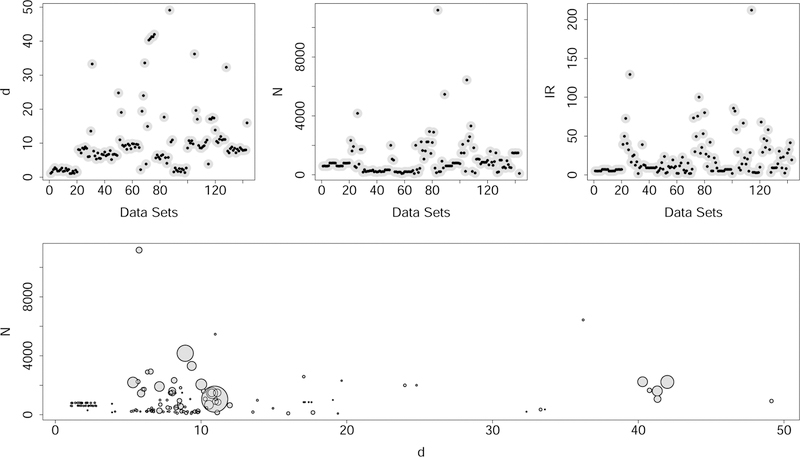
Figure 1:
Summary of 143 benchmark imbalanced data sets. Top figures display dimension of feature space d, sample size N, and imbalance ratio IR. Bottom figure displays d versus N with symbol size displaying value of IR. This identifies several interesting data sets with large IR values, with some of these having larger d.
4.1.1. Performance metrics: the G-mean
In assessing performance, we used TNR, TPR, and the G-mean. The G-mean is the geometric mean of TNR and TPR, i.e., G-mean = (TNR × TPR)1/2 and it is meant to replace misclassification rate in imbalanced data settings, since an overall accuracy close to 1 can be achieved by classifying all data points as majority class labels for heavily imbalanced data as previously noted. By way of contrast, the tt-mean is close to 1 only when both the true negative and true positive rates are close to 1 and the difference between the two is small [46].
4.1.2. The q*-classifier appears to optimize the G-mean
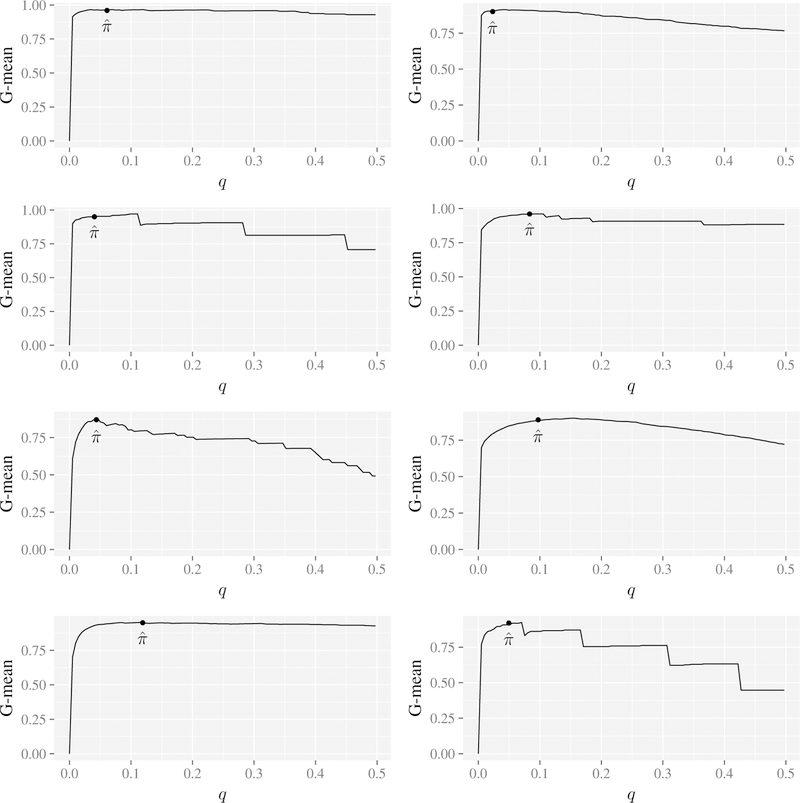
Before discussing the results, it is worth noting that even though the q*-classifier was not specifically developed to maximize the G-mean, we observed that by applying random forests q-classification under different values of q, that the maximum G-mean is achieved when q is approximately (i.e., the G-mean appears to be maximized by RFQ). This is illustrated in Figure 2 using 8 selected benchmark data sets. This is strong evidence that TNR+TPR-optimality is a useful property for a classifier.
Figure 2:
G-mean from random forests q-classification using various q for thresholding for 8 different bench-mark data sets. Notice that the maximum value is near in all instances.
4.1.3. Results
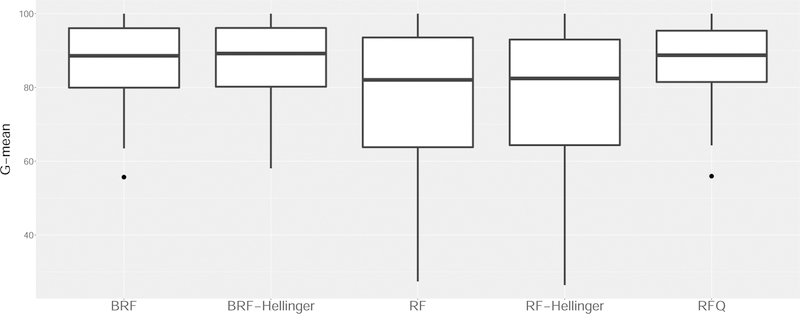
For the analysis of the 143 benchmark data sets, we used 10-fold cross-validation repeated 250 times. The G-mean for each procedure is reported in Figure 3. We observe that RFQ and BRF outperform all other methods. We also observe that using Hellinger distance as the splitting criterion instead of the Gini index does not noticeably improve performance, and thus we did not include it in further experiments.
Figure 3:
G-mean performance of different classifiers across 143 benchmark imbalanced data sets. (BRF=Balanced Random Forests; RF=Random Forests; RFQ = Random Forests q*-classifier).
5. Analyzing performance differences between RFQ and BRF
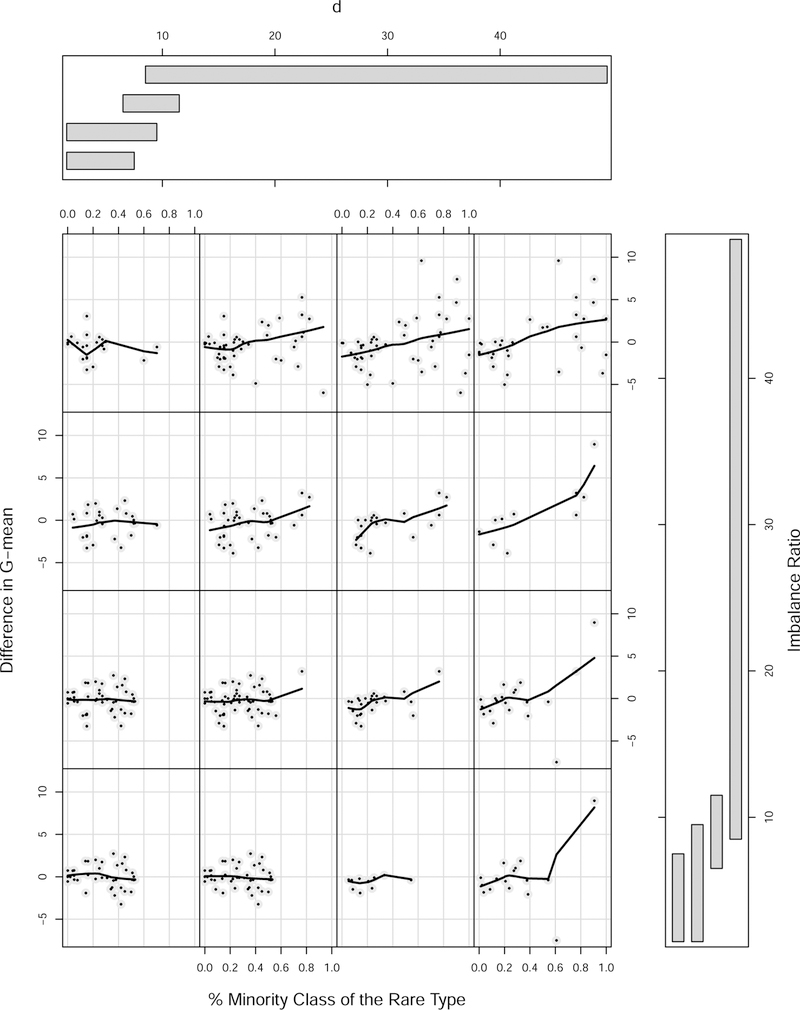
From Figure 3 it appears that RFQ and BRF have roughly similar performance overall. However, upon further investigation (Figure 4), we found that when the imbalance ratio is high, and when the percent of minority class examples of the rare type is high, and when d is high, RFQ outperformed BRF. We investigate this effect further in this Section.
Figure 4:
A closer look at difference in G-mean performance of RFQ and BRF for benchmark data sets. Vertical axis plots difference in G-mean as a function of % rare minority class examples, feature dimension d, and imbalance ratio IR. There is an increasing trend upwards (thus favoring RFQ) as % rare minority class examples increases with increasing d and increasing IR.
5.1. An explanation of why RFQ is better
As we have noted previously, while RFQ and BRF both possess the TNR+TPR optimality property, in practice the difference between the two methods is that RFQ utilizes all N data points, whereas BRF uses the smaller sample size of 2N1, which it must in order to balance the data.
We suggested that the reduced sample size of BRF reduces its efficiency in estimating unknown model parameters. We now provide a more detailed explanation of how this affects BRF’s performance for the scenarios described above. We consider a simple logistic regression setting where the true conditional class probability function is
By (4), δq* (x) = 1 if log(∆D(x)) ≥ 0. Hence x is classified as a minority class instance if
Under the logistic model this simplifies to α + βT x ≥ γ, where γ = log(π/(1 − π)) (as comparison, the Bayes rule, δB(x), classifies x as a minority class sample if log(∆B(x)) ≥ 0, which simplifies to α + βT x ≥ 0). To gain more insight into δq* (x), first note the following identity for π:
Now in the setting of marginal imbalance, since p(x) ≈ 0, we must have α ≪ 0, and therefore,
Combining this with π ≈ 0, deduce that
The q*-classifier classifies x as a minority class instance if α + βT x ≥ γ. Hence, δq* (x) = 1 if
For example, if X ∼ N(µ, Σ)
Therefore,
The above represents the theoretical boundary for achieving TNR+TPR optimality, but RFQ and BRF must classify the data according to an estimated δq*. Suppose the two procedures directly estimate δq* (x) by estimating θ = (β, βT µ, βT Σβ) (i.e., instead of indirectly estimating p(x)). Then RFQ will have an advantage because estimating θ uses data across both classes and RFQ uses all N data points whereas BRF uses a sample size of 2N1 evenly split across the two classes. Furthermore, performance differences will become magnified as the imbalance ratio increases (since 2N1 becomes even smaller compared with N ) and when the dimension d increases (since estimation becomes more difficult). This also explains why RFQ is better in rare instance settings. Recall from Definition 2 that a minority class example x is rare if 4 to 5 of its nearest neigbors are majority class examples. We can imagine a setting where rare instances are a by product of indistinguishable conditional densities. That is, for x′ close to x we have f (x′|1) = f (x′|0). If this region has positive measure, then by the identification of finite mixtures of multivariate normals, f (x|1) = f (x|0) = f (x) almost everywhere. This shows data from both classes are important for estimating all components of θ, thus further favoring RFQ.
5.2. Performance comparisons on simulated data
To provide further evidence for the above, we converted five simulations from the mlbench R-package [55] into imbalanced data in addition to simulating imbalanced data directly using caret package [56] as detailed in Table 2. We repeated each experiment 250 times with forests of 5000 trees grown on each training data set (nodesize=1, mtry=d/3). We compared the performance of RFQ to BRF and standard random forests (RF) and obtained the following results reported in Table 3. The results are consistent with what we observed across the 143 benchmark data sets. Clearly, RFQ outperforms BRF (as well as RF) with respect to the G-mean, across all of the six simulated high-dimensional imbalanced data models (Wilcoxon signed rank test p-value = 0.03). This shows RFQ can offer significant improvement for complex imbalanced data in high-dimensional settings.
Table 2:
Simulated data sets.
| Na | Signalb | Noisec | IRd | % Raree | |
|---|---|---|---|---|---|
| Two Normf | 1250 | 20 | 500 | 49 | 99% |
| Waveformg | 1250 | 21 | 500 | 49 | 99% |
| TwoClassSimh | 1250 | 20 | 500 | 9 | 92% |
| Friedman 1i | 1250 | 5 | 500 | 49 | 99% |
| Friedman 2i | 1250 | 4 | 500 | 48 | 99% |
| Friedman 3i | 1250 | 4 | 500 | 49 | 99% |
The sample size for training data and test data.
The number of signal (true) variables.
The number of resampled noise variables.
As defined in Definition 1.
As defined in Definition 2.
Class 2 is randomly downsampled to 25 instances.
Classes 1 + 2 form the majority class; class 3 is randomly down-sampled to 25 instances.
Intercept =16 and 100 of the 500 noise variables correlated with ρ = 0.7.
Where y ≥ yq=0.98 are classified as 1 and 0 otherwise.
Table 3:
Performance comparisons on simulated data sets.
| RFQ | BRF | RF | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TPR | TNR | G-mean | TPR | TNR | G-mean | TPR | TNR | G-mean | |
| Two Norm | 86.71 | 58.88 | 71.34 | 13.55 | 100 | 35.37 | 1.96 | 100 | 14.00 |
| Waveform | 91.58 | 56.04 | 71.56 | 53.98 | 94.29 | 70.87 | 1.96 | 100 | 14.00 |
| TwoClassSim | 84.73 | 56.32 | 69.00 | 7.38 | 99.79 | 26.28 | 1.86 | 99.98 | 11.49 |
| Friedman 1 | 68.20 | 54.13 | 60.46 | 2.93 | 99.97 | 16.43 | 2.00 | 100 | 14.12 |
| Friedman 2 | 95.54 | 56.26 | 73.27 | 11.35 | 99.89 | 31.99 | 1.98 | 100 | 14.02 |
| Friedman 3 | 48.71 | 54.84 | 51.33 | 2.06 | 100 | 14.26 | 2.01 | 100 | 14.14 |
5.3. Cognitive impairment data
We chose the Alzheimers Disease CSF Data from the AppliedPredictiveModeling R-package [57] to further explore performance of RFQ in difficult settings. This data set is a modified version of the data in [58]. There are N = 333 observations with d = 130 predictors; the outcome is presence/absence of cognitive impairment with N0 = 242 controls and N1 = 91 impaired, for an IR of 2.66. We explored the relationship among performance, dimensionality, and IR by adding progressively more noise variables (obtained by resampling the predictor variables) and by progressively subsampling the minority class, where each smaller subsample of the minority class was randomly sampled from the subsample of the previous iteration (i.e., nested subsamples). Table 4 contains the results of 10-fold cross-validation repeated 250 times under the various scenarios with forests of 5000 trees grown on each training data set with nodesize=1, mtry=d/3 for each scenario.
Table 4:
Performance on cognitive impairment data.
| RFQ | BRF | RF | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TPR | TNR | G-mean | TPR | TNR | G-mean | TPR | TNR | G-mean | |
| Scenario 1a | 88.78 | 71.48 | 79.34 | 75.82 | 88.14 | 81.33 | 49.66 | 96.42 | 68.34 |
| Scenario 2b | 89.72 | 69.21 | 78.50 | 65.83 | 89.82 | 76.35 | 27.83 | 98.86 | 50.93 |
| Scenario 3c | 89.09 | 66.89 | 76.87 | 59.11 | 90.84 | 72.64 | 14.45 | 99.64 | 36.19 |
| Scenario 4d | 87.92 | 62.58 | 73.78 | 48.67 | 92.44 | 66.24 | 8.23 | 100 | 27.57 |
| Scenario 5e | 88.82 | 65.87 | 76.13 | 65.68 | 89.78 | 76.19 | 13.79 | 99.59 | 35.14 |
| Scenario 6f | 89.37 | 60.48 | 73.11 | 52.82 | 92.55 | 69.09 | 7.27 | 99.99 | 26.06 |
| Scenario 7g | 88.94 | 55.19 | 69.56 | 39.03 | 94.53 | 59.36 | 5.75 | 100 | 23.68 |
| Scenario 8h | 88.83 | 47.01 | 64.01 | 22.25 | 97.11 | 44.33 | 5.27 | 100 | 22.92 |
| Scenario 9i | 84.54 | 62.10 | 71.97 | 56.98 | 89.57 | 70.61 | 6.85 | 99.95 | 25.38 |
| Scenario 10j | 84.94 | 53.33 | 66.73 | 38.26 | 94.42 | 58.45 | 5.46 | 100 | 23.23 |
| Scenario 11k | 85.17 | 45.42 | 61.56 | 20.99 | 97.14 | 42.53 | 5.23 | 100 | 22.86 |
| Scenario 12l | 84.46 | 36.85 | 55.01 | 9.46 | 99.24 | 28.80 | 5.21 | 100 | 22.83 |
| Scenario 13m | 78.76 | 61.26 | 68.88 | 49.17 | 88.42 | 64.65 | 5.55 | 100 | 23.37 |
| Scenario 14n | 78.33 | 51.41 | 62.84 | 25.64 | 95.35 | 46.63 | 5.22 | 100 | 22.84 |
| Scenario 15o | 79.15 | 43.84 | 58.21 | 12.10 | 98.41 | 31.94 | 5.21 | 100 | 22.83 |
| Scenario 16p | 78.96 | 36.10 | 52.63 | 6.49 | 99.74 | 24.68 | 5.21 | 100 | 22.83 |
Original data
Original data + 200 noise variables
Original data + 500 noise variables
Original data + 1000 noise variables
Subsampled data with 40 cases randomly selected and all controls
Subsampled data with 40 cases randomly selected and all controls + 200 noise variables
Subsampled data with 40 cases randomly selected and all controls + 500 noise variables
Subsampled data with 40 cases randomly selected and all controls + 1000 noise variables
Subsampled data with 20 cases randomly selected and all controls
Subsampled data with 20 cases randomly selected and all controls + 200 noise variables
Subsampled data with 20 cases randomly selected and all controls + 500 noise variables
Subsampled data with 20 cases randomly selected and all controls + 1000 noise variables
Subsampled data with 10 cases randomly selected and all controls
Subsampled data with 10 cases randomly selected and all controls + 200 noise variables
Subsampled data with 10 cases randomly selected and all controls + 500 noise variables
Subsampled data with 10 cases randomly selected and all controls + 1000 noise variables
Even though the unaltered cognitive impairment data features a modest IR of 2.66, standard random forests (RF) only classifies slightly more than half of the patients with cognitive impairment correctly and its performance rapidly deteriorates with the addition of noise and increasing IR through subsampling the minority class. While BRF tends to perform well on the unaltered data and under increasing IR, its performance rapidly deteriorates in higher dimensions (i.e., with increasing noise) to the point that its performance is not much better than RF and significantly inferior to RFQ. In contrast, RFQ outperforms BRF (and RF) with respect to the G-mean under all scenarios considered except for the unaltered data with no noise and the data with 40 of the 91 cognitively impaired patients randomly selected with no noise (Wilcoxon signed rank test p-value < 0.001). Under all scenarios the performance of RFQ over the minority class remains constant and is superior to BRF and RF, although with the cost of an increased FPR with increasing dimensions.
5.4. Customer churn data
As another example, we looked at the Customer Churn Data from the C50 R-package [59]. This is artificial customer churn data modeled on real world data where the outcome is customer churn yes/no. The data is already split into training and test data, so no cross-validation is required. In the training data there are N = 3333 observations of which N1 = 483 are instances of customer churn, for an IR of 5.90.
As with the cognitive impairment data, we progressively add more noise variables and progressively subsample the minority class. Table 5 contains the results of running the test data through the forests under the various scenarios with forests of 5000 trees grown on each training data set with nodesize=1, mtry=d/3 for each scenario.
Table 5:
Performance on customer churn data.
| RFQ | BRF | RF | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TPR | TNR | G-mean | TPR | TNR | G-mean | TPR | TNR | G-mean | |
| Scenario 1a | 86.19 | 90.02 | 88.09 | 83.52 | 95.64 | 89.37 | 73.72 | 99.79 | 85.77 |
| Scenario 2b | 89.31 | 75.55 | 82.14 | 64.37 | 94.11 | 77.83 | 30.07 | 100 | 54.83 |
| Scenario 3c | 91.09 | 71.60 | 80.76 | 42.98 | 96.33 | 64.35 | 18.49 | 100 | 42.99 |
| Scenario 4d | 90.20 | 69.80 | 79.34 | 34.08 | 97.64 | 57.68 | 6.46 | 100 | 25.41 |
| Scenario 5e | 87.08 | 87.88 | 87.48 | 82.18 | 93.21 | 87.52 | 61.69 | 99.93 | 78.52 |
| Scenario 6f | 88.42 | 71.46 | 79.49 | 47.44 | 94.94 | 67.11 | 9.13 | 100 | 30.22 |
| Scenario 7g | 88.42 | 67.16 | 77.06 | 35.41 | 97.02 | 58.61 | 2.45 | 100 | 15.65 |
| Scenario 8h | 85.75 | 62.94 | 73.46 | 21.16 | 99.17 | 45.81 | 0.22 | 100 | 4.72 |
| Scenario 9i | 81.29 | 84.55 | 82.91 | 73.27 | 91.83 | 82.03 | 46.55 | 100 | 68.23 |
| Scenario 10j | 80.40 | 67.93 | 73.90 | 31.85 | 95.98 | 55.29 | 2.00 | 100 | 14.16 |
| Scenario 11k | 81.29 | 64.81 | 72.58 | 25.61 | 97.37 | 49.94 | 0.22 | 100 | 4.72 |
| Scenario 12l | 78.62 | 59.82 | 68.58 | 8.69 | 99.58 | 29.41 | 0.22 | 100 | 4.72 |
| Scenario 13m | 82.63 | 84.34 | 83.48 | 65.26 | 91.69 | 77.35 | 12.25 | 100 | 35.00 |
| Scenario 14n | 80.85 | 62.11 | 70.86 | 30.96 | 96.19 | 54.57 | 2.90 | 100 | 17.02 |
| Scenario 15o | 79.51 | 59.33 | 68.69 | 13.59 | 98.96 | 36.67 | 2.45 | 100 | 15.65 |
| Scenario 16p | 80.40 | 56.01 | 67.11 | 2.00 | 99.86 | 14.15 | 0.22 | 100 | 4.72 |
Original data
Original data + 200 noise variables
Original data + 500 noise variables
Original data + 1000 noise variables
Subsampled data with 240 cases randomly selected and all controls
Subsampled data with 240 cases randomly selected and all controls + 200 noise variables
Subsampled data with 240 cases randomly selected and all controls + 500 noise variables
Subsampled data with 240 cases randomly selected and all controls + 1000 noise variables
Subsampled data with 120 cases randomly selected and all controls
Subsampled data with 120 cases randomly selected and all controls + 200 noise variables
Subsampled data with 120 cases randomly selected and all controls + 500 noise variables
Subsampled data with 120 cases randomly selected and all controls + 1000 noise variables
Subsampled data with 60 cases randomly selected and all controls
Subsampled data with 60 cases randomly selected and all controls + 200 noise variables
Subsampled data with 60 cases randomly selected and all controls + 500 noise variables
Subsampled data with 60 cases randomly selected and all controls + 1000 noise variables
We observe exactly the same pattern of performance with the customer churn data as with the cognitive impairment data. As before, the performance of RF rapidly deteriorates with the addition of noise and increasing IR; BRF performs decently on the unaltered data and under increasing IR but its performance rapidly deteriorates in higher dimensions; RFQ outperforms BRF and RF with respect to the G-mean under all scenarios except for the unaltered data with no noise and the data with 240 of the 483 instances of customer churn randomly selected with no noise (Wilcoxon signed rank test p-value < 0.001). Under all scenarios the performance of RFQ over the minority class remains constant and is superior to BRF and RF but with increased FPR in higher dimensions.
6. Variable importance
We claim that the standard variable importance (VIMP) measure in random forests introduced by Breiman and Cutler [1, 4], called Breiman-Cutler importance [6], is inappropriate for RFQ in the presence of significantly imbalanced data due to the fact that almost all nodes in an individual tree will con-tain 0’s. We propose instead to assess variable importance using the G-mean combined with Ishwaran-Kogalur importance [15, 54], the latter being an ensemble rather than tree-based measure.
In Breiman-Cutler permutation importance, a variable’s OOB (out-of-bag) data is permuted and run down the tree. The original OOB prediction error is subtracted from the resulting OOB prediction error, resulting in tree importance. Averaging this value over the forest yields permutation importance. This type of importance, which is tree-based, is appropriate for BRF because each tree is a reasonably good classifier, therefore making prediction error a reasonable way to assess a variable’s contribution to the model.
For RFQ this will not be a good measure because RFQ’s good prediction performance arises from converting a random forest ensemble classifier into a random forest ensemble q-classifier. Therefore, we will instead use Ishwaran-Kogalur importance [15, 54], an ensemble-based measure, defined as the prediction error for the original ensemble subtracted from the prediction error for the new ensemble obtained by permuting a variable’s data. For RFQ, performance is measured by the G-mean. Thus, we apply Ishwaran-Kogalur importance using G-mean prediction error. Ensembles were defined in blocks of 20 trees. For BRF, we also use G-mean for prediction error, but with Breiman-Cutler importance. We also compare results to standard random forests (RF) using Breiman-Cutler importance calculated using classification error (the standard approach).
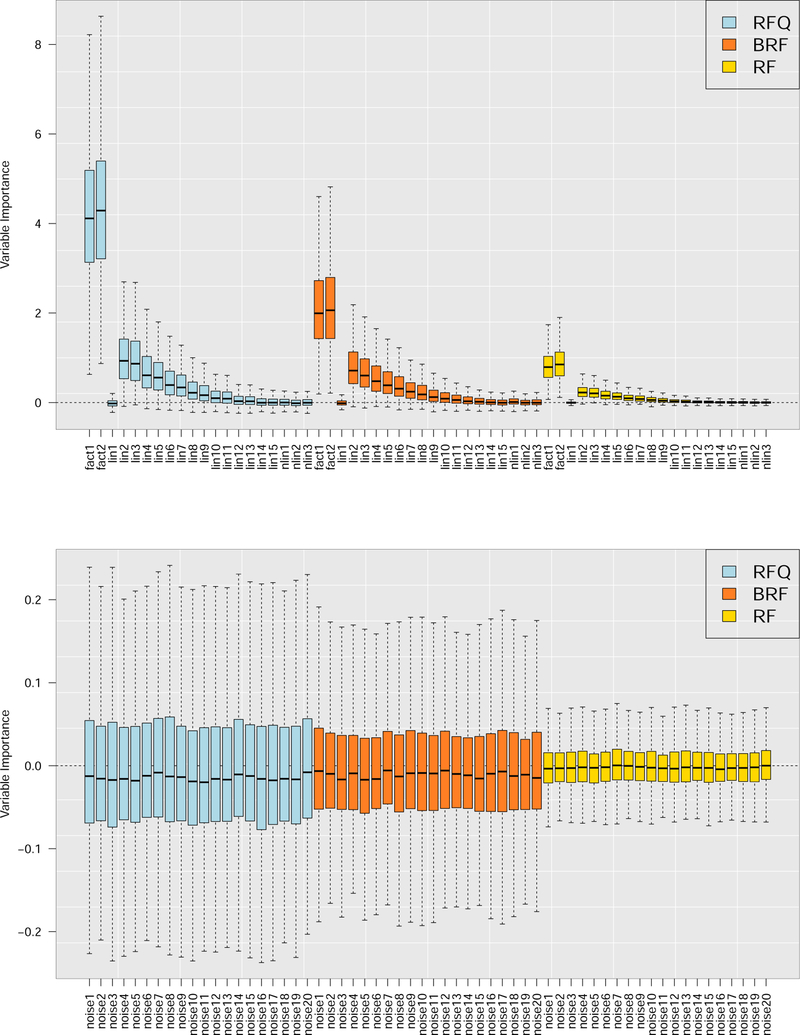
To assess performance of the proposed variable importance measures, we used the twoClassSim function from the caret package [56]: 2 factors, 15 linear variables, 3 non-linear variables, and 20 noise variables. Sample size was N = 1000 with IR = 6 which was induced by downsampling class 2. Results averaged from 1000 runs are displayed in Figure 5. The results show RFQ outperforms BRF which, in turn, outperforms RF.
Figure 5:
Variable importance (VIMP) for RFQ, BRF and RF from 1000 runs using simulated imbalanced data. There are 2 factors, 15 linear variables, 3 non-linear variables, and 20 noise variables (no signal). Top panel displays signal variables, bottom panel are noisy variables.
7. Multiclass imbalanced data
In this Section we explore the performance of RFQ, BRF and RF in the multiclass imbalanced data setting. We accomplish this by decomposing the multiclass imbalanced data into K(K-1)/2 two-class data sets, where K is the number of classes, obtaining classifiers on each and then taking a majority vote over the results. The empirical results that follow are based on forests of 5000 trees grown on each training data set with nodesize=1, mtry=d/3 and 50 resampled noise variables.
7.1. Waveform simulations
As a preliminary exploration of the more challenging multiclass imbalance data setting [60], we chose the waveform data simulation from the mlbench R-package [55], which produces three classes of (approximately) equal size. We generated N = 1000 samples for the training (initially) and test data sets. To obtain multiclass imbalanced data, we subsampled the second and third classes to obtain different class ratios. For each of the three class imbalanced data sets derived from the waveform simulation, we adopted the approach of [61] and trained RFQ, BRF and RF on two-class data sets. The multiclass classifier was obtained by taking a majority vote over the three predicted class labels for the test data. We compared the performance of the RFQ, BRF and RF multiclass classifier using Friedman’s one-vs-one approach using the true positive rate for each of the three classes and the G-Mean. This we did 250 times, averaging the results, which are listed in Table B.1 of Appendix B, where the “true positive rates” (i.e., the performance metrics within each class) are denoted by TPR1, TPR2 and TPR3, respectively.
The (unweighted) G-mean is not necessarily the appropriate metric for measuring classification performance in the multiclass imbalanced data setting, especially in cases of extreme imbalance. For the imbalanced data sets with the ratios 100:25:1, 100:10:1 and 100:5:1 the G-means for RFQ and BRF are similar but TPR for the third class with the fewest instances is in the range 85–86 for RFQ whereas the range is 42–44 for BRF. Granted, TPR over the first class with far more instances than the other classes is approximately 52–55 for RFQ whereas it is approximately 86–87 for BRF, but in real-world settings the cost associated with misclassifying the class with the fewest instances is likely to be far higher, as with two class imbalanced data. To account for this, we also looked at the G-means for two classes at a time, denoted as G-meankk’’ for classes k and k’’, as well as the weighted G-mean, which in the three class setting we define as
Specifically, in Table B.2 of Appendix B, we looked at the weighted G-mean with β1 = 1/2 and β2 = β3 = 1, which is not necessarily ideal because it does not take into account the imbalance between the second and third classes; nevertheless, it is sufficient to illustrate our point.
In Table B.2 we see a pronounced difference in the weighted G-mean with β1 = 1/2, β2 = 1, and β3 = 1 for the highly imbalanced data sets with the ratios 100:25:1, 100:10:1 and 100:5:1. To see why this is appropriate we look at the two class G-means. The performance of RFQ is superior to BRF with respect to G-mean13 and G-mean23, whereas BRF is superior to RFQ only with respect to G-mean12. Even though the performance of RFQ is superior with respect to two of the three two class G-means and the difference in G-mean23 in favor of RFQ is approximately the same as the difference in G-mean12 in favor of BRF, the unweighted G-mean is insensitive to this. For this reason, we believe that the weighted G-mean in especially appropriate in the multiclass imbalanced data setting.
7.2. Cassini simulations
As another example, we chose the cassini data simulation from the mlbench R-package [55], which produces three classes of in the ratio 2:2:1. As with the waveform simulation, we generated N = 1000 samples for the training (initially) and test data sets with noise variables and then subsampled the second and third classes to obtain different class ratios.
The averaged results from 250 repetitions are listed in Table B.3 of Appendix B. In contrast to the waveform imbalanced data sets, for the cassini data sets with the most extreme class imbalance, i.e., 100:50:1, 100:25:1, 100:10:1 and 100:5:1, RFQ is clearly superior to BRF with respect to the unweighted G-mean.
We then looked at the weighted G-mean with β1 = 1/2 and β2 = β3 = 1. In Table B.4 of Appendix B, we see an even more pronounced difference in performance between RFQ and BRF with respect to the weighted G-mean with β1 = 1/2, β2 = 1, and β3 = 1 for the extremely imbalanced data sets with the ratios 100:50:1, 100:25:1, 100:10:1 and 100:5:1 and essentially identical performance over most of the other imbalanced data sets.
It should be noted that these results are limited in that we only considered three class imbalanced data, which is a special case in that the number of competent classifiers (i.e., classifiers trained on a given class) outnumber non-competent classifiers (i.e., classifiers that were not trained on the class in question); for three class imbalanced data using one-vs-one there are exactly two competent classifiers and one non-competent classifier, so classification by majority vote works. However, the ratio of competent to non-competent classifiers becomes 1:1 for data with four classes and monotonically decreases in favor of non-competent classifiers as the number of classes increases. In these imbalanced multiclass settings, a more sophisticated approach using some form of weighted voting should be used instead [60, 62].
8. Comparison to boosting
Gradient boosting is another machine learning method known to possess state of the art classification performance. Therefore we sought to compare performance of RFQ to boosting. For boosting procedures, we used boosted parametric splines using binomial loss (Spline Boost). For nonparametric boosting, we boosted trees using binomial loss (Tree Boost) and Huber loss (Tree Hboost). Parametric spline boosting was implemented using the R-package mboost [63] and tree boosting by the R-package gbm [64]. In both cases, 1000 trees were boosted with regularization parameter 0.1. Depth of trees was set to three interactions and spline bases were set to default values used by mboost.
As an enhancemnent to RFQ we also considered an extension using variable selection. Using a preliminary RF, we calculated Ishwaran-Kogalur importance using G-mean prediction error as in Section 6. Variables were then removed if they were deemed non-significant at the 5% level, where level of significance was obtained using asymptotic confidence regions calculated using random forest variable importance subsampling [6]. Using the remaining non-filtered variables, RFQ was then run as before. We call this method RFQvsel.
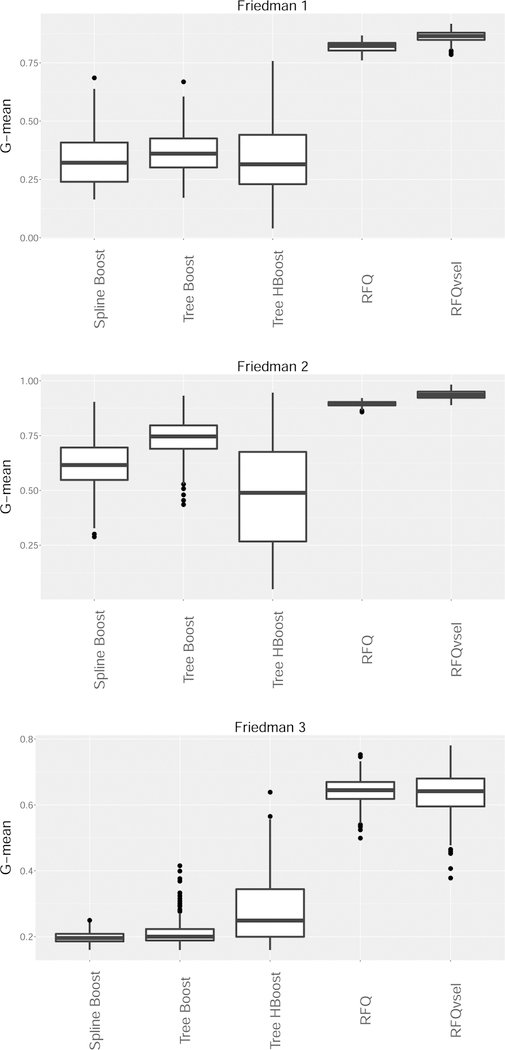
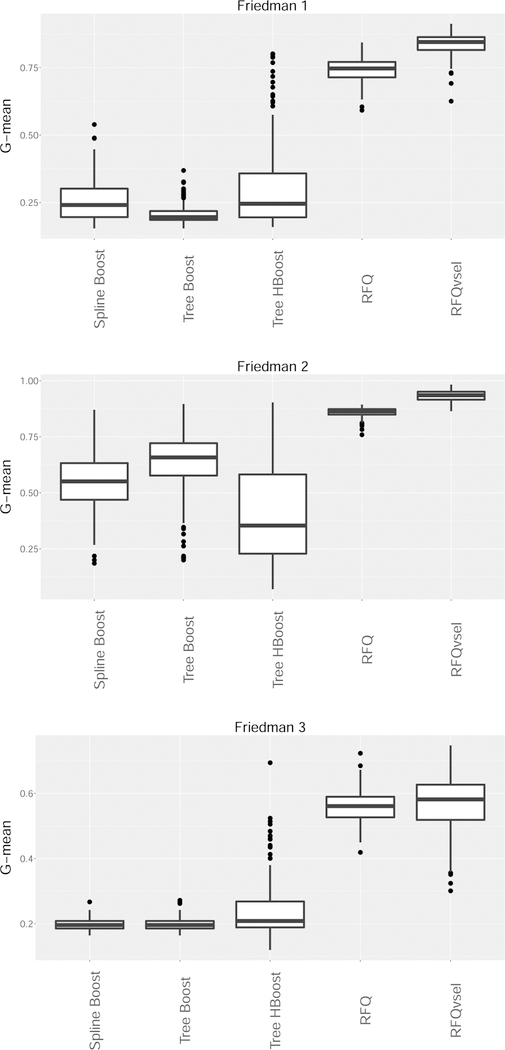
We used the three Friedman simulations to test performance. Sample size was set to N = 1250 with G-mean performance assessed on a test set of the same size. Low dimensional simulations with 25 noise features and high dimensional simulations with 250 noise features were used. All experiments were repeated independently 250 times.
Figures 6 and 7 display the test set G-mean performance values for the low and high dimensional simulation scenarios, respectively. Overall, the results are very encouraging for RFQ procedures which are overwhelmingly superior to boosting procedures. Interestingly, the dimension reduction used by RFQvsel performed very well, especially in the high dimensional simulations. For example, in the Friedman 1 simulation performance of RFQvsel is more robust to increasing dimension than RFQ. In terms of the boosting procedures there appears to be no overall consensus. Sometimes Huber loss for trees is better than binomial loss. There is also no clear winner between parametric and nonparametric boosting.
Figure 6:
G-mean performance of boosting classifiers versus RFQ for Friedman low dimensional simulations. (Spline Boost, Tree Boost are boosted splines and boosted trees using binomial loss; Tree HBoost are boosted trees with Huber loss; RFQvsel is RFQ with variable selection filtering).
Figure 7:
G-mean performance of boosting classifiers versus RFQ for Friedman high dimensional simulations. (Spline Boost, Tree Boost are boosted splines and boosted trees using binomial loss; Tree HBoost are boosted trees with Huber loss; RFQvsel is RFQ with variable selection filtering).
9. Discussion
We introduced a classifier based on the ratio of data densities for learning imbalanced data and showed this resulted in a q-classifier with the property that its threshold q = q* yielded TNR+TPR-optimality. We called this the q*-classifier and implemented q*-classification using random forests. We coined this method RFQ and showed RFQ to be highly competitive with the current and widely used balanced random forests (BRF) method of undersampling the majority class (used by the randomForest R-package for example). In our experiments with 143 imbalanced benchmark data sets, we observed that while BRF significantly improves classification with respect to the minority class, and unquestionably outperforms the standard random forests algorithm, its performance is roughly the same as RFQ on standard imbalanced data sets, but generally inferior in the difficult setting of high-complexity, high-imbalancedness, and high-dimensionality. This was further confirmed by in depth experiments on simulated and real world data sets. Furthermore, we demonstrated that RFQ is better at selecting variables across imbalanced data using G-mean as the performance criterion with Ishwaran-Kogalur importance than BRF with Breiman-Cutler importance (the standard method used in random forest analyses). In the multiclass imbalanced setting, we showed that RFQ also outperforms BRF over extremely imbalanced data sets.
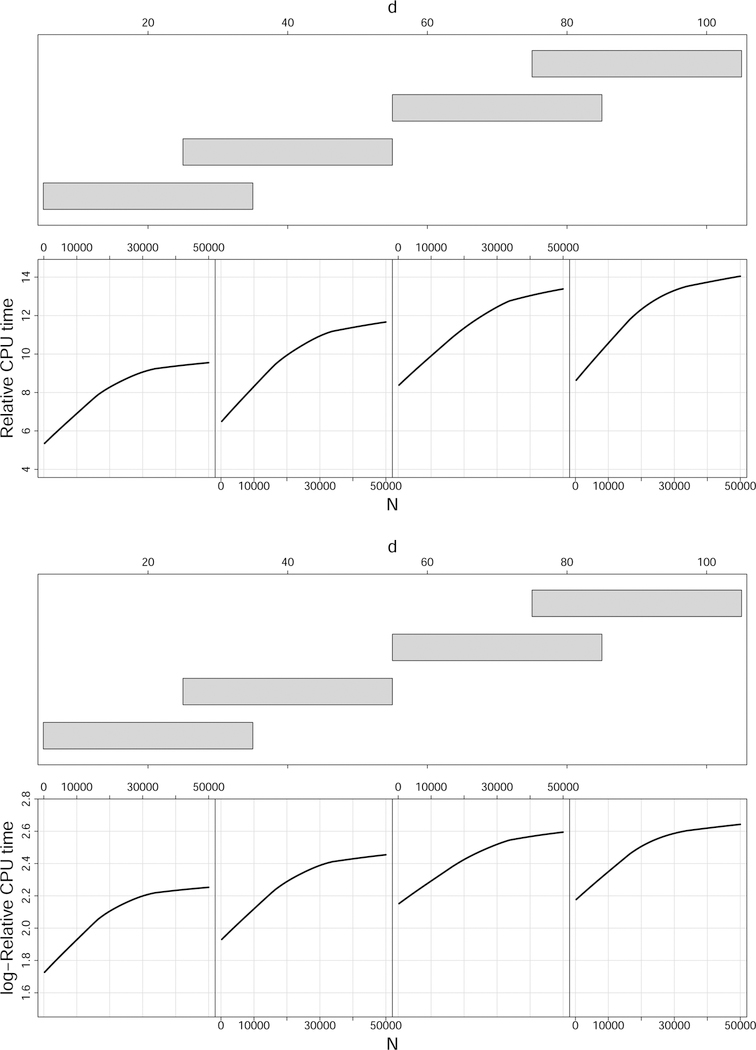
However one advantage of BRF is that it is computationally faster due to the low sample size used to construct its trees. At the same time, this advantage does not appear to be large. Figure 8 displays relative CPU times and log-relative CPU times for RFQ versus BRF for the Friedman 1 simulation as N and d are varied. Even when d = 100 and N = 50, 000, the relative CPU time is only 14. We also observe that as N increases, relative CPU times asymptote which suggests that in big data settings these differences may not be insurmountable. In fact, Theorem 4 suggests subsampling could be used as a simple remedy for RFQ in big N settings. Recall that Theorem 4 shows as long as the data is subsampled according to a response based sampling scheme, RFQ will continue to maintain its TNR+TPR optimality property. Subsampling will greatly reduce computational time and importantly the sampling can be devised so that the majority class label cardinality is much larger than the value of N1 used by BRF, thereby also ensuring good classification performance.
Figure 8:
Computational times for RFQ and BRF for Friedman 1 simulation for different sample sizes N and feature dimension d. Top plot is relative CPU time for RFQ versus BRF. Bottom plot is log-relative CPU time.
Supplementary Material
Highlights.
The new classifier jointly optimizes true positive and true negative rates for imbalanced data while simultaneously minimizing weighted risk.
It outperforms the existing random forests method in complex settings of rare minority instances, high dimensionality and highly imbalanced data.
Its performance is superior with respect to variable selection for imbalanced data.
The classifier is also highly competitive for multiclass imbalanced data.
Acknowledgments
We would like to acknowledge support for this project from the National Institutes of Health (NIGMS grant R01GM125072).
Appendix A: Proof of Theorem 1
Although Theorem 1 is well known we provide a proof here for the convenience of the reader.
Proof. We will show that
| (A.1) |
which implies that The first term in the sum (A.1) when j = 0 equals
Similarly, the second term in the sum (A.1) when j = 1 is
Taking the expectation of Y conditional on X and L of the sum in (A.1), yields
where recall that . We will show that the above sum is greater than or equal to zero. Taking the expectation over X and ℒ completes the argument.
When p(X) we have
If , we have . If , we have (ℓ0 + ℓ1)p(X) − ℓ0 ≥ 0.
When p(X) we have
If , we have . If , we have .
This establishes (A.1). To complete the proof, we have to show
The proof above reveals that r(δWB, l0, l1) is the expected value of
where the last line follows because □
Appendix B: Results from Multiclass Imbalanced Data
Section 7 explored the performance of RFQ, BRF, and RF in the multiclass imbalanced data setting. This was accomplished by decomposing the multiclass imbalanced data into 3 two-class data sets, obtaining classifiers on each and then taking a majority vote over the results. Here we list the tables from the empirical analysis which were based on forests of 5000 trees grown on each training data set with nodesize=1, mtry=d/3 and 50 resampled noise variables.
Table B.1:
Performance comparisons on simulated 3 class imbalanced data sets derived from waveform.
| RFQ | BRF | RF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class Ratios | TPR1 | TPR2 | TPR3 | G-mean | TPR1 | TPR2 | TPR3 | G-mean | TPR1 | TPR2 | TPR3 | G-mean |
| 5:4:1 | 64.65 | 84.46 | 94.51 | 80.16 | 80.20 | 90.80 | 79.87 | 83.41 | 90.38 | 91.28 | 37.84 | 67.72 |
| 10:5:1 | 60.12 | 83.86 | 94.18 | 77.94 | 82.52 | 88.44 | 77.46 | 82.61 | 96.10 | 81.83 | 19.46 | 53.18 |
| 10:1:1 | 53.25 | 90.26 | 91.79 | 76.03 | 83.75 | 81.63 | 81.40 | 82.15 | 99.92 | 36.03 | 35.78 | 50.24 |
| 20:5:1 | 55.12 | 83.26 | 92.48 | 75.05 | 83.65 | 87.11 | 74.10 | 81.32 | 99.26 | 64.88 | 6.71 | 33.47 |
| 20:1:1 | 51.84 | 87.39 | 89.97 | 74.00 | 84.08 | 78.20 | 78.42 | 80.03 | 99.99 | 17.95 | 18.57 | 30.41 |
| 50:25:1 | 57.77 | 76.55 | 87.45 | 72.72 | 83.74 | 89.53 | 59.34 | 76.05 | 95.98 | 82.10 | 0.02 | 0.68 |
| 50:5:1 | 52.78 | 78.06 | 88.90 | 71.35 | 85.97 | 83.66 | 61.61 | 75.94 | 99.97 | 41.09 | 0.25 | 4.25 |
| 100:25:1 | 54.74 | 71.80 | 85.38 | 69.28 | 86.25 | 87.99 | 43.94 | 68.54 | 99.27 | 65.01 | 0.00 | 0.00 |
| 100:10:1 | 52.27 | 73.24 | 86.09 | 68.84 | 86.68 | 85.10 | 42.24 | 66.77 | 99.96 | 40.93 | 0.00 | 0.00 |
| 100:5:1 | 51.63 | 74.82 | 84.82 | 68.67 | 86.98 | 81.04 | 43.05 | 66.16 | 99.99 | 20.46 | 0.00 | 0.00 |
Table B.2:
Performance comparisons on simulated 3 class imbalanced data sets derived from waveform with respect to two class (unweighted) G-means and the weighted G-mean with β1 = 1/2 and β2 = β3 = 1 (cf. Table B.1).
| RFQ | BRF | |||||||
|---|---|---|---|---|---|---|---|---|
| Class Ratios | G-mean12 | G-mean13 | G-mean23 | Wt. G-mean | G-mean12 | G-mean13 | G-mean23 | Wt. G-mean |
| 5:4:1 | 73.84 | 78.13 | 89.32 | 86.22 | 85.29 | 79.98 | 85.13 | 86.55 |
| 10:5:1 | 70.92 | 75.19 | 88.84 | 84.86 | 85.38 | 79.88 | 82.71 | 85.31 |
| 10:1:1 | 69.25 | 69.83 | 90.97 | 84.48 | 82.60 | 82.49 | 81.45 | 84.64 |
| 20:5:1 | 67.64 | 71.31 | 87.71 | 82.91 | 85.30 | 78.63 | 80.27 | 83.80 |
| 20:1:1 | 67.19 | 68.18 | 88.59 | 82.61 | 80.97 | 81.07 | 78.19 | 82.40 |
| 50:25:1 | 66.38 | 70.97 | 81.73 | 79.71 | 86.53 | 70.22 | 72.60 | 78.35 |
| 50:5:1 | 64.01 | 68.34 | 83.21 | 79.43 | 84.71 | 72.49 | 71.53 | 77.91 |
| 100:25:1 | 62.51 | 68.23 | 78.10 | 76.65 | 87.04 | 60.76 | 61.44 | 70.29 |
| 100:10:1 | 61.66 | 66.95 | 79.22 | 76.75 | 85.77 | 59.53 | 59.00 | 68.42 |
| 100:5:1 | 61.94 | 66.03 | 79.43 | 76.72 | 83.85 | 60.20 | 58.10 | 67.75 |
Table B.3:
Performance comparisons on simulated 3 class imbalanced data sets derived from cassini.
| RFQ | BRF | RF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class Ratios | TPR1 | TPR2 | TPR3 | G-mean | TPR1 | TPR2 | TPR3 | G-mean | TPR1 | TPR2 | TPR3 | G-mean |
| 10:5:1 | 90.66 | 97.41 | 97.55 | 95.13 | 98.58 | 100 | 94.76 | 97.73 | 99.52 | 100 | 92.93 | 97.40 |
| 25:5:1 | 78.98 | 86.91 | 95.10 | 86.69 | 97.22 | 100 | 84.54 | 93.57 | 99.66 | 100 | 72.17 | 89.36 |
| 50:25:1 | 73.67 | 73.41 | 91.15 | 78.87 | 96.49 | 100 | 55.95 | 80.82 | 99.50 | 100 | 1.41 | 17.62 |
| 50:10:1 | 72.60 | 70.85 | 92.32 | 77.90 | 96.33 | 99.99 | 57.54 | 81.63 | 99.67 | 100 | 1.86 | 21.83 |
| 50:5:1 | 68.59 | 74.98 | 91.48 | 77.62 | 95.92 | 99.95 | 58.45 | 81.95 | 99.81 | 100 | 2.28 | 22.64 |
| 50:2:1 | 62.54 | 85.06 | 89.03 | 77.75 | 95.43 | 97.66 | 63.48 | 83.47 | 99.75 | 99.98 | 4.50 | 29.32 |
| 100:50:1 | 70.67 | 70.17 | 86.59 | 75.23 | 95.49 | 99.61 | 24.06 | 59.22 | 99.52 | 100 | 0.00 | 0.00 |
| 100:25:1 | 70.24 | 65.88 | 86.77 | 73.54 | 95.08 | 99.53 | 24.69 | 59.93 | 99.60 | 100 | 0.00 | 0.00 |
| 100:10:1 | 66.12 | 60.27 | 86.95 | 69.93 | 94.56 | 99.12 | 24.54 | 59.40 | 99.76 | 100 | 0.00 | 0.00 |
| 100:5:1 | 60.64 | 59.64 | 86.63 | 67.55 | 93.87 | 97.37 | 26.52 | 60.67 | 99.76 | 100 | 0.00 | 0.00 |
Table B.4:
Performance comparisons on simulated 3 class imbalanced data sets derived from cassini with respect to two class (unweighted) G-means and the weighted G-mean with β1 = 1/2 and β2= β3 = 1 (cf. Table B.3).
| RFQ | BRF | |||||||
|---|---|---|---|---|---|---|---|---|
| Class Ratios | G-mean12 | G-mean13 | G-mean23 | Wt. G-mean | G-mean12 | G-mean13 | G-mean23 | Wt. G-mean |
| 10:5:1 | 93.97 | 94.03 | 97.46 | 96.70 | 99.28 | 96.63 | 97.33 | 97.97 |
| 25:5:1 | 82.82 | 86.62 | 90.87 | 90.17 | 98.60 | 90.55 | 91.84 | 94.01 |
| 50:25:1 | 73.51 | 81.81 | 81.68 | 83.00 | 98.23 | 72.86 | 74.20 | 81.31 |
| 50:10:1 | 71.67 | 81.76 | 80.76 | 82.17 | 98.14 | 73.92 | 75.33 | 82.14 |
| 50:5:1 | 71.64 | 79.10 | 82.66 | 82.66 | 97.91 | 74.37 | 75.94 | 82.53 |
| 50:2:1 | 72.85 | 74.40 | 86.79 | 84.09 | 96.53 | 77.36 | 78.23 | 84.13 |
| 100:50:1 | 70.39 | 78.00 | 77.71 | 79.71 | 97.52 | 46.21 | 47.28 | 59.71 |
| 100:25:1 | 67.96 | 77.86 | 75.33 | 78.00 | 97.28 | 46.99 | 48.14 | 60.46 |
| 100:10:1 | 63.02 | 75.58 | 72.03 | 74.92 | 96.81 | 46.53 | 47.71 | 59.97 |
| 100:5:1 | 59.95 | 72.30 | 71.42 | 73.43 | 95.59 | 48.42 | 49.33 | 61.34 |
Footnotes
Conflict of interest
None declared.
References
- [1].Breiman L, Random forests, Machine Learning 45 (1) (2001) 5–32. [Google Scholar]
- [2].Verikas A, Gelzinis A, Bacauskiene M, Mining data with random forests: A survey and results of new tests, Pattern Recognition 44 (2) (2011) 330–349. [Google Scholar]
- [3].Biau G, Scornet E, A random forest guided tour, Test 25 (2) (2016) 197–227. [Google Scholar]
- [4].Breiman L, Manual on Setting up, Using, and Understanding Random Forests V3 1, 2002. [Google Scholar]
- [5].Ishwaran H, Variable importance in binary regression trees and forests, Electronic Journal of Statis-tics 1 (2007) 519–537. [Google Scholar]
- [6].Ishwaran H, Lu M, Standard errors and confidence intervals for variable importance in random forest regression, classification, and survival (To appear in Statistics in Medicine), Tech. Rep, Division of Biostatistics, Miller School of Medicine, University of Miami, FL, USA, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Malley JD, Malley KG, Pajevic S, Statistical Learning for Biomedical Data, Cambridge University Press, 2011. [Google Scholar]
- [8].Rodriguez-Galiano V, Sanchez-Castillo M, Chica-Olmo M, Chica-Rivas M, Machine learning predictive models for mineral prospectivity: An evaluation of neural networks, random forest, regression trees and support vector machines, Ore Geology Reviews 71 (2015) 804–818. [Google Scholar]
- [9].Li B, Yang G, Wan R, Dai X, Zhang Y, Comparison of random forests and other statistical methods for the prediction of lake water level: a case study of the Poyang Lake in China, Hydrology Research 47 (S1) (2016) 69–83. [Google Scholar]
- [10].Bouts MJ, Tiebosch IA, Van Der Toorn A, Viergever MA, Wu O, Dijkhuizen RM, Early identification of potentially salvageable tissue with MRI-based predictive algorithms after experi-mental ischemic stroke, Journal of Cerebral Blood Flow and Metabolism 33 (7) (2013) 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tremoulet AH, Dutkowski J, Sato Y, Kanegaye JT, Ling XB, Burns JC, P. E. M. K. D. R. Group, et al. , Novel data-mining approach identifies biomarkers for diagnosis of Kawasaki disease, Pediatric Research [DOI] [PMC free article] [PubMed]
- [12].Greenstein D, Weisinger B, Malley JD, Clasen L, Gogtay N, Using multivariate machine learning methods and structural MRI to classify childhood onset schizophrenia and healthy controls, Frontiers in Psychiatry 3 (2012) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lahouar A, Slama JBH, Day-ahead load forecast using random forest and expert input selection, Energy Conversion and Management 103 (2015) 1040–1051. [Google Scholar]
- [14].Marin J, Va´zquez D, Lo´pez AM, Amores J, Leibe B, Random forests of local experts for pedestrian detection, in: Proceedings of the IEEE International Conference on Computer Vision, 2592–2599, 2013. [Google Scholar]
- [15].Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS, Random survival forests, The Annals of Applied Statistics (2008) 841–860.
- [16].Ishwaran H, Gerds TA, Kogalur UB, Moore RD, Gange SJ, Lau BM, Random survival forests for competing risks, Biostatistics 15 (4) (2014) 757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ishwaran H, Blackstone EH, Apperson-Hansen C, Rice TW, A novel approach to cancer staging: application to esophageal cancer, Biostatistics 10 (4) (2009) 603–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rice TW, Rusch VW, Ishwaran H, Blackstone EH, Cancer of the esophagus and esopha-gogastric junction, Cancer 116 (16) (2010) 3763–3773. [DOI] [PubMed] [Google Scholar]
- [19].Khalilia M, Chakraborty S, Popescu M, Predicting disease risks from highly imbalanced data using random forest, BMC Medical Informatics and Decision Making 11 (1) (2011) 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Williams JK, Using random forests to diagnose aviation turbulence, Machine Learning 95 (1) (2014) 51–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Galar M, Fernandez A, Barrenechea E, Bustince H, Herrera F, A review on ensembles for the class imbalance problem: bagging-, boosting-, and hybrid-based approaches, IEEE Transactions on Systems, Man, and Cybernetics, Part C (Applications and Reviews) 42 (4) (2012) 463–484. [Google Scholar]
- [22].Yang Q, Wu X, 10 challenging problems in data mining research, International Journal of Information Technology & Decision Making 5 (04) (2006) 597–604. [Google Scholar]
- [23].He H, Garcia EA, Learning from imbalanced data, IEEE Transactions on Knowledge & Data Engineering (9) (2008) 1263–1284.
- [24].Sun Y, Wong AK, Kamel MS, Classification of imbalanced data: A review, International Journal of Pattern Recognition and Artificial Intelligence 23 (04) (2009) 687–719. [Google Scholar]
- [25].Lo´pez V, Ferna´ndez A, Moreno-Torres JG, Herrera F, Analysis of preprocessing vs. cost-sensitive learning for imbalanced classification. Open problems on intrinsic data characteristics, Expert Systems with Applications 39 (7) (2012) 6585–6608. [Google Scholar]
- [26].Lo´pez V, Ferna´ndez A, Garc´ıa S, Palade V, Herrera F, An insight into classification with imbalanced data: Empirical results and current trends on using data intrinsic characteristics, Information Sciences 250 (2013) 113–141. [Google Scholar]
- [27].Krawczyk B, Learning from imbalanced data: open challenges and future directions, Progress in Artificial Intelligence 5 (4) (2016) 221–232. [Google Scholar]
- [28].Haixiang G, Yijing L, Shang J, Mingyun G, Yuanyue H, Bing G, Learning from class-imbalanced data: Review of methods and applications, Expert Systems with Applications 73 (2017) 220–239. [Google Scholar]
- [29].Das S, Datta S, Chaudhuri BB, Handling data irregularities in classification: Foundations, trends, and future challenges, Pattern Recognition 81 (2018) 674–693. [Google Scholar]
- [30].Breiman L, Chen C, Liaw A, Using random forest to learn imbalanced data, Tech. Rep, University of California, Berkeley, 2004. [Google Scholar]
- [31].Liaw A, Wiener M, Classification and Regression by randomForest, R News 2 (3) (2002) 18–22, URL http://CRAN.R-project.org/doc/Rnews/. [Google Scholar]
- [32].Stefanowski J, Dealing with data difficulty factors while learning from imbalanced data, in: Challenges in Computational Statistics and Data Mining, Springer, 333–363, 2016. [Google Scholar]
- [33].Mease D, Wyner AJ, Buja A, Boosted classification trees and class probability/quantile estimation, Journal of Machine Learning Research 8 (Mar) (2007) 409–439. [Google Scholar]
- [34].Japkowicz N, Stephen S, The class imbalance problem: A systematic study, Intelligent Data Analysis 6 (5) (2002) 429–449. [Google Scholar]
- [35].Jo T, Japkowicz N, Class imbalances versus small disjuncts, ACM Sigkdd Explorations Newsletter 6 (1) (2004) 40–49. [Google Scholar]
- [36].Napierala K, Stefanowski J, Types of minority class examples and their influence on learning classifiers from imbalanced data, Journal of Intelligent Information Systems 46 (3) (2016) 563–597. [Google Scholar]
- [37].Lyon R, Brooke J, Knowles J, Stappers B, Hellinger distance trees for imbalanced streams, arXiv preprint arXiv:1405.2278
- [38].Blaszczynski J, Stefanowski J, Neighbourhood sampling in bagging for imbalanced data, Neuro-computing 150 (2015) 529–542. [Google Scholar]
- [39].Fithian W, Hastie T, Local case-control sampling: Efficient subsampling in imbalanced data sets, Quality Control and Applied Statistics 60 (3) (2015) 187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kubat M, Matwin S, Addressing the curse of imbalanced training sets: one-sided selection, in: ICML, vol. 97, 179–186, 1997. [Google Scholar]
- [41].Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP, SMOTE: synthetic minority over-sampling technique, Journal of Artificial Intelligence Research (2002) 321–357.
- [42].Ferna´ndez A, Garcia S, Herrera F, Chawla NV, SMOTE for Learning from Imbalanced Data: Progress and Challenges, Marking the 15-year Anniversary, Journal of Artificial Intelligence Re-search 61 (2018) 863–905. [Google Scholar]
- [43].Chawla NV, Lazarevic A, Hall LO, Bowyer KW, SMOTEBoost: Improving prediction of the minority class in boosting, in: European conference on principles of data mining and knowledge discovery, Springer, 107–119, 2003. [Google Scholar]
- [44].Seiffert C, Khoshgoftaar TM, Van Hulse J, Napolitano A, RUSBoost: A hybrid approach to alleviating class imbalance, IEEE Transactions on Systems, Man, and Cybernetics-Part A: Systems and Humans 40 (1) (2010) 185–197. [Google Scholar]
- [45].Galar M, Ferna´ndez A, Barrenechea E, Herrera F, EUSBoost: Enhancing ensembles for highly imbalanced data-sets by evolutionary undersampling, Pattern Recognition 46 (12) (2013) 3460–3471. [Google Scholar]
- [46].Kubat M, Holte R, Matwin S, Learning when negative examples abound, in: European Conference on Machine Learning, Springer, 146–153, 1997. [Google Scholar]
- [47].Cieslak DA, Hoens TR, Chawla NV, Kegelmeyer WP, Hellinger distance decision trees are robust and skew-insensitive, Data Mining and Knowledge Discovery 24 (1) (2012) 136–158. [Google Scholar]
- [48].Datta S, Das S, Near-Bayesian Support Vector Machines for imbalanced data classification with equal or unequal misclassification costs, Neural Networks 70 (2015) 39–52. [DOI] [PubMed] [Google Scholar]
- [49].Go´nzalez S, Garc´ıa S, La´zaro M, Figueiras-Vidal AR, Herrera F, Class Switching according to Nearest Enemy Distance for learning from highly imbalanced data-sets, Pattern Recognition 70 (2017) 12–24. [Google Scholar]
- [50].Breiman L, Randomizing outputs to increase prediction accuracy, Machine Learning 40 (3) (2000) 229–242. [Google Scholar]
- [51].Friedman JH, On bias, variance, 0/1–loss, and the curse-of-dimensionality, Data Mining and Knowledge Discovery 1 (1) (1997) 55–77. [Google Scholar]
- [52].Elkan C, The foundations of cost-sensitive learning, in: International Joint Conference on Artificial Intelligence, vol. 17, Lawrence Erlbaum Associates Ltd, 973–978, 2001. [Google Scholar]
- [53].R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, URL https://www.R-project.org/, 2018. [Google Scholar]
- [54].Ishwaran H, Kogalur UB, Random Forests for Survival, Regression, and Classification (RF-SRC), URL https://cran.r-project.org/package=randomForestSRC, R pack-age version 2.5.0, 2018.
- [55].Leisch F, Dimitriadou E, mlbench: Machine Learning Benchmark Problems, URL http://cran.r-project.org/package=mlbench, R package version 2.1–1, 2010.
- [56].Max K, caret: Classification and Regression Training, URL https://cran.r-project.org/package=caret, R package version 6.0–77, 2017.
- [57].Max K, Kjell J, AppliedPredictiveModeling: Functions and Data Sets for Ap-plied Predictive Modeling, URL https://cran.r-project.org/package=AppliedPredictiveModeling, R package version 1.1–7, 2018.
- [58].Craig-Schapiro R, Kuhn M, Xiong C, Pickering EH, Liu J, Misko TP, Perrin RJ, Bales KR, Soares H, Fagan AM, et al. , Multiplexed immunoassay panel identifies novel CSF biomarkers for Alzheimer’s disease diagnosis and prognosis, PloS One 6 (4) (2011) e18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Max K, Steve W, Nathan C, Mark C, C50: C5.0 Decision Trees and Rule-Based Models, URL https://cran.r-project.org/package=C50, R package version 0.1.2, 2018.
- [60].Zhang Z, Krawczyk B, Garcia S, Rosales-Perez A, Herrera F, Empowering one-vs-one decom-position with ensemble learning for multi-class imbalanced data, Knowledge-Based Systems 106 (2016) 251–263. [Google Scholar]
- [61].Friedman J, Another approach to polychotomous classification, Tech. Rep, Department of Statistics, Stanford University, 1996. [Google Scholar]
- [62].Santhanam V, Morariu VI, Harwood D, Davis LS, A non-parametric approach to extending generic binary classifiers for multi-classification, Pattern Recognition 58 (2016) 149–158. [Google Scholar]
- [63].Bu¨hlmann P, Hothorn T, Boosting algorithms: regularization, prediction and model fitting, Statistical Science 22 (4) (2007) 477–505. [Google Scholar]
- [64].Ridgeway G, The state of boosting, Computing Science and Statistics 31 (1999) 172–181. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.