Abstract
Objective Development of computational approaches and tools to effectively integrate multidomain data is urgently needed for the development of newly targeted cancer therapeutics.
Methods We proposed an integrative network-based infrastructure to identify new druggable targets and anticancer indications for existing drugs through targeting significantly mutated genes (SMGs) discovered in the human cancer genomes. The underlying assumption is that a drug would have a high potential for anticancer indication if its up-/down-regulated genes from the Connectivity Map tended to be SMGs or their neighbors in the human protein interaction network.
Results We assembled and curated 693 SMGs in 29 cancer types and found 121 proteins currently targeted by known anticancer or noncancer (repurposed) drugs. We found that the approved or experimental cancer drugs could potentially target these SMGs in 33.3% of the mutated cancer samples, and this number increased to 68.0% by drug repositioning through surveying exome-sequencing data in approximately 5000 normal-tumor pairs from The Cancer Genome Atlas. Furthermore, we identified 284 potential new indications connecting 28 cancer types and 48 existing drugs (adjusted P < .05), with a 66.7% success rate validated by literature data. Several existing drugs (e.g., niclosamide, valproic acid, captopril, and resveratrol) were predicted to have potential indications for multiple cancer types. Finally, we used integrative analysis to showcase a potential mechanism-of-action for resveratrol in breast and lung cancer treatment whereby it targets several SMGs (ARNTL, ASPM, CTTN, EIF4G1, FOXP1, and STIP1).
Conclusions In summary, we demonstrated that our integrative network-based infrastructure is a promising strategy to identify potential druggable targets and uncover new indications for existing drugs to speed up molecularly targeted cancer therapeutics.
Keywords: cancer genome, significantly mutated genes, network-based drug repositioning, drug-gene signatures, precision cancer medicine
INTRODUCTION
The past several decades have seen massive advances in many scientific, technological, and managerial efforts to raise the efficiency of drug discovery and development. However, the number of new United States Food and Drug Administration (US FDA) approved drugs has nearly halved since 1950, and the cost of drug discovery and development has grown fairly steadily.1 On the other hand, massive amounts of drug and biological data have been released to the community. Hence, scientists are seeking innovative technologies and approaches to reduce the cost and raise the efficiency of drug discovery and development. Recent advances in biotechnologies, such as massively parallel high-throughput sequencing and genome-wide association studies (GWAS), provide unprecedented opportunities in identifying potential targets for drug discovery and development. A recent study revealed that human genetic data generated from GWAS provide a valuable resource to select the best drug targets and indications in the development of new drugs.2
Several national and international cancer genomics projects, such as The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium, have enabled investigators to comprehensively characterize the somatic mutational landscape and mutational signatures in a large number of tumor samples.3–5 These abundant data not only allow investigators to identify significantly mutated genes (SMGs) in cancer through both computational and experimental approaches,3,6,7 but also provide novel druggable targets and opportunities for developing new molecularly targeted cancer therapies that target SMGs.8 Griffith et al.9 developed a Drug-Gene Interaction database to mine existing resources to generate hypotheses for the mutated genes that might be therapeutic targets or prioritized for anticancer drug development. The development of new computational methods that efficiently integrate massive amounts of next-generation sequencing data from TCGA and International Cancer Genome Consortium projects provides new opportunities for the timely development of molecularly targeted treatments, one of the major areas in cancer precision medicine.
Tumor initiation, progression, and resistance to certain therapeutic agents are often attributed to the accumulation of one or multiple driver or actionable mutations that activate oncogenes or inactivate tumor suppressor genes (TSGs), as well as their signaling pathways.10 Traditional cytotoxic agents targeting cell division and DNA replication have achieved great success in molecular cancer therapy, but they often have severe side effects. Molecularly targeted agents (e.g., kinase inhibitors) that target oncogenes or oncoproteins that drive tumor initiation or progression show high selectivity for tumor cells, hence reducing side effects. Therefore, targeting driver mutations (e.g., oncogenic mutations) is expected to provide valuable opportunities for precision cancer medicine.11 For example, p.Arg132His on isocitrate dehydrogenase type I is the most frequent mutation in a subgroup of gliomas and various other types of cancer.12 Schumacher et al.13 identified a new vaccine that selectively targets mutant isocitrate dehydrogenase type I (p.Arg132His), induces antitumor immunity, and further halts growth of gliomas. Another example is somatic mutation BRAF p.Val600Glu, which occurs in ∼50% of melanoma; vemurafenib can effectively target this specific mutation.14 However, most targeted agents often have some pitfalls; for example, all kinase inhibitors eventually lead to drug resistance.15 Moreover, the attempt to activate TSGs that have been inactivated by somatic mutations or deletions is more challenging than targeting oncogenes. One of the reasons is that mutations in TSGs are often truncated mutations scattered in the coding regions but with low recurrence. An alternative approach is to target pathways or subnetworks (e.g., neighbors in the protein interaction network [PIN]) perturbed by the inactivation mutations in TSGs.16 The development of innovative computational methods via targeting oncogenic mutations in oncoproteins or proteins regulated by inactivation mutations in TSGs is urgently needed for the development of targeted therapeutic agents.
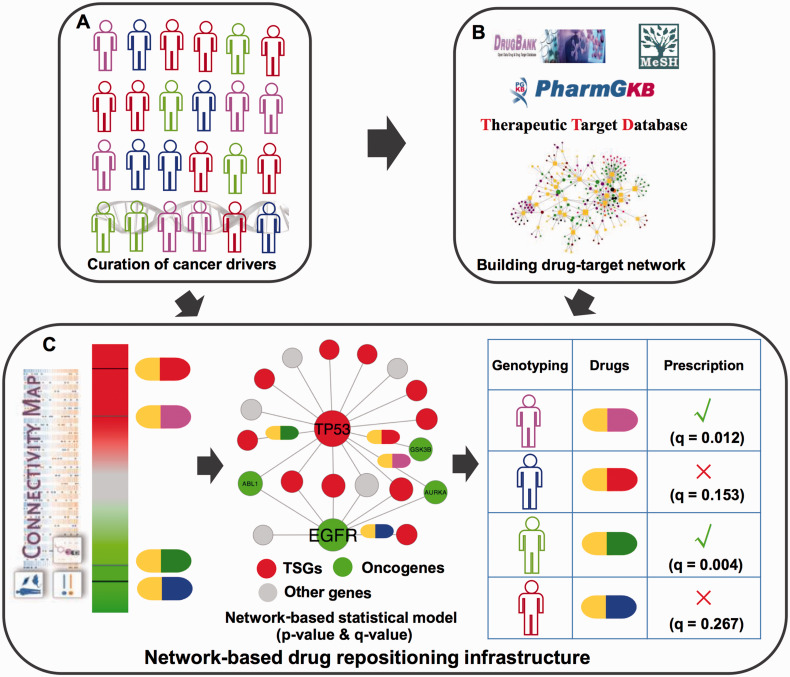
In this study, we developed an integrative network-based infrastructure to prioritize druggable targets or potential new indications for existing drugs by directly targeting SMGs or their neighbors in the PIN. Specifically, we assembled and curated 693 SMGs derived from large-scale cancer genomics datasets in public domains and TCGA projects for 29 cancer types. We found that 121 druggable proteins encoded by SMGs were targeted by known anticancer drugs or repurposed drugs through integrating the drug-target interactions from 3 well-known public databases. Furthermore, we developed a network-based statistical approach to prioritize new anticancer indications for the existing drugs by integrating SMGs discovered in TCGA and public domains and drug-gene signatures from the Connectivity Map (Figure 1). Collectively, this study highlights the potential value of network-based approaches in identifying potential druggable targets and new anticancer indications for existing drugs by utilizing large-scale cancer somatic mutations to aid in the timely development of precision cancer medicine.
Figure 1:
Diagram of network-based infrastructure for prioritizing potential druggable targets and searching for the existing drugs targeting significantly mutated genes (SMGs) in cancer genomes. This infrastructure is designed for better development of newly targeted cancer therapies. (A) Manual curation of SMGs discovered in large-scale cancer genome data generated from The Cancer Genome Atlas and public domains (see the Materials and Methods section). (B) Searching for new potential druggable anticancer targets via mapping the curated SMGs in A into drug-target interaction network collected from public databases: DrugBank, PharmGKB, and Therapeutic Target Database. (C) Development of a network-based drug repositioning approach to prioritize new potential anticancer indications for existing drugs. It incorporates drug-gene signatures from the Connectivity Map, the manually curated SMGs, and the protein-protein interaction network using a statistical approach (see the Materials and Methods section). TSGs: tumor suppressor genes.
MATERIALS AND METHODS
Manual curation of the significantly mutated genes discovered in cancer genomes
We manually curated and assembled high-quality SMGs from 15 large-scale TCGA cancer genome analysis projects and other public domains as briefly described below.11,17–29 For example, Lawrence et al.19 identified 224 SMGs from 4742 human cancer genomes across 21 cancer types using the MutSig method. Rubio-Perez et al.11 identified 459 SMGs acting in one or multiple cancer types by analyzing 6792 tumor samples. The details are provided in online Supplementary Table S1. We annotated all SMGs using gene Entrez ID, chromosome location, and the official gene symbols from the National Center for Biotechnology Information database.30 This process resulted in a total of 693 manually curated, unique SMGs across 29 cancer types (online Supplementary Table S1).
Creation of protein interaction network
We used a large-scale, high-quality PIN by integrating 4 types of protein-protein interactions (PPIs) with complementary biological information: physical interactions, PPIs with 3-dimensional structural data, innate immunity-related PPIs, and kinase-substrate interactions mediating phosphorylation reactions, as described in our previous studies.31,32 In total, we compiled 29 475 PPIs connecting 625 proteins encoded by SMGs and 7249 neighbors of SMG-encoded proteins in PIN.
Construction of drug-target interaction network
We collected drug-target interactions from 3 public databases: Therapeutic Target Database,33 DrugBank (v3.0),34 and PharmGKB.35 Drugs are grouped using Anatomical Therapeutic Chemical classification system codes36–38 and are further annotated using the Unified Medical Language System and Medical Subject Headings vocabularies.39 All protein coding genes were mapped and annotated using the gene Entrez ID and official gene symbol from the National Center for Biotechnology Information database.30 In total, we obtained 17 490 drug-target interactions connecting 4059 US FDA-approved or experimental drugs and 2746 human proteins. After mapping the proteins encoded by 693 SMGs into the above global network, we found 1396 drug-target interactions connecting 121 proteins encoded by SMGs and 636 drugs, including 342 US FDA-approved drugs and 267 experimental drugs under preclinical or clinical studies (online Supplementary Table S2).
Collection and preparation of drug-gene signatures
We compiled drug-gene signatures from the Connectivity Map (CMap, build 02).40 The CMap was composed of over 7000 gene expression profiles from 4 human cultured cancer cell lines treated with 1309 bioactive compounds covering 6100 individual instances at different concentrations. A measure amplitude (a) of the extent of differential expression for a given probe set was defined in the original study,40 briefly described below:
where t is the scaled and thresholded average difference value (a measure of the relative level of a transcript monitored by a particular probe set) for a compound treatment group and c is the thresholded average difference value for the matching control group. The average difference values were scaled and thresholded based on an MAS 5.0 (Affymetrix) approach.40 Based on the above definition, a = 0 indicates no differential expression, a > 0 indicates increased expression (up-regulation) by the drug treatment, and a < 0 indicates decreased expression (down-regulation) by the treatment. In CMap, an amplitude of 0.67 denotes a 2-fold induction. According to the original study,40 we defined a gene signature as being up-regulated if its amplitude was greater than 0.67 and down-regulated if its amplitude was less than −0.67. Finally, we mapped probe sets into SMGs. In total, we compiled 380 969 up- or down-regulated drug-gene pairs from the CMap connecting 6092 instances and 7476 SMGs (170) or their coding protein neighbors (7306) in PIN.
Predicting new anticancer indications for existing drugs
For each cancer type, we manually curated its SMGs from TCGA and public domains. We also collected the up-/down-regulated genes for each drug from CMap. Then, we assumed that a drug would have a high potential for a specific anticancer indication if its up-/down-regulated genes from CMap tended to be SMGs or their neighbors in protein interaction network. For each drug-cancer pair, we conducted Fisher’s exact test to calculate the significance of the enrichment of SMGs in up-/down-regulated genes. The p-values were corrected by using Benjamini-Hochberg false discovery rate41 for each drug-cancer pair. Finally, we used the adjusted P-value (q) threshold < 0.05 to identify the significant drug-cancer pairs and prioritize new potential anticancer indications for existing drugs.
Collection and preparation of somatic mutations
We downloaded the somatic mutation data from 3 sources: (1) Sanger website: (ftp://ftp.sanger.ac.uk/pub/cancer/AlexandrovEtAl, December 16, 2013); (2) Elledge’s Laboratory website at Harvard University (http://elledgelab.med.harvard.edu/?page_id=689; accessed in March 2014)42; and (3) COSMIC: Catalogue of Somatic Mutations in Cancer (v69) database (http://cancer.sanger.ac.uk/cosmic). Here, we only used nonsynonymous somatic mutations with TCGA tumor-normal matched sample IDs to ensure the quality of somatic mutation data. In total, we compiled 808 042 nonsynonymous mutations, including missense mutations and small inserts/deletions (indels) in 5003 cancer genomes for 16 cancer types. The detailed descriptions of somatic data collection and preparation are provided in our previous study.43
Kaplan-Meier survival analysis
We used the online tool PREdiction of Clinical Outcomes from Genomic profiles (https://precog.stanford.edu) to investigate the relationship between gene expression and survival rate. Currently, PREdiction of Clinical Outcomes from Genomic consists of the overall survival data of approximately 18 000 patients and measures the relationship between gene expression and survival rate using a univariate Cox regression. The statistical associations were calculated using z-scores, where a > 1.96 corresponds to a 2-sided P < .05. The details are provided in a previous study.44
Network and statistical analysis
All statistical analysis was performed using the R platform (v3.01, http://www.r-project.org/). Networks were visualized using Cytoscape (v2.8.1).45
RESULTS
Overview of a network-based infrastructure
We proposed a network-based infrastructure to prioritize potential druggable targets and identify potential new indications of existing drugs for precision cancer medicine by identifying those that may specifically target SMGs or their neighbors in PIN. Previous studies have revealed that most drugs have polypharmacological features.36,37,46 The hypothesis of our network-based drug repositioning approach is that if a set of up- or down-regulated genes perturbed by a drug of interest is overrepresented as SMGs or their neighbors in the PIN for a particular cancer type, this drug has a high potential of having a new indication for this cancer type. As shown in Figure 1, for oncogenes or oncoproteins, we proposed to prioritize potential drugs (e.g., inhibitors) to inhibit the function or expression of oncogenes. For TSGs, we proposed to prioritize potential drugs targeting their neighbors in the PIN. We defined a significant drug-cancer pair as one with a q < .05. Finally, we performed network analysis and survival rate analysis to explore the potential mechanism-of-action (MOA) of new predicted anticancer indications for existing drugs.
A global drug-target interaction network through targeting SMGs
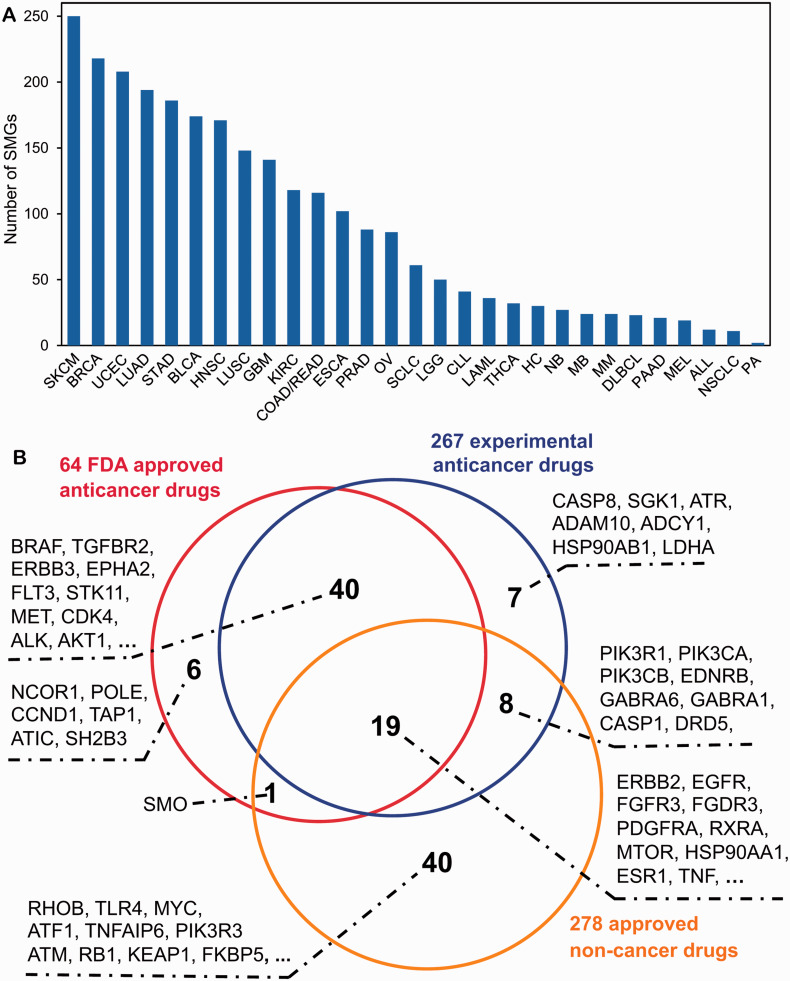
In this study, we curated and assembled 693 SMGs across 29 cancer types (online Supplementary Table S1): acute lymphocytic leukemia (ALL), bladder carcinoma (BLCA), breast carcinoma (BRCA), chronic lymphocytic leukemia (CLL), colon or rectum adenocarcinoma (COAD/READ), diffuse large B-cell lymphoma (DLBCL), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), hepatocarcinoma (HC), head and neck squamous carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), acute myeloid leukemia (LAML), low grade glioma (LGG), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), medulloblastoma (MB), melanoma (MEL), multiple myeloma (MM), neuroblastoma (NB), non-small cell lung cancer (NSCLC), ovarian serous cystadenocarcinoma (OV), pilocytic astrocytoma (PA), pancreatic adenocarcinoma, prostate adenocarcinoma (PAAD), small cell lung cancer (SCLC, cutaneous melanoma (SKCM or CM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrioid (UCEC). Figure 2A shows that the average number of SMGs per cancer type is 90, with ranges from 2 SMGs observed in PA to 250 SMGs observed in SKCM. We next examined how many US FDA-approved drugs or experimental drugs may target SMGs by integrating drug pharmacological data from 3 public databases: DrugBank,34 Therapeutic Target Database,33 and PharmGKB35 (see the Materials and Methods section). As shown in Figure 2B, we found 64 FDA-approved anticancer agents (drugs with Anatomical Therapeutic Chemical first class code “L”) targeting 66 SMGs, 267 experimental anticancer agents under preclinical or clinical studies targeting 75 SMGs, and 278 FDA approved noncancer drugs targeting 54 SMGs. In total, there are 121 unique SMGs targeted by US FDA-approved drugs or experimental drugs, indicating a potential for cancer genomics data to aid in the development of molecularly targeted cancer therapeutics. The detailed lists of approved or experimental drugs targeting SMGs in different cancer types are provided in online Supplemental Table S2.
Figure 2:
Survey of significantly mutated genes (SMGs) and druggable targets in various types of cancer. (A) Distribution of the number of SMGs across 29 cancer types (see online Supplementary Table S1). (B) Venn diagram showing the druggable proteins encoded by SMGs targeted by US FDA-approved anticancer drugs, experimental anticancer drugs, and noncancer drugs. Due to space limitations, when the number of genes is more than 10 in Venn diagram, only the top 10 cancer targets with the largest number of approved drugs are listed. The detailed lists of genes shown in Figure 2 are provided in online Supplementary Table S2. The abbreviations for cancer types are provided in the main text.
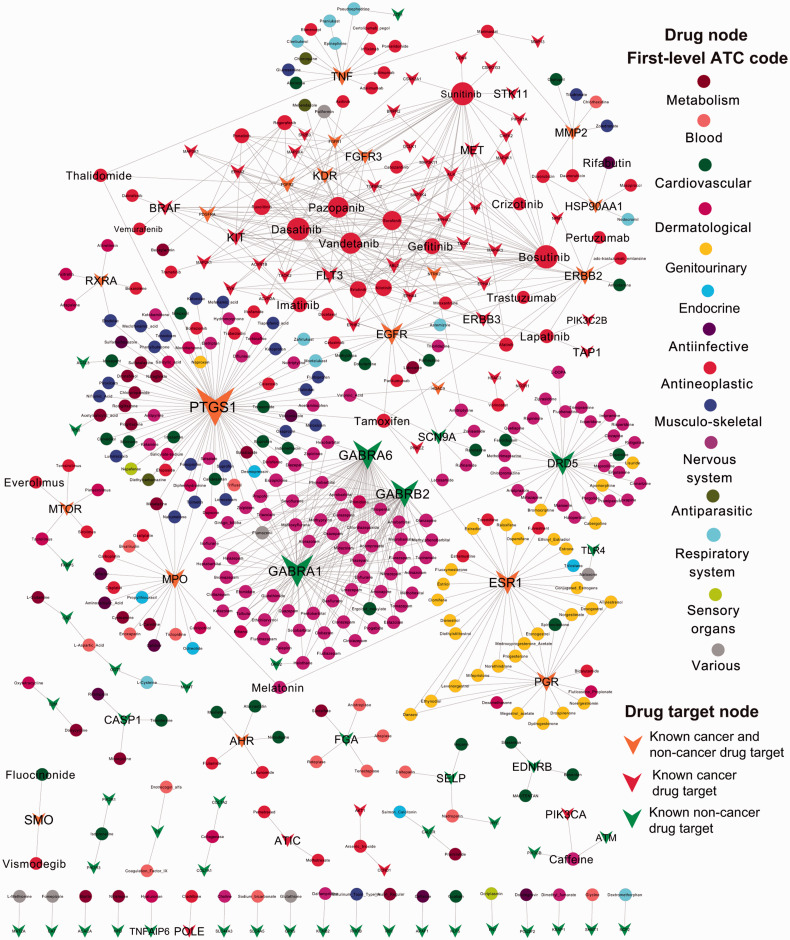
We further built a bipartite drug-target interaction network to visualize the MOA of 64 approved anticancer drugs and 278 approved noncancer drugs targeting 113 SMGs (Figure 3). We found that approved anticancer drugs, such as pazopanib, dasatinib, vandetanib, and bosutinib, target multiple SMGs, consistent with the polypharmacological profiles of targeted agents (e.g., kinase inhibitors).47 In addition, we found that several SMGs (such as TLR4 and SCN9A) can potentially be targeted by FDA-approved noncancer drugs. For example, TLR4, encoding toll-like receptor 4, was significantly mutated in the ER+/HER2− subgroup of breast cancer,18 and recent studies further revealed that TLR4 plays critical roles in breast cancer.48Figure 3 reveals that a specific opiate drug (naloxone) inhibits TLR4 signaling,49 which may be potentially useful in breast cancer chemoprevention and treatment. In addition, SCN9A, encoding sodium channel protein type 9 subunit alpha, was previously reported to be mutated in glioblastoma.21Figure 3 reveals that several approved noncancer drugs, such as ranolazine50 and zonisamide,51 block SCN9A function. This suggests that ranolazine and zonisamide may provide potential indications for glioblastoma chemoprevention and targeted therapy.
Figure 3:
Global drug-target interaction network connecting 113 proteins (denoted by Vee shape) encoded by SMGs and 342 US FDA-approved drugs (denoted by circle, 64 anticancer drugs and 278 noncancer drugs). All drugs were grouped using the Anatomical Therapeutic Chemical classification system codes, as described in a previous study.35 Drug targets are denoted by Vee shapes: (i) orange Vees denote both known cancer and repurposed (noncancer) drug targets (SMGs targeted by both known anticancer drugs and noncancer drugs); (ii) red Vees denotes known cancer drug targets (SMGs targeted by known anticancer drugs); (iii) green Vees denote repurposed (noncancer) cancer drug targets (SMGs targeted by known noncancer drugs). The size of nodes (circles and Vees) reflects their degree (connectivity) in the network.
Surveying benefit rates by the currently targeted cancer therapy
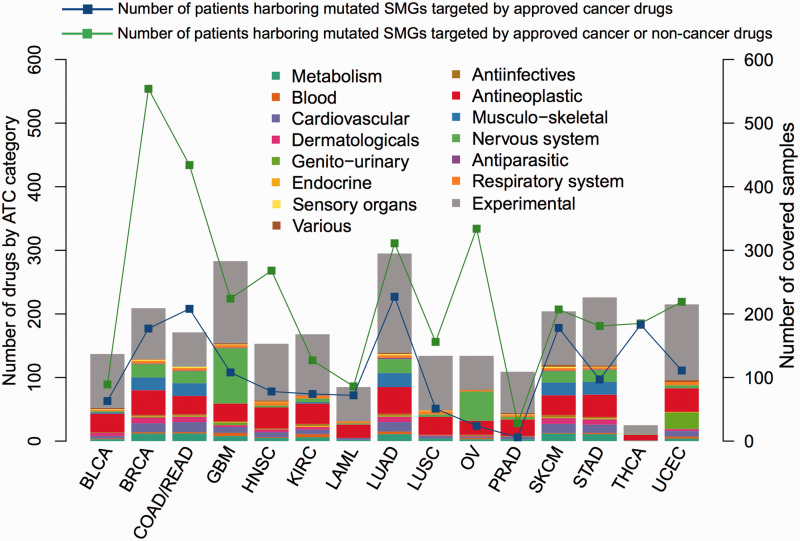
We examined how many cancer patients could potentially benefit from the currently targeted molecular therapies by surveying 5003 tumor genomes with unique TCGA sample IDs harboring nonsynonymous mutations in specific SMGs across 16 cancer types. Figure 4 reveals that the approved or experimental cancer drugs could potentially target SMGs in 33.3% mutated cancer samples. For example, breast cancer and lung cancer have a large number of SMGs, such as EGFR and ERBB2, that are targeted by many available US FDA-approved targeted agents (e.g., afatinib, lapatinib, gefitinib), as shown in Figure 3. The number would increase to 68.0% if we utilized drug repositioning by including noncancer (repurposed) drugs. Collectively, this information makes the currently targeted agents more promising for cancer therapeutics. It is worth noting that our analysis of simply using percentage patients for benefit rate in targeted cancer therapies may be biased toward the frequency of SMGs and cancer types due to data incompleteness.
Figure 4:
Distribution of the available agents targeting significantly mutated genes (SMGs) and the number of mutated patients harboring nonsynonymous mutations in drug-targeting SMGs across 15 cancer types from The Cancer Genome Atlas. One cancer type (PAAD: pancreatic adenocarcinoma) did not have any SMGs targeted by approved drugs, so it was not included. All drugs were grouped using the Anatomical Therapeutic Chemical (ATC) classification system codes, as labeled in the figure. The X-axis shows the type of cancer. The abbreviations for cancer types are provided in the main text.
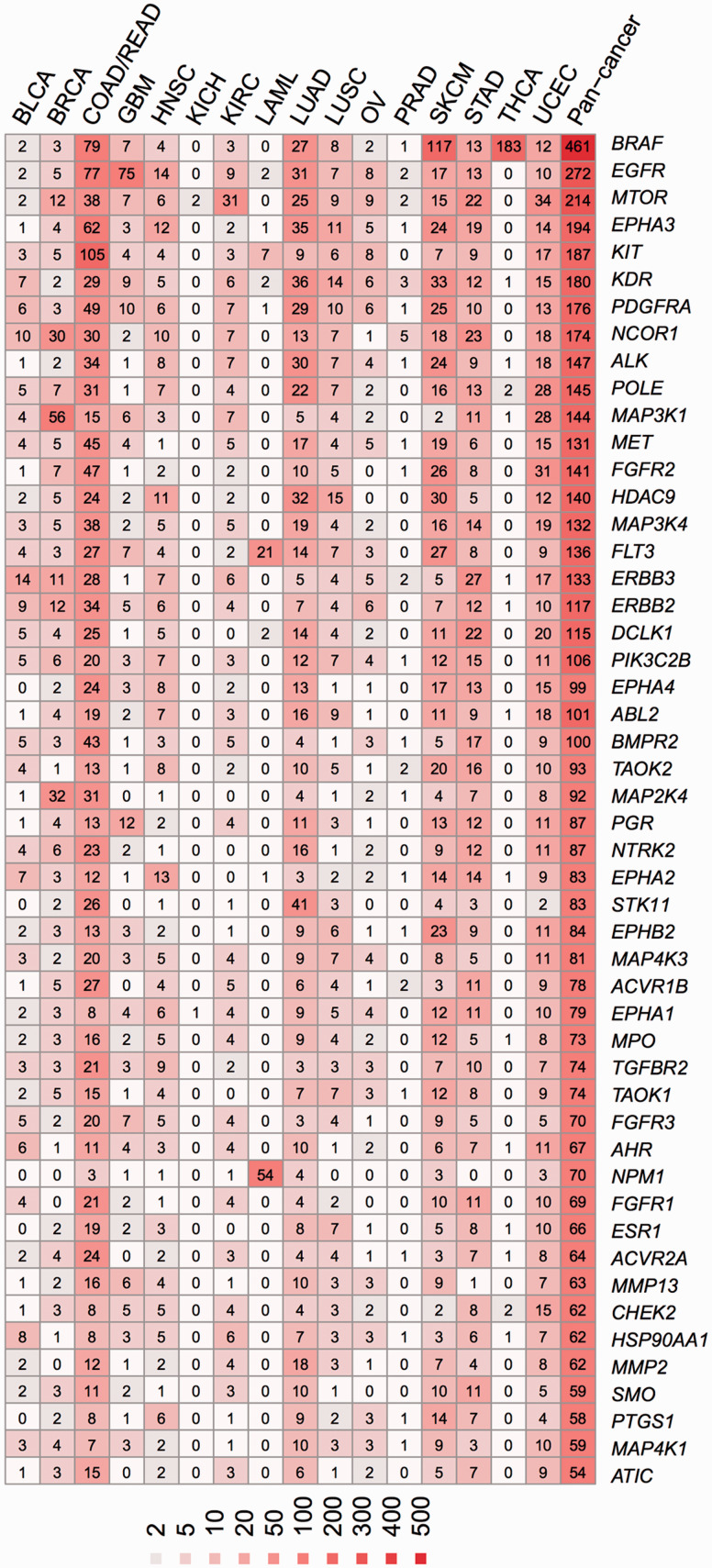
Figure 5 shows the nonsynonymous mutation spectrum for 66 druggable SMGs targeted by US FDA-approved anticancer agents. Those 66 SMGs were selected for presentation based on their high frequencies in the total samples. The 5 most commonly mutated druggable SMGs are BRAF, EGFR, MTOR, EPHA3, and KIT. For example, 45% of the patients (117/259) in SKCM harbored a mutation in BRAF. Specifically, among 325 patients, 183 (56%) TCHA patients had the mutation at the BRAF V600 site. Although there are only a few targeted agents available for thyroid carcinoma (Figure 4), some approved drugs (e.g., vemurafenib) targeting BRAF V600E may provide new off-label indications for thyroid cancer therapy.52 In addition, several new SMGs, such as SMO, have been investigated or used for targeted cancer therapy. For example, vismodegib, developed by Genentech, an SMO inhibitor halting the hedgehog signaling pathway (Figure 3), was approved for adult basal cell carcinoma treatment on January 30, 2012.34 Collectively, Figure 5 may provide useful information for the development and application of off-label uses for cancer therapies.
Figure 5:
Frequencies of nonsynonymous mutations in 66 selected significantly mutated genes (SMGs) targeted by US FDA-approved anticancer agents across 15 cancer types in approximately 5000 cancer genomes. These 66 SMGs were presented here because they harbored the largest number of nonsynonymous somatic mutations in pancancer samples collected from The Cancer Genome Atlas. One cancer type (PAAD: pancreatic adenocarcinoma) was not included, as explained in Figure 4’s legend. The color keys (heatmap) reflect the number of nonsynonymous somatic mutations counted in each cancer type or pancancer samples. The abbreviations for cancer types are provided in the main text.
Prioritizing new anticancer indications through targeting SMGs
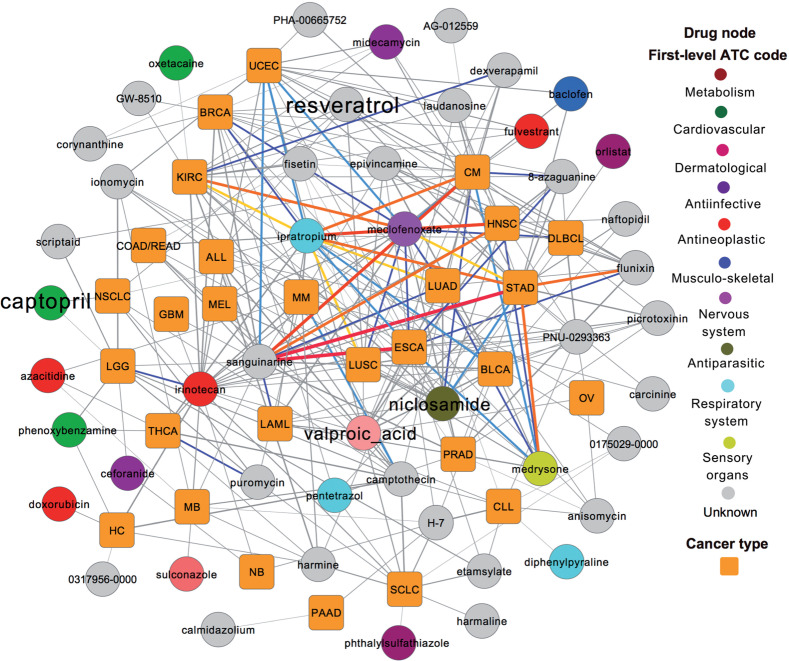
Here, we proposed a network-based statistical approach to prioritize new potential indications for existing drugs by incorporating drug-gene signatures from CMap into 693 manually curated SMGs and their neighbors in the PIN across 29 cancer types (Figure 1). Based on the notion of systems pharmacology, our network-based statistical approach prioritizes existing agents that target oncoproteins or network neighbors regulated by TSGs for further development of targeted cancer therapy (Figure 1). Using the threshold of q < .05, we identified 284 significant drug-cancer pairs connecting 28 cancer types (gold squares) with 48 existing drugs (circles) in Figure 6 and online Supplementary Table S3. We did not find any significant drug-cancer pairs for 1 rare cancer type (pilocytic astrocytoma), which had only 2 SMGs (Figure 2B). To verify the performance of predicted results, we systematically searched existing literature for the predicted indications on these 48 drugs. Among the 48 predicted drugs, 32 drugs (32/48 = 66.7% success rate) were previously reported to have potential anticancer indications in literature data (online Supplementary Table S4). In the next paragraphs, we selected 4 drugs (niclosamide, valproic acid, captopril, and resveratrol) that have more experimentally validated data in the literature as example drugs to illustrate their anticancer profiles and MOA.
Figure 6:
Drug-cancer indication network for 284 predicted drug-cancer pairs (adjusted P-value [q] < .05, online Supplementary Table S3) connecting 48 existing drugs (circles) and 28 cancer types (orange squares). The thickness of links represents the q values calculated by our statistical approach (online Supplementary Table S3). The color of links represents significance categories of q values: red, q < 10−9; orange, 10−9 < q < 10−8; yellow, 10−8 < q < 10−7; blue, 10−7 < q < 10−6; cyan, 10−6 < q < 10−5; and gray, 10−5 < q < 0.05. The text size of 4 drugs (resveratrol, captopril, niclosamide, and valproic acid) was made larger because they were selected as example drugs and discussed in the main text. The abbreviations for cancer types are provided in the main text.
Niclosamide is approved for the treatment of tapeworm infection and acts by inhibiting oxidative phosphorylation, glucose uptake, and anaerobic metabolism.53Figure 6 shows that niclosamide is predicted to have potential indications for multiple cancer types, such as STAD (q = 1.1 × 10−6), SKCM (q = 2.1 × 10−5), LUAD (q = 4.1 × 10−5), and colon or rectum adenocarcinoma (q = 6.9 × 10−3). Two recent studies suggested that niclosamide had high tumor suppression activity in human sporadic colorectal cancer54 and glioblastoma55 by inhibiting the Wnt/beta-catenin pathway. Valproic acid, a fatty acid with an anticonvulsant profile, is approved for the treatment of epilepsy. Figure 6 shows that valproic acid is predicted to have potential anticancer indications for several cancer types, such as esophageal carcinoma (q = 1.1 × 10−4), bladder carcinoma (q = 1.8 × 10−4), and LUAD (q = 2.7 × 10−4). Recent studies reported that valproic acid has anticancer (e.g., lung cancer) and anti-virus properties as an HDAC inhibitor.56–58 Captopril, a competitive inhibitor of angiotension-converting enzyme, was approved for the treatment of hypertension. In our analysis, we predicted that captopril has new indications for NSCLC (q = 0.021) and low-grade glioma (q = 0.048). A recent study reported that captopril is a promising agent for inhibiting lung tumor growth and metastasis in a xenografts model.59 Collectively, our network-based approach demonstrated high performance in prioritizing new anticancer indications for existing drugs (e.g., niclosamide, valproic acid, and captopril), as indicated in Figure 6.
Case study: discovery of potential anticancer mechanism-of-action for resveratrol
We next investigated the potential MOA of resveratrol for its predicted anticancer indications. Resveratrol is a naturally derived stilbene that exists in various foods and beverages. In 1997, Jang et al.60 first reported the possible anticancer indication of resveratrol in a 2-stage mouse skin model. Now, resveratrol has been widely used and tested in preclinical or clinical studies to examine its chemopreventive and antitumor activities.61,62 So far, the molecular mechanism of resveratrol’s anticancer indication remains to be elucidated prior to its clinical approval as an anticancer agent.62 In this study, resveratrol was predicted to have multiple anticancer indications (Figure 6): SKCM (q = 4.2 × 10−4), LUSC (q = 1.2 × 10−3), STAD (q = 1.5 × 10−3), kidney renal clear cell carcinoma (q = 2.0 × 10−3), head and neck squamous carcinoma (q = 4.0 × 10−3), uterine corpus endometrioid (q = 5.8 × 10−3), BRCA (q = 8.9 × 10−3), acute lymphocytic leukemia (q = 0.011), and NSCLC (q = 0.011). This is consistent with the previous studies,61,62 but our work provides detailed information on a total of 9 cancer types.
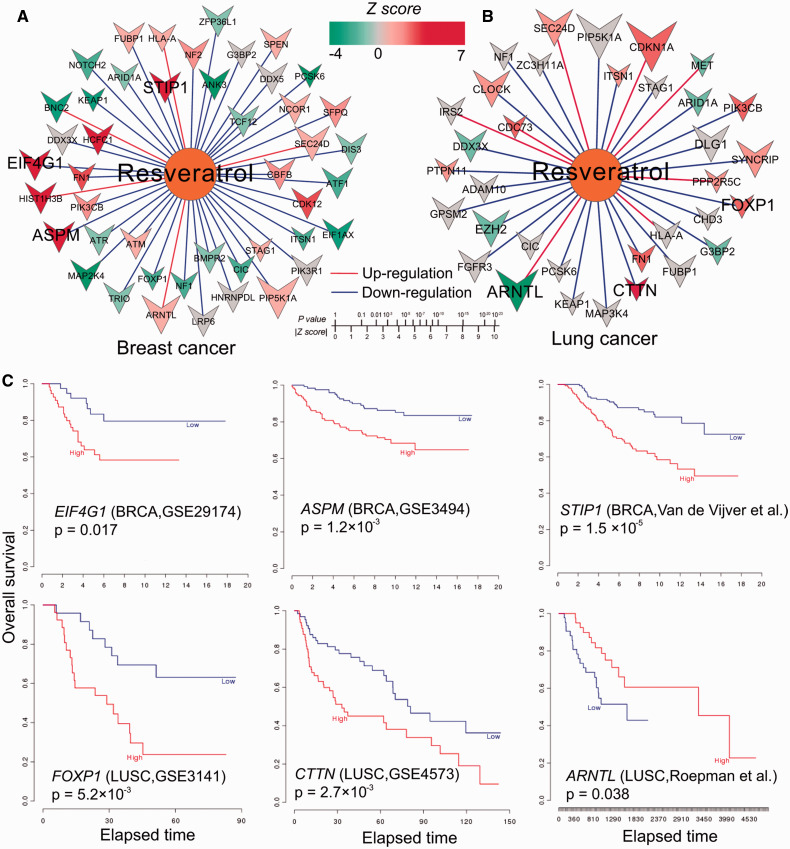
We constructed a resveratrol-gene network to investigate the MOA of resveratrol targeting SMGs in beast cancer (Figure 7A) and lung cancer (Figure 7B) by integrating network analysis and survival analysis. As shown in Figure 7, the color of the node represents the z-score of association between gene expression and survival rate measured by univariate Cox regression, as described in a previous work.44 The size of the gene node represents the amplitude (a) of differential expression with resveratrol, as described in CMap.40 As shown in Figure 7, we asserted that if the high expression (or low expression) of a down-regulated gene (or up-regulated gene) by resveratrol was associated with poor survival rate in a particular cancer type, this gene would be more likely to explain the MOA for its anticancer therapeutic effect by resveratrol, based on a previous study.63Figure 7A reveals that several SMGs (such as ASPM, EIF4G1, and STIP1) in BRCA are down-regulated by resveratrol. Furthermore, high expression of EIF4G1 (P = .017, log-rank test, GES2917464), ASPM (P = 1.2 × 10−3, GES349465), and STIP1 (P = 1.5 × 10−5, Van de Vijver et al.66) was significantly associated with poor survival rate in breast cancer. EIF4G1, encoding eukaryotic translation initiation factor gamma 1, plays crucial roles in breast cancer.67 Previous studies reported that resveratrol inhibited human breast cancer cell migration and invasion,68 so down-regulation of EIF4G1, ASPM, and STIP1 by resveratrol may provide a potential MOA for its chemoprevention and therapeutic properties in breast cancer.
Figure 7:
Potential mechanism-of-action (MOA) for resveratrol’s predicted anticancer indications in breast and lung cancers. (A and B) The networks connecting resveratrol and significantly mutated genes in breast (A) and lung (B) cancer, respectively. In (A) and (B), the color of nodes represents the z-score of the association between gene expression and survival rate calculated by univariate Cox regression as described in a previous study.44 The size of gene nodes denotes the amplitude (a) value of differential expression by the treatment of resveratrol using the Connectivity Map data.40 The red edges denote up-regulation and blue edges denote down-regulation. (C) Kaplan-Meier survival curves show 6 overlapped genes (selected based on the most significant z-scores) between the significantly mutated genes in breast and lung cancer and the resveratrol down/up-regulated genes from the Connectivity Map. The curves were prepared by an online tool, PREdiction of Clinical Outcomes from Genomic profiles: https://precog.stanford.edu.
As shown in Figure 7B, resveratrol down-regulated FOXP1 gene expression, while high expression of FOXP1 was significantly associated with poor survival in LUSC (P = 5.2 × 10−3, GES314169). FOXP1, encoding forkhead box P1, is a member of the forkhead box transcription factor family and plays important roles in lung cancer.70 In addition, resveratrol up-regulated ARNTL expression, but down-regulated CTTN expression (Figure 7B). Interestingly, low expression of ARNTL was significantly associated with poor survival (P = .038, Roepman et al.71), while high expression of CTTN was significantly associated with poor survival (P = 5.2 × 10−3, GES457372) in LUSC. A recent study showed that ARNTL plays a potential tumor suppressor role in ovarian cancer.73CTTN, which encodes cortactin, was previously reported to mediate progression of NSCLC74 and other cancer types.75 Taken together, down-regulation of CTTN and FOXP1 and up-regulation of ARNTL may be a potential MOA for the anti–lung cancer effects of resveratrol.61,62 The aforementioned analysis lends itself to proper experimental validation in the future.
DISCUSSION
Massive amounts of next-generation sequencing data generated from thousands of tumor genomes enabled the development of precision cancer medicine by the identification of new drugs or existing drugs via targeting cancer driver events or SMGs discovered in cancer genomes. In this study, we manually curated 693 SMGs from large-scale cancer genomics studies and found 121 druggable proteins encoded by SMGs. We found that 33.1% of patients could potentially benefit from current US FDA-approved targeted agents, and this number increased to 68.0% when utilizing drug repositioning strategies, suggesting that cancer genomic data forms a strong foundation for precision cancer medicine in the future. Furthermore, we proposed a network-based drug repositioning statistical framework with moderately high accuracy (66.7% success rate) to prioritize new potential indications for existing drugs by integrating SMGs, drug-gene signatures, and the PPI network. We also identified a potential MOA for the putative anticancer effects of resveratrol in breast and lung cancer. In summary, this study demonstrated the potential application of our network-based approach in identifying existing drugs for precision cancer medicine by integrating large-scale cancer genomics and drug pharmacological data.
Limitations and future work
There are several possible limitations in the current network-based infrastructure. First, the data is still incomplete and biased. For example, we examined potential molecular mechanisms of resveratrol’s anticancer effects in breast cancer and lung cancer by integrating drug-gene signatures and clinical analysis (Figure 7). However, CMap data, while innovative and powerful, suffer from the lack of control on the selection of the optimal drug dose, which should be subtoxic to produce informative expression profile data.40 Second, the current CMap 2.0 version included drug-gene signatures identified in only 4 different cancer cell lines.40 However, we studied more than 20 different cancer types in this work. We could not perform individual cancer type analyses using 4 different cancer cell lines versus over 20 different cancer types, but this would be possible when a future version of CMap is released to cover many cancer types. Third, compounds used at higher concentrations can lead to widespread effects due to off-target and secondary effects that are difficult to control in a high-throughput setting. Hence, experimental validations of the predicted MOA are warranted in the future. In addition, we could integrate high-quality, comprehensive drug-gene signatures from LINCSCLOUD (http://www.lincscloud.org) into our network-based approach for more precise anticancer drug repositioning in the future. Fourth, cancer is highly heterogeneous and each cancer may have its subtypes with unique molecular signatures. We will further explore these limitations in the future. Despite the above limitations, our work effectively utilizes the largest-ever cancer genomic dataset, thus providing a robust framework to identify potential existing drug candidates for drug repositioning in cancer.
As shown in Figure 1, we hypothesized that if SMGs or their coding protein neighbors in the PIN were overrepresented in the set of genes up- or down-regulated by a drug of interest for a particular cancer type, this drug would have a high potential of a new indication for this cancer type. This strategy is effective because it weights not only mutational effect but also related biological regulation in the molecular network.76 However, accurately distinguishing oncogenes from TSGs in particular cancer types is a very difficult task. In the current study, we combined oncogenes and TSGs as a union of SMGs, which may cause some data bias or potential false positive discoveries. In the future, we will build more reliable network-based approaches by considering agonists for TSGs to restore gene function or inhibitors for oncogenes to inhibit their oncogenic potential. In addition, we will implement more robust statistical algorithms, such as Kullback-Leibler divergence, for prioritizing new drugs to inhibit the function of neighbors of TSGs in a transcriptional regulatory network by integrating comprehensive gene expression profiles perturbed by drug treatment77 using the data identified from functional RNAi and CRISPR-Cas9 screens.78,79 In addition to SMGs derived from cancer genomics data, we may integrate some cancer susceptibility genes generated from GWAS2 or phenome-wide association studies,80 like the data from GWAS Catalog81 and phenome-wide association studies Catalog,82 into our network-based framework to identify new potential druggable targets or new indications for existing drugs, which will aid in the timely development of precision medicine in broad phenotypes.
CONCLUSION
In this study, we proposed an integrative network-based infrastructure that has the potential to advance the field of precision cancer medicine by prioritizing druggable targets and new indications for existing drugs through targeting cancer driver genes. Based on large-scale cancer genomics data across over 5000 cancer genomes from TCGA, we found that 33.1% of patients could benefit from currently targeted agents, and the benefit rate increased to 68.0% when utilizing a drug repositioning approach. Moreover, we developed a statistical framework for anticancer drug repositioning to integrate drug-gene signatures, manually curated cancer drivers, and the PPI network. We identified 284 potential indications connecting 28 cancer types and 48 existing drugs with a 66.7% success rate validated by literature data. Finally, we showcased a potential MOA for resveratrol anticancer effects through targeting several SMGs (e.g., ARNTL, ASPM, CTTN, EIF4G1, FOXP1, and STIP1) in breast and lung cancers via integrative network analysis and survival rate analysis. In summary, we demonstrated a promising network-based infrastructure to identify potential targets and anticancer drug repositioning candidates for precision cancer medicine by targeting cancer driver genes that can be discovered in massive amounts of cancer genomics data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the two reviewers for their valuable comments that helped us improve the manuscript.
CONTRIBUTORS
F.C. and Z.Z. conceived and designed the study. F.C. carried out experiments and most of the data analysis. J.Z. and M.F. participated in data analysis. F.C. and Z.Z. wrote the manuscript.
FUNDING
This work was partially supported by National Institutes of Health grants (R01LM011177, P50CA095103, P50CA098131, and P30CA068485), The Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation, and Ingram Professorship Funds. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1. Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11(3):191–200. [DOI] [PubMed] [Google Scholar]
- 2. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47(8):856–860. [DOI] [PubMed] [Google Scholar]
- 3. Cheng F, Zhao J, Zhao Z. Advances in computational approaches for prioritizing driver mutations and significantly mutated genes in cancer genomes. Brief Bioinform. 2015; doi: 10.1093/bib/bbv068. First published online: August 24, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng F, Liu C, Lin C, et al. A gene gravity model for the evolution of cancer genomes: a study of 3,000 cancer genomes across 9 cancer types. PLoS Comput Biol. 2015;11(9):e1004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jia P, Wang Q, Chen Q, et al. MSEA: detection and quantification of mutation hotspots through mutation set enrichment analysis. Genome Biol. 2014;15(10):489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jia P, Pao W, Zhao Z. Patterns and processes of somatic mutations in nine major cancers. BMC Med Genomics. 2014;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vuong H, Cheng F, Lin CC, et al. Functional consequences of somatic mutations in cancer using protein pocket-based prioritization approach. Genome Med. 2014;6(10):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10(12):1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–511. [DOI] [PubMed] [Google Scholar]
- 11. Rubio-Perez C, Tamborero D, Schroeder MP, et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell. 2015;27(3):382–396. [DOI] [PubMed] [Google Scholar]
- 12. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schumacher T, Bunse L, Pusch S, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. [DOI] [PubMed] [Google Scholar]
- 14. Bollag G, Tsai J, Zhang J, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11(11):873–886. [DOI] [PubMed] [Google Scholar]
- 15. Cheng F, Jia P, Wang Q, et al. Quantitative network mapping of the human kinome interactome reveals new clues for rational kinase inhibitor discovery and individualized cancer therapy . Oncotarget. 2014;5(11):3697–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobbelstein M, Moll U. Targeting tumour-supportive cellular machineries in anticancer drug development. Nat Rev Drug Discov. 2014;13(3):179–196. [DOI] [PubMed] [Google Scholar]
- 17. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573–582. [DOI] [PubMed] [Google Scholar]
- 30. Corrdinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41(Database issue):D8–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng F, Jia P, Wang Q, et al. Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome. Mol Biol Evol. 2014;31(8):2156–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim P, Cheng F, Zhao J, et al. ccmGDB: a database for cancer cell metabolism genes. Nucleic Acids Res. 2015; 44(Database issue):D959-D968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu F, Shi Z, Qin C, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Knox C, Law V, Jewison T, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hernandez-Boussard T, Whirl-Carrillo M, Hebert JM, et al. The pharmacogenetics and pharmacogenomics knowledge base: accentuating the knowledge. Nucleic Acids Res. 2008;36(Database issue):D913–D918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng F, Liu C, Jiang J, et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012;8(5):e1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng F, Li W, Wu Z, et al. Prediction of polypharmacological profiles of drugs by the integration of chemical, side effect, and therapeutic space. J Chem Inf Model. 2013;53(4):753–762. [DOI] [PubMed] [Google Scholar]
- 38. Cheng F, Zhao Z. Machine learning-based prediction of drug-drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J Am Med Inform Assoc. 2014;21(e2):e278–e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. [DOI] [PubMed] [Google Scholar]
- 41. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 42. Davoli T, Xu AW, Mengwasser KE, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao J, Cheng F, Wang Y, et al. Systematic prioritization of druggable mutations in ∼5,000 genomes across 16 cancer types using a structural genomics-based approach. Mol Cell Proteomics. 2016;15(2):642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. [DOI] [PubMed] [Google Scholar]
- 47. Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10(2):130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haricharan S, Brown P. TLR4 has a TP53-dependent dual role in regulating breast cancer cell growth. Proc Natl Acad Sci USA. 2015;112(25):E3216–E3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lewis SS, Loram LC, Hutchinson MR, et al. (+)-naloxone, an opioid-inactive toll-like receptor 4 signaling inhibitor, reverses multiple models of chronic neuropathic pain in rats. J Pain. 2012;13(5):498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rajamani S, Shryock JC, Belardinelli L. Block of tetrodotoxin-sensitive, Na(V)1.7 and tetrodotoxin-resistant, Na(V)1.8, Na+ channels by ranolazine. Channels (Austin). 2008;2(6):449–460. [DOI] [PubMed] [Google Scholar]
- 51. Kothare SV, Kaleyias J. Zonisamide: review of pharmacology, clinical efficacy, tolerability, and safety. Expert Opin Drug Metab Toxicol. 2008;4(4):493–506. [DOI] [PubMed] [Google Scholar]
- 52. Kim KB, Cabanillas ME, Lazar AJ, et al. Clinical responses to vemurafenib in patients with metastatic papillary thyroid cancer harboring BRAF(V600E) mutation. Thyroid. 2013;23(10):1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weinbach EC, Garbus J. Mechanism of action of reagents that uncouple oxidative phosphorylation. Nature. 1969;221(5185):1016–1018. [DOI] [PubMed] [Google Scholar]
- 54. Osada T, Chen M, Yang XY, et al. Antihelminth compound niclosamide downregulates Wnt signaling and elicits antitumor responses in tumors with activating APC mutations. Cancer Res. 2011;71(12):4172–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wieland A, Trageser D, Gogolok S, et al. Anticancer effects of niclosamide in human glioblastoma. Clin Cancer Res. 2013;19(15):4124–4136. [DOI] [PubMed] [Google Scholar]
- 56. Kramer OH, Zhu P, Ostendorff HP, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22(13):3411–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ylisastigui L, Archin NM, Lehrman G, et al. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18(8):1101–1108. [DOI] [PubMed] [Google Scholar]
- 58. Shirsath N, Rathos M, Chaudhari U, et al. Potentiation of anticancer effect of valproic acid, an antiepileptic agent with histone deacetylase inhibitory activity, by the cyclin-dependent kinase inhibitor P276-00 in human non-small-cell lung cancer cell lines. Lung Cancer. 2013;82(2):214–221. [DOI] [PubMed] [Google Scholar]
- 59. Attoub S, Gaben AM, Al-Salam S, et al. Captopril as a potential inhibitor of lung tumor growth and metastasis. Ann N Y Acad Sci. 2008;1138:65–72. [DOI] [PubMed] [Google Scholar]
- 60. Jang M, Cai L, Udeani GO, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275(5297):218–220. [DOI] [PubMed] [Google Scholar]
- 61. Fulda S. Resveratrol and derivatives for the prevention and treatment of cancer. Drug Discov Today. 2010;15(17-18):757–765. [DOI] [PubMed] [Google Scholar]
- 62. Subramanian L, Youssef S, Bhattacharya S, et al. Resveratrol: challenges in translation to the clinic–a critical discussion. Clin Cancer Res. 2010;16(24):5942–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Amelio I, Gostev M, Knight RA, et al. DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis. 2014;5:e1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Farazi TA, Horlings HM, Ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102(38):13550–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. [DOI] [PubMed] [Google Scholar]
- 67. Badura M, Braunstein S, Zavadil J, et al. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci USA. 2012;109(46):18767–18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tang FY, Su YC, Chen NC, et al. Resveratrol inhibits migration and invasion of human breast-cancer cells. Mol Nutr Food Res. 2008;52(6):683–691. [DOI] [PubMed] [Google Scholar]
- 69. Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. [DOI] [PubMed] [Google Scholar]
- 70. Feng J, Zhang X, Zhu H, et al. High expression of FoxP1 is associated with improved survival in patients with non-small cell lung cancer. Am J Clin Pathol. 2012;138(2):230–235. [DOI] [PubMed] [Google Scholar]
- 71. Roepman P, Jassem J, Smit EF, et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res. 2009;15(1):284–290. [DOI] [PubMed] [Google Scholar]
- 72. Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66(15):7466–7472. [DOI] [PubMed] [Google Scholar]
- 73. Yeh CM, Shay J, Zeng TC, et al. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int J Oncol. 2014;45(5):2101–2107. [DOI] [PubMed] [Google Scholar]
- 74. Noh SJ, Baek HA, Park HS, et al. Expression of SIRT1 and cortactin is associated with progression of non-small cell lung cancer. Pathol Res Pract. 2013;209(6):365–370. [DOI] [PubMed] [Google Scholar]
- 75. MacGrath SM, Koleske AJ. Cortactin in cell migration and cancer at a glance. J Cell Sci. 2012;125(Pt 7):1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sun J, Zhao M, Jia P, et al. Deciphering signaling pathway networks to understand the molecular mechanisms of metformin action. PLoS Comput Biol. 2015;11(6):e1004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Woo JH, Shimoni Y, Yang WS, et al. Elucidating compound mechanism of action by network perturbation analysis. Cell. 2015;162(2):441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schramek D, Sendoel A, Segal JP, et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343(6168):309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.