Abstract
MEF2C is a member of the highly conserved MEF2 family of transcription factors and is a key regulator of cardiovascular development. In mice, Mef2c is expressed in the developing heart and vasculature, including the endothelium. Loss of Mef2c function in germline knockout mice leads to early embryonic demise and profound developmental abnormalities in the cardiovascular system. Previous attempts to uncover the cause of embryonic lethality by specifically disrupting Mef2c function in the heart or vasculature failed to recapitulate the global Mef2c knockout phenotype and instead resulted in relatively minor defects that did not compromise viability or result in significant cardiovascular defects. However, previous studies examined the requirement of Mef2c in the myocardial and endothelial lineages using Cre lines that begin to be expressed after the expression of Mef2c has already commenced. Here, we tested the requirement of Mef2c in the myocardial and endothelial lineages using conditional knockout approaches in mice with Cre lines that deleted Mef2c prior to onset of its expression in embryonic development. We found that deletion of Mef2c in the early myocardial lineage using Nkx2–5Cre resulted in cardiac and vascular abnormalities that were indistinguishable from the defects in the global Mef2c knockout. In contrast, early deletion of Mef2c in the vascular endothelium using an Etv2::Cre line active prior to the onset of Mef2c expression resulted in viable offspring that were indistinguishable from wild type controls with no overt defects in vascular development, despite nearly complete early deletion of Mef2c in the vascular endothelium. Thus, these studies support the idea that the requirement of MEF2C for vascular development is secondary to its requirement in the heart and suggest that the observed failure in vascular remodeling in Mef2c knockout mice results from defective heart function.
Keywords: MEF2C, heart development, vascular development, endothelial cells, endothelium, vascular remodeling, morphogenesis, mouse
1. Introduction
MEF2C is a member of the myocyte enhancer factor 2 family of transcription factors. MEF2 proteins are defined by the presence of amino terminal MADS and MEF2 domains through which they dimerize and bind DNA in a sequence specific manner (Black and Olson, 1998). Like other MEF2 proteins, MEF2C functions as a signal-responsive transcriptional regulator and has extensive interactions with other transcription factors, co-activators, and co-repressors. It can either activate or repress transcription depending on posttranslational modifications and co-factor interactions (Black and Cripps, 2010; Dong et al., 2017; McKinsey et al., 2002). MEF2C is widely appreciated for its role in cardiovascular development (Black and Cripps, 2010; Potthoff and Olson, 2007). Global disruption of Mef2c function in mice leads to profound defects in the heart and vasculature and results in embryonic lethality between E9.5 and E10.5 (Lin et al., 1997). In Mef2c-null mutants, cardiomyocytes are specified, but the heart tube does not elongate sufficiently for proper looping, leading to the formation of only one common ventricle (Lin et al., 1997). Similarly, vascular endothelial cells are specified in Mef2c mutants and form a primitive vascular network, but the vasculature fails to remodel properly leading to severe defects (Bi et al., 1999; Lin et al., 1998; Lin et al., 1997).
Mef2c is expressed in myocardial progenitors within the cardiac crescent by embryonic day (E) 7.5 and is detectable in vascular endothelial cells by E8.5 (Dodou et al., 2004; Edmondson et al., 1994). Interestingly, numerous, discrete enhancers in the Mef2c locus control its transcription in the heart and the vasculature (Agarwal et al., 2011; De Val et al., 2004; De Val et al., 2008; Dodou et al., 2004; von Both et al., 2004). Detailed studies of those enhancers demonstrate that Mef2c activation is controlled by different upstream regulatory pathways in the myocardial and endothelial lineages and suggest that Mef2c has autonomous requirements in cardiomyocytes and endothelial cells (De Val et al., 2004; De Val et al., 2008; Dodou et al., 2004; von Both et al., 2004). However, no study designed to test cell autonomous requirements has recapitulated the germline phenotype. Conditional knockout of Mef2c in cardiomyocytes caused only mild or no deficiencies in the embryonic heart (Vong et al., 2005). Deletion of Mef2c from the endothelial lineage has been reported to result in defective branching in embryonic development and postnatally, but animals survive and have no overt defects (Sacilotto et al., 2016; Xu et al., 2012). One potential explanation for why these prior studies did not recapitulate the germline knockout phenotype is that Mef2c was deleted in those studies using Cre lines driven by enhancers of cardiac or vascular differentiation genes that are activated after the initiation of Mef2c expression (Agah et al., 1997; Chen et al., 1998; Dodou et al., 2004; Edmondson et al., 1994; Koni et al., 2001; Moses et al., 2001; Wang et al., 2010). Expression of Mef2c prior to Cre-mediated deletion may thus limit the severity of the observed phenotypes by allowing initiation of MEF2C-dependent gene regulatory networks.
Here, we tested whether Mef2c function is required in the heart and vasculature by creating lineage specific knockouts prior to the onset of Mef2c transcription in either tissue. When Mef2c was deleted specifically in early stage cardiac precursors using Nkx2–5Cre, we found that embryos recapitulate the germline knockout phenotype, including vascular deficiencies, and die around E9.5. In contrast, Mef2c deletion in the endothelium using an Etv2::Cre line, which is active prior to the onset of Mef2c expression, resulted in viable offspring with no overt defects in early vascular development despite complete excision of Mef2c from the endothelium prior to E8.5. These data strongly support the idea that the vascular defects, which occur as a result of loss of Mef2c function, are secondary to the loss of Mef2c in the heart and are a likely consequence of failed vascular remodeling due to defective cardiac development and function.
2. Results and discussion
2.1. Early deletion of Mef2c in the heart causes embryonic demise at midgestation
To delete Mef2c specifically in the cardiac lineage prior to onset of Mef2c expression, we utilized the well-characterized Nkx2–5Cre line (Moses et al., 2001). In this knock-in line, the expression of Cre accurately mirrors the temporospatial pattern of expression of the Nkx2-5 gene, one of the earliest lineage markers of cardiac fate (Lints et al., 1993). Importantly, Nkx2–5 expression commences prior to assembly of the linear heart tube at around E7, which precedes Mef2c expression. The Nkx2–5Cre expression domain encompasses all cardiac progenitors of the first and second heart field, including the cells of the myocardium and endocardium (Ma et al., 2008; Moses et al., 2001).
In crosses of Nkx2–5Cre/+;Mef2c+/− with Mef2cflox/flox mice, the Nkx2–5Cre cardiac conditional knockout mice (Mef2cflox/-; Nkx2–5Cre/+) died between E9.5 and E10.5, just as Mef2c germline KO animals (Fig. 1). At E9.5, Mef2cflox/-; Nkx2–5Cre/+ embryos were clearly smaller than their control littermates and exhibited an overt phenotype that is essentially identical to that of germline Mef2c−/− knockout mice (Fig. 1C and G). Nkx2–5Cre cardiac conditional knockout embryos were not detected at E14.5, but control littermates of all other genotypes were present in expected ratios (Table 1A). This is in contrast to previous cardiac-specific deletions of Mef2c with MLC-2vCre [Myl2tm1(cre)Krc; (Chen et al., 1998)] or αMyHC-Cre [Tg(Myh6-cre)2182Mds; (Agah et al., 1997)], which did not adversely affect embryonic development (Vong et al., 2005). Cre recombinase activity in MLC-2vCre and αMyHC-Cre mice begins significantly later than in Nkx2–5Cre mice and after the onset of Mef2c expression in cardiac progenitors (Agah et al., 1997; Chen et al., 1998; Dodou et al., 2004; Edmondson et al., 1994; Moses et al., 2001). We interpret the recapitulation of the germline Mef2c knockout phenotype in Nkx2–5Cre cardiac conditional knockout mice to be due to the earlier excision of Mef2c in cardiac progenitors.
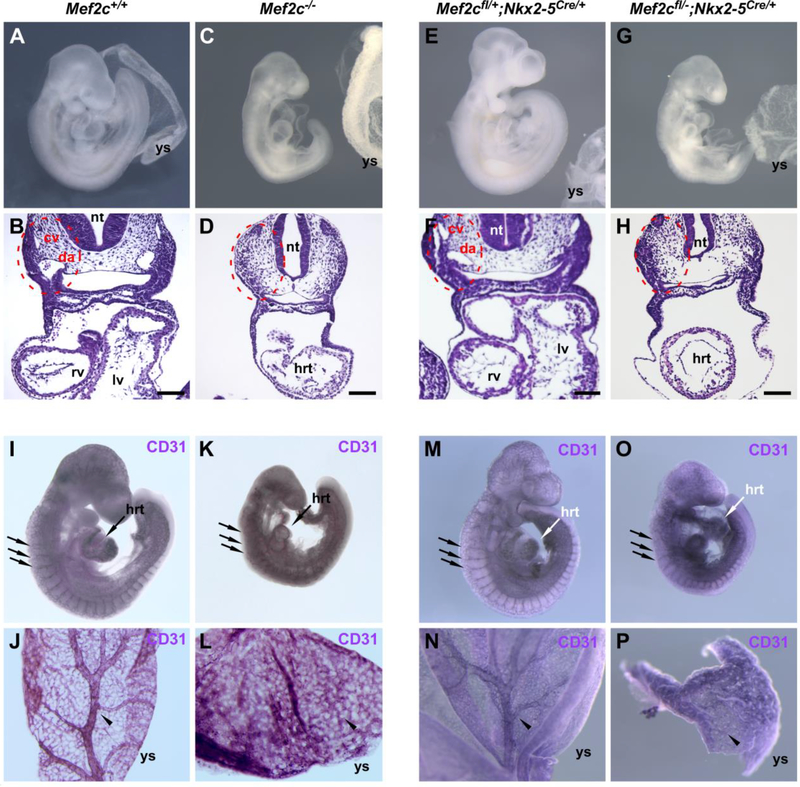
Fig. 1.
Cardiac-specific deletion of Mef2c, using Nkx2–5Cre/+, recapitulates the global Mef2c knockout phenotype. Embryos were harvested at embryonic day 9.5, inspected for gross appearance (A, C, E, G) or sectioned and hematoxylin and eosin (H&E) stained (B, D, F, H). Embryos with global or cardiacspecific loss of Mef2c function (C, G) were noticeably smaller than control littermates (A, E). The main blood vessels in the trunk were present in cross sections of control embryos (red dashed circles in B, F), but absent in global or conditional knockouts (red dashed circles in D, H). Immunohistochemical staining with anti-CD31 antibody in whole mount embryos (I, K, M, O) or yolk sacs (J, L, N, P) shows that the blood vasculature initially forms in all embryos (unlabeled arrows in I, K, M, O indicate intersomitic vessels), but fails to remodel in global and Nkx2–5Cre cardiac-specific knockout embryos. Yolk sacs of control embryos show arborization (arrowheads in J, N), but it remained as an unremodeled plexus in global and Nkx2–5Cre conditional knockouts (arrowheads in L, P). cv, cardinal vein; da, dorsal aorta; hrt, heart; lv, left ventricle; nt, neural tube; rv, right ventricle; ys, yolk sac. Scale bars, 100 μm.
Table 1.
Genotype frequencies of progeny from crosses to generate (A) cardiac or (B) endothelial-specific Mef2c knockout mice. Expected and observed progeny of each genotype from (A) cardiac Nkx2–5Cre/+; Mef2c+/− × Mef2cflox/flox crosses, collected at E14.5; or (B) Etv2::CreTg/0; Mef2c+/− × Mef2cflox/flox crosses, collected at P10. CKO, conditional knockout; df, degrees of freedom in χ2 test.
| A | Genotype | % Expected | # Observed |
| Mef2cflox/+ (wt) | 25 | 8 | |
| Mef2cflox/+; Nkx2–5Cre/+ (het) | 25 | 13 | |
| Mef2cflox/− | 25 | 9 | |
| Mef2cflox/−;Nkx2–5Cre/+ (CKO) | 25 | 0 | |
| χ2=11.867, 3 df; P=0.0079 | |||
| B | Genotype | % Expected | # Observed |
| Mef2cflox/+ (wt) | 25 | 23 | |
| Mef2cflox/+;Etv2::CreTg/0 (het) | 25 | 18 | |
| Mef2cflox/− | 25 | 14 | |
| Mef2cflox/−;Etv2::CreTg/0 (CKO) | 25 | 16 | |
| χ2=2.521, 3 df; P=0.4715 (not signif.) |
2.2. Early deletion of Mef2c in the heart leads to cardiac and vascular defects
The first step of cardiac morphogenesis is the formation of a linear heart tube. In Mef2c germline knockout mice, the linear heart tube is hypoplastic compared to controls and fails to undergo rightward looping, effectively abolishing the formation of the right ventricle (Lin et al., 1997; Vong et al., 2006); (Fig. 1, compare panels B and D). Similarly, conditional deletion of Mef2c with Nkx2–5Cre resulted in an unlooped, hypoplastic heart tube at E9.5 (Fig. 1H), essentially mimicking the phenotype observed in global knockout mice (Fig. 1, compare panels D and H). The Nkx2–5Cre line was generated by insertion of the Cre sequence into the Nkx2–5 locus, thereby disrupting Nkx2-5 function from the targeted allele (Moses et al., 2001). However, Mef2cflox/+; Nkx2–5Cre/+ littermates have normal heart morphology (Fig. 1, compare panels A and B to panels E and F), confirming that disruption of one functional Nkx2–5 allele and one functional Mef2c allele did not result in a structural heart phenotype. Additionally, Mef2cflox/+; Nkx2–5Cre/+ mice were born at predicted Mendelian frequency (Table 1A), further supporting the idea that the loss of one Nkx2–5 allele did not contribute to the observed Mef2c phenotype in Mef2c conditional knockout mice.
There are profound deficiencies in the anatomy of the blood vasculature in Mef2c germline knockout mice (Bi et al., 1999; Lin et al., 1998; Lin et al., 1997; Fig. 1D, K, and L). At E9.5, the major blood vessels of the trunk, the dorsal aorta and cardinal vein, had clearly formed in controls, but were not visible in transverse sections of mutant embryos (Fig. 1B and D). Nkx2–5Cre is not expressed in vascular endothelium (Moses et al., 2001), thus sparing the Mef2c locus from excision in endothelial cells of the vasculature in Mef2cflox/-; Nkx2–5Cre/+ conditional knockout embryos. Nevertheless, profound vascular defects were still present in Mef2cflox/-; Nkx2–5Cre/+ conditional knockout embryos. In the embryo proper, this included absence of the dorsal aorta and cardinal vein and disrupted or absent intersomitic vessels (Fig. 1H and O). In the yolk sac, the vascular plexus, which was clearly remodeled to an arborized structure in control embryos, failed to properly remodel in cardiac-restricted conditional knockouts (Fig. 1N and P). Importantly, these defects in vascular development and remodeling were essentially identical to those observed in global Mef2c knockout embryos (Fig. 1D, K, and L).
CD31 (PECAM-1) is a marker of endothelial cells (Garlanda and Dejana, 1997). The presence of CD31 expression in both germline and Nkx2–5Cre cardiac conditional knockout embryos suggests that endothelial cells are properly specified in the absence of MEF2C and that the vascular defects in Mef2c germline knockout mice are secondary to primary defects in heart development and function. The absence of major blood vessels in the embryo and the failure of the vasculature to remodel is likely due to the loss of cardiac contraction and the resultant hemodynamic forces, which are required for vessel remodeling and stabilization (Lucitti et al., 2007; Udan et al., 2013).
2.3. Etv2::Cre results in early recombination in endothelial and hematopoietic lineages and derivatives
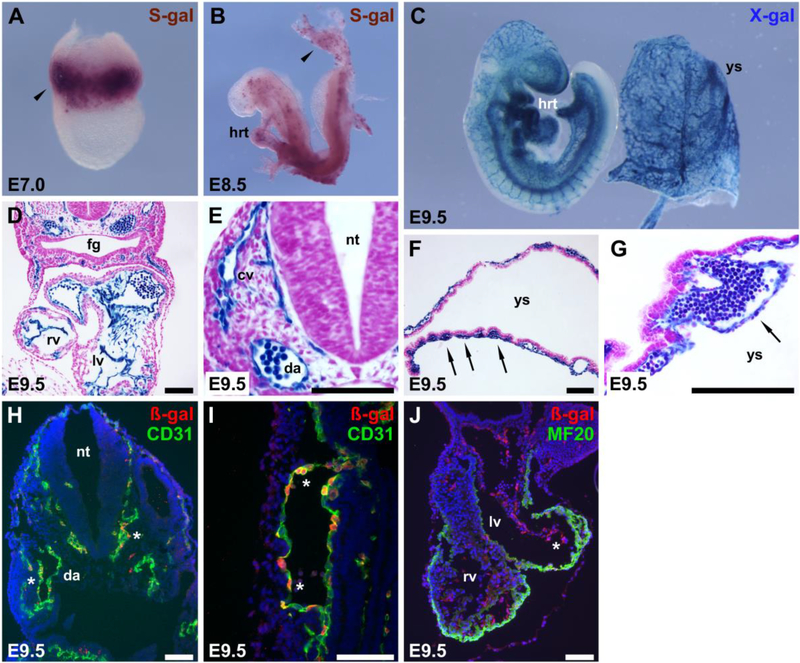
Etv2 is an Ets transcription factor and one of the earliest lineage determinants of endothelial and blood cells in embryonic development. In Etv2 mutants, no endothelial or blood cells are specified, and embryos die by E9.5 due to severe vascular and hematopoietic defects (Ferdous et al., 2009; Lee et al., 2008). Etv2 binds directly to an early endothelial-specific enhancer in the Mef2c locus to activate Mef2c expression in vivo (De Val et al., 2008). Because Etv2 is upstream of Mef2c, we reasoned that inactivation of Mef2c by Cre expression under the control of early regulatory elements from the Etv2 gene would allow excision of Mef2c prior to its expression. Therefore, we cloned a 3.3 kb-fragment of the upstream control region of Etv2, including the endogenous promoter and transcription start site, into a Cre expression vector and generated stable transgenic mouse lines. To define the lineage marked by the activity of this Etv2 promoter and enhancer, we crossed this Etv2::Cre transgenic line to Rosa26LacZ/LacZ Cre-dependent lacZ reporter mice (Soriano, 1999). Cre-mediated recombination occurred in endothelial and hematopoietic progenitor cells in the yolk sac and embryo proper prior to E7 (Fig. 2A). By E8.5, the entire endothelial lineage was marked by the activity of the Etv2::Cre transgene (Fig. 2B), and robust X-gal staining at E9.5 was observed in endothelial and blood cells throughout the developing embryo and in the extra-embryonic vasculature (Fig. 2C-J). Although the entire endothelium was marked by the activity of the Etv2::Cre transgene, no ß-galactosidase activity or expression was observed in the myocardium (Fig. 2D and J), indicating that little or no excision was occurring in cardiac myocytes. Importantly, Cre activity from this Etv2::Cre transgenic line precedes Mef2c expression (Dodou et al., 2004; Edmondson et al., 1994), allowing excision prior to any expression of Mef2c in the vasculature. In a previously published study, Rasmussen et al. described a very similar Etv2::Cre transgenic line (Er71-Cre) made with a 3.9-kb Etv2 proximal fragment, including the minimal promoter and transcriptional start site (Rasmussen et al., 2011). That study reported essentially identical early activity of that independently-generated Etv2::Cre line when crossed to Rosa26LacZ/LacZ reporter mice to the Cre activity observed here (Rasmussen et al., 2011), supporting our findings that Etv2::Cre acts very early in embryonic development with activity throughout all endothelium and blood.
Fig. 2.
Genetic fate mapping of Etv2::Cre shows that Cre activity is restricted to hematopoietic and endothelial cells and/or their precursors. Etv2::CreTg/0 mice were crossed to Rosa26lacZ/lacZ mice, and embryos were collected at E7.0 (A), E8.5 (B), or E 9.5 (C–G), and stained with either Salmon-gal (S-gal) or X-gal to detect ß-galactosidase activity from the Rosa26lacZ/lacZ reporter. Embryos and yolk sacs were analyzed either as whole mounts (A–C) or sectioned and counterstained with neutral fast red (D– G). Arrowheads in (A, B) indicate staining in extra-embryonic blood and blood vessel-forming regions. At E9.5, the reporter was active exclusively in endocardium (D), vascular endothelial cells of the embryo and yolk sac (arrows in F, G), and blood cells contained within patent vessels. (H–J) ßgalactosidase expression (red) overlapped with CD31 expression (green) in vascular endothelium (asterisks in H, I). (J) MF20 antibody detects myosin heavy chain (Myosin 4), expressed in cardiomyocytes (red) and did not colocalize with endocardial reporter expression (green). Asterisk in (J) marks Etv2::Cre ß-gal activity in the endocardium. cv, cardinal vein; da, dorsal aorta; ec, endocardium; fg, foregut; hrt, heart; lv, left ventricle; nt, neural tube; rv, right ventricle; ys, yolk sac. Scale bars, 100 μm.
2.4. Early deletion of Mef2c in the endothelium does not result in an overt phenotype and does not negatively affect viability
In crosses of Etv2::CreTg/0;Mef2c+/− with Mef2cflox/flox mice, the endothelial-specific conditional knockout mice (Etv2::CreTg/0;Mef2cflox/-) were born and survived at Mendelian-predicted frequencies (Table 1B) and were viable and fertile with no overt defects. During embryonic development, Etv2::Cre endothelial-specific conditional knockouts were indistinguishable from controls at all stages examined (Fig. 3A-H). The hearts of endothelial-specific conditional knockout embryos looped normally and had normal morphology (Fig. 3A and B). Likewise, vascular development appeared normal: dorsal aortae and cardinal veins were properly assembled and patent, and all other vessels in the conditional knockout animals appeared normal and functional (Fig. 3).
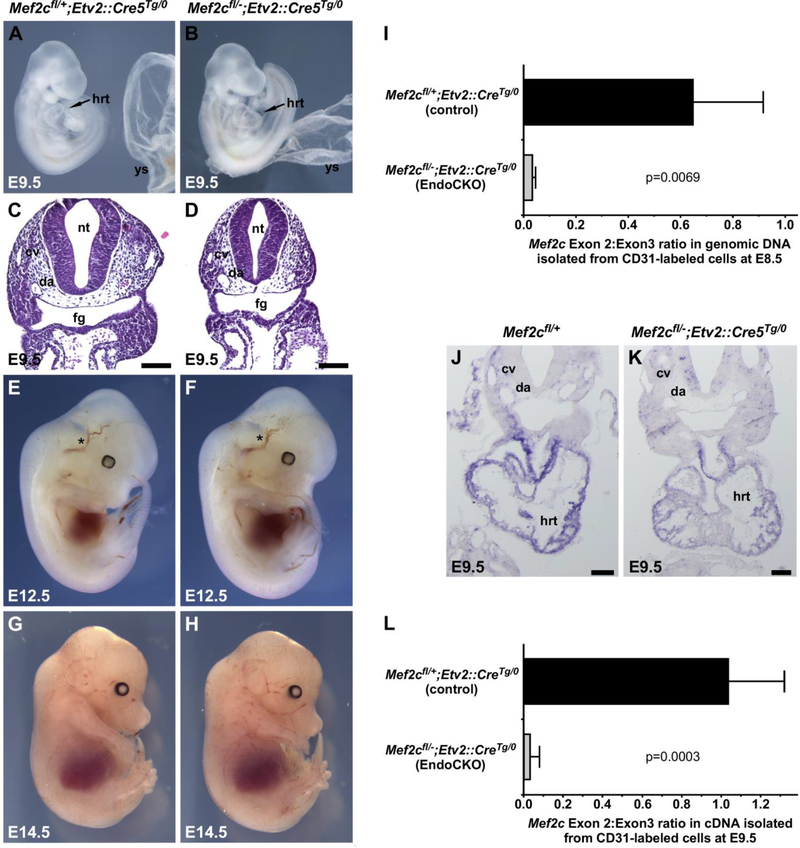
Fig. 3.
Complete endothelium-specific deletion of Mef2c prior to its expression in the vasculature does not impair formation of the cardiovascular system. (A-H, J, K) Wild type or Etv2::Cre endothelial-specific Mef2c knockout embryos were harvested at E9.5 (A–D, J, K), E12.5 (E, F), or E14.5 (G, H) and examined for gross appearance or sectioned and H&E stained (C, D). In all cases and at all time points Etv2::Cre endothelial-specific Mef2c knockout embryos were indistinguishable from control littermates. Asterisks mark the carotid artery at E12.5 (E, F). (I) Quantification of exons 2 and 3 in genomic DNA by quantitative PCR revealed efficient excision of exon 2 in endothelial cells by E8.5 in Mef2cflox/–; Etv2::CreTg/0 conditional knockout embryos. P-values were calculated by Student’s t-test; p=0.0069; n=5. (J, K) In situ hybridization for exon 2 in the Mef2c transcript shows loss of detectable Mef2c transcript in the vasculature at E9.5 in Etv2::Cre endothelial-specific Mef2c knockout embryos; transcripts were still clearly evident in the heart. (L) Exon 2-containing Mef2c transcripts were reduced by 97% at E9.5 in Etv2::Cre endothelial-specific Mef2c knockout embryos compared to controls. Pvalues were calculated by Student’s t-test; p=0.0003; n=6. cv, cardinal vein; da, dorsal aorta; fg, foregut; hrt, heart; nt, neural tube; ys, yolk sac. Scale bars, 100 μm.
To ensure that Mef2c exon 2 was deleted in vascular endothelial cells, we isolated CD31+ cells from endothelial-specific conditional knockout and control embryos at E8.5 by FACS sorting and determined the prevalence of Mef2c coding exons 2 and 3 in genomic DNA (Fig. 3I). Coding exon 2 is the floxed exon in Mef2cflox/flox mice (Vong et al., 2005); exon 3 serves as an internal control in these analyses. Deletion of exon 2 still allows a Mef2c transcript (lacking the deleted exon) to be produced, but it does not encode a functional protein (Barnes et al., 2016; Vong et al., 2005). Importantly, Etv2::Cre expression led to a nearly complete deletion of Mef2c exon 2 by E8.5 (Fig. 3I). Consistent with complete deletion of Mef2c from endothelial cell genomic DNA by E8.5, expression of exon 2 was undetectable in Mef2c transcripts in blood vessels and endocardium of Mef2cflox/-;Etv2::CreTg/0 knockout embryos by RNA in situ hybridization under conditions in which it was easily detectable in control embryos (Fig. 3J and K). Mef2c exon 2-containing transcript was readily detectable in the myocardium of conditional knockout embryos (Fig. 3J and K). To quantify Mef2c expression in endothelial cells specifically, we again used FACS sorting to isolate CD31+ cells and assessed the prevalence of exon 2 and exon 3 in Mef2c transcripts by quantitative reverse transcriptase-PCR (Fig. 3L). We observed a 97% decrease in exon 2-containing, functional Mef2c transcripts isolated from Etv2::Cre conditional knockouts compared to controls (Fig. 3L), indicating essentially complete loss of Mef2c in endothelial cells with no evident phenotype.
Previous studies of Mef2c global knockout mice suggested a possible cell autonomous requirement for Mef2c in endothelial cells to support proper vascular development (Lin et al., 1998). However, the lack of an apparent phenotype in endothelial-specific knockout embryos using Etv2::Cre, an early and efficient Cre line, suggests instead that there is not an absolute cell-autonomous requirement for Mef2c in endothelial cells for embryonic development and viability. Rather, our data strongly suggest that the observed early vascular defects are secondary to impaired heart function and are a consequence of the loss of Mef2c in the cardiac lineage. Interestingly, other studies deleting Mef2c specifically in endothelial cells have revealed later, postnatal roles for MEF2C in angiogenesis and in the interplay of endothelial cells with surrounding vascular smooth muscle cells (Lu et al., 2017; Maiti et al., 2008; Sacilotto et al., 2016; Xu et al., 2012). The lack of an early developmental vascular phenotype in the absence of Mef2c specifically in the vascular endothelium may reflect the fact that MEF2 proteins may act redundantly (Desjardins and Naya, 2016). Indeed, this notion is supported by a recent report of impaired angiogenic sprouting in embryonic development that required the simultaneous knockout of Mef2c and Mef2a in the vasculature (Sacilotto et al., 2016).
Mef2c is widely appreciated as a marker of early endothelial cells, where its expression is controlled by multiple, deeply conserved endothelial-specific enhancers (De Val et al., 2004; De Val et al., 2008). The observation that Mef2c is not required cell autonomously for vascular development raises interesting questions about why Mef2c is expressed in endothelial cells during early development and why early, highly-specific, conserved endothelial enhancers of Mef2c exist. It will be interesting in future studies to determine if early-acting Mef2c endothelial enhancers are required for later expression of Mef2c, when it is involved in postnatal endothelial cell functions.
3. Materials and Methods
3.1. Transgenic and mutant mice
Mef2cflox/flox,[Mef2ctm1Jjs; MGI:3603182], Mef2c+/− [Mef2ctm1Eno; MGI:1857491], Rosa26LacZ/LacZ [Gt(ROSA)26Sortm1Sor; MGI:1861932]; Rosa26mTmG/mTmG [Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo; MGI:3716464], and Nkx2–5Cre/+ [Nkx2–5tm1(cre)Rjs; MGI:2654594] mice have each been described previously (Lin et al., 1997; Moses et al., 2001; Muzumdar et al., 2007; Soriano, 1999; Vong et al., 2005). Cre-dependent recombination of the Mef2cflox allele leads to removal of Mef2c coding exon 2, corresponding to amino acids 18–86, resulting in a null allele but leaving the remainder of the Mef2c transcript intact (Vong et al., 2005). To generate Etv2::Cre transgenic mice a 3,309-bb fragment of the Etv2 upstream control region (Mm10, chr7:30636190–30639498), including the endogenous transcription start site, was amplified from mouse genomic and cloned into a promoterless Cre transgenic vector described previously (Verzi et al., 2005), followed by purification of the transgene fragment and generation of transgenic mice by oocyte microinjection as described previously (Anderson et al., 2004). Genotyping was performed on DNA isolated from yolk sacs or from tail biopsies by Southern blot or PCR. All mouse lines used in these studies were maintained on an outbred CD-1/FVB mixed background for 10+ generations; no variable penetrance was observed. All experiments using animals were approved by the UCSF Institutional Animal Care and Use Committee and complied with federal and institutional guidelines.
3.2. Histology, immunofluorescence, immunohistochemistry, and in situ hybridization
Section and whole-mount RNA in situ hybridization and whole-mount X-gal or Salmon-gal staining to detect ß-galactosidase activity was performed as previously described (Anderson et al., 2004; Kishigami et al., 2006; Rojas et al., 2005). For Mef2c in situ hybridization, we used a 208-bp probe that corresponds to coding exon 2 as described elsewhere (Anderson et al., 2015). Lineage analysis using Etv2::Cre and Rosa26LacZ/LacZ mice was performed as described previously (Verzi et al., 2005). For whole mount immunohistochemistry, rat anti- mouse CD31 (1:250, BD Biosciences, #553370) and horse radish peroxidase (HRP)-conjugated goat anti-rat secondary antibody (1:250, Abcam, ab7097) were used and detected with the DAB substrate kit (Vector, SK-4100), as described previously (Barnes et al., 2010). Immunofluorescence detection of ß-galactosidase, CD31, and myosin heavy chain (MF20) was performed as described previously (Schachterle et al., 2012). The following primary antibodies were used at a 1:100 dilution: rat anti- mouse CD31 (BD Biosciences, #553370), mouse anti-chicken MYH1E (DSHB, MF20), and chicken anti-ß-galactosidase (Abcam, ab9361). The following secondary antibodies were used at a 1:300 dilution: goat anti-rat AF488 (Abcam, ab150157), goat anti-mouse AF488 (Abcam, ab150113), and goat anti-chicken AF594 (Abcam, ab150172).
3.3. Flow cytometry and quantitative PCR
For analysis of recombination efficiency, Etv2::CreTg/0; Mef2c+/− mice were crossed to Rosa26mTmG/mTmG; Mef2cflox/flox mice, and embryos were collected at E8.5 or E9.5 and dissociated to create a single cell suspension for fluorescent-activated cell sorting (FACS), as described previously (Hu et al., 2015). At E8.5, cells were for sorted for GFP; at E9.5 embryos were co-stained with an APC-conjugated anti-CD31 antibody (BD Pharmingen, clone #553370) and sorted for GFP followed by CD31 sorting (Lizama et al., 2015). DNA or RNA of individual embryos was isolated from sorted cells with the Qiagen Allprep Micro Kit according to the manufacturer’s instructions. Reverse transcription was conducted with the BioRad iScript reagent and quantitative PCR was performed as described previously (Anderson et al., 2017). For quantification of excision, we used primers against targeted exon 2 (E2-F, 5′-gtgctgtgcgactgtgagat-3′; E2-R, 5′-tctgagtttgtccggctctc-3′) or exon 3 (E3-F, 5′-ttgccttccctgtccatacc-3′; E3-R, 5′-acccttgcctgcttacttca-3′) that is not excised and serves as the control. Genotypes were confirmed by PCR using individual DNA samples.
Highlights.
MEF2C is an essential transcriptional regulator of cardiovascular development
Deletion of Mef2c in early cardiac progenitors causes heart and vascular defects
Deletion of Mef2c in early endothelial progenitors results in no overt phenotype
Early vascular defects in Mef2c-null mice are secondary to heart defects
Acknowledgements
We thank Shan-Mei Xu for assistance with generating transgenic animals, and Carlos Lizama for help with cell sorting.
Funding
This work was supported by grants HL064658 and HL136182 from the National Heart, Lung, and Blood Institute. T.S. was supported by a postdoctoral fellowship from the American Heart Association, Western States Affiliate (16POST30740016). S.C.M. was supported in part by CIRM training grant TG2–01153.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD, 1997. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest 100, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P, Verzi MP, Nguyen T, Hu J, Ehlers ML, McCulley DJ, Xu SM, Dodou E, Anderson JP, Wei ML, Black BL, 2011. The MADS box transcription factor MEF2C regulates melanocyte development and is a direct transcriptional target and partner of SOX10. Development 138, 2555–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CM, Hu J, Barnes RM, Heidt AB, Cornelissen I, Black BL, 2015. Myocyte enhancer factor 2C function in skeletal muscle is required for normal growth and glucose metabolism in mice. Skelet Muscle 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CM, Hu J, Thomas R, Gainous TB, Celona B, Sinha T, Dickel DE, Heidt AB, Xu SM, Bruneau BG, Pollard KS, Pennacchio LA, Black BL, 2017. Cooperative activation of cardiac transcription through myocardin bridging of paired MEF2 sites. Development 144, 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Dodou E, Heidt AB, De Val SJ, Jaehnig EJ, Greene SB, Olson EN, Black BL, 2004. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol Cell Biol 24, 3757–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, Conway SJ, Vincentz JW, Firulli AB, 2010. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev Dyn 239, 3086–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Harris IS, Jaehnig EJ, Sauls K, Sinha T, Rojas A, Schachterle W, McCulley DJ, Norris RA, Black BL, 2016. MEF2C regulates outflow tract alignment and transcriptional control of Tdgf1. Development 143, 774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Drake CJ, Schwarz JJ, 1999. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol 211, 255–267. [DOI] [PubMed] [Google Scholar]
- Black BL, Cripps RM, 2010. Myocyte Enhancer Factor 2 Transcription Factors in Heart development and Disease, in: Rosenthal N, Harvey RP (Eds.), Heart Development and Regeneration. Academic Press, Oxford, pp. 673–699. [Google Scholar]
- Black BL, Olson EN, 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annual review of cell and developmental biology 14, 167–196. [DOI] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Minamisawa S, Price RL, Becker KD, Hickey R, Ross J Jr., Chien KR, 1998. Selective requirement of myosin light chain 2v in embryonic heart function. The Journal of biological chemistry 273, 1252–1256. [DOI] [PubMed] [Google Scholar]
- De Val S, Anderson JP, Heidt AB, Khiem D, Xu SM, Black BL, 2004. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol 275, 424–434. [DOI] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL, 2008. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135, 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Naya FJ, 2016. The Function of the MEF2 Family of Transcription Factors in Cardiac Development, Cardiogenomics, and Direct Reprogramming. Journal of cardiovascular development and disease 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL, 2004. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 131, 3931–3942. [DOI] [PubMed] [Google Scholar]
- Dong C, Yang XZ, Zhang CY, Liu YY, Zhou RB, Cheng QD, Yan EK, Yin DC, 2017. Myocyte enhancer factor 2C and its directly-interacting proteins: A review. Progress in biophysics and molecular biology 126, 22–30. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF, Olson EN, 1994. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120, 1251–1263. [DOI] [PubMed] [Google Scholar]
- Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson EN, Kyba M, Garry DJ, 2009. Nkx2–5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proceedings of the National Academy of Sciences of the United States of America 106, 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlanda C, Dejana E, 1997. Heterogeneity of endothelial cells. Specific markers. Arteriosclerosis, thrombosis, and vascular biology 17, 1193–1202. [DOI] [PubMed] [Google Scholar]
- Hu J, Verzi MP, Robinson AS, Tang PL, Hua LL, Xu SM, Kwok PY, Black BL, 2015. Endothelin signaling activates Mef2c expression in the neural crest through a MEF2C-dependent positive-feedback transcriptional pathway. Development 142, 2775–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Komatsu Y, Takeda H, Nomura-Kitabayashi A, Yamauchi Y, Abe K, Yamamura K, Mishina Y, 2006. Optimized beta-galactosidase staining method for simultaneous detection of endogenous gene expression in early mouse embryos. Genesis 44, 57–65. [DOI] [PubMed] [Google Scholar]
- Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA, 2001. Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. The Journal of experimental medicine 193, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K, 2008. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Lu J, Yanagisawa H, Webb R, Lyons GE, Richardson JA, Olson EN, 1998. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development 125, 4565–4574. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN, 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP, 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 969. [DOI] [PubMed] [Google Scholar]
- Lizama CO, Hawkins JS, Schmitt CE, Bos FL, Zape JP, Cautivo KM, Borges Pinto H, Rhyner AM, Yu H, Donohoe ME, Wythe JD, Zovein AC, 2015. Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nature communications 6, 7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YW, Lowery AM, Sun LY, Singer HA, Dai G, Adam AP, Vincent PA, Schwarz JJ, 2017. Endothelial Myocyte Enhancer Factor 2c Inhibits Migration of Smooth Muscle Cells Through Fenestrations in the Internal Elastic Lamina. Arteriosclerosis, thrombosis, and vascular biology 37, 1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucitti JL, Jones EA, Huang C, Chen J, Fraser SE, Dickinson ME, 2007. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development 134, 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Zhou B, Pu WT, 2008. Reassessment of Isl1 and Nkx2–5 cardiac fate maps using a Gata4based reporter of Cre activity. Dev Biol 323, 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti D, Xu Z, Duh EJ, 2008. Vascular endothelial growth factor induces MEF2C and MEF2dependent activity in endothelial cells. Investigative ophthalmology & visual science 49, 3640–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN, 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends in biochemical sciences 27, 40–47. [DOI] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ, 2001. Embryonic expression of an Nkx2–5/Cre gene using ROSA26 reporter mice. Genesis 31, 176–180. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L, 2007. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN, 2007. MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140. [DOI] [PubMed] [Google Scholar]
- Rasmussen TL, Kweon J, Diekmann MA, Belema-Bedada F, Song Q, Bowlin K, Shi X, Ferdous A, Li T, Kyba M, Metzger JM, Koyano-Nakagawa N, Garry DJ, 2011. ER71 directs mesodermal fate decisions during embryogenesis. Development 138, 4801–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL, 2005. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development 132, 3405–3417. [DOI] [PubMed] [Google Scholar]
- Sacilotto N, Chouliaras KM, Nikitenko LL, Lu YW, Fritzsche M, Wallace MD, Nornes S, Garcia-Moreno F, Payne S, Bridges E, Liu K, Biggs D, Ratnayaka I, Herbert SP, Molnar Z, Harris AL, Davies B, Bond GL, Bou-Gharios G, Schwarz JJ, De Val S, 2016. MEF2 transcription factors are key regulators of sprouting angiogenesis. Genes Dev 30, 2297–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachterle W, Rojas A, Xu SM, Black BL, 2012. ETS-dependent regulation of a distal Gata4 cardiac enhancer. Dev Biol 361, 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P, 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Udan RS, Vadakkan TJ, Dickinson ME, 2013. Dynamic responses of endothelial cells to changes in blood flow during vascular remodeling of the mouse yolk sac. Development 140, 4041–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL, 2005. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol 287, 134–145. [DOI] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL, 2004. Foxh1 is essential for development of the anterior heart field. Dev Cell 7, 331–345. [DOI] [PubMed] [Google Scholar]
- Vong L, Bi W, O’Connor-Halligan KE, Li C, Cserjesi P, Schwarz JJ, 2006. MEF2C is required for the normal allocation of cells between the ventricular and sinoatrial precursors of the primary heart field. Dev Dyn 235, 1809–1821. [DOI] [PubMed] [Google Scholar]
- Vong LH, Ragusa MJ, Schwarz JJ, 2005. Generation of conditional Mef2cloxP/loxP mice for temporal- and tissue-specific analyses. Genesis 43, 43–48. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U, Barberis A, Benjamin LE, Makinen T, Nobes CD, Adams RH, 2010. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486. [DOI] [PubMed] [Google Scholar]
- Xu Z, Gong J, Maiti D, Vong L, Wu L, Schwarz JJ, Duh EJ, 2012. MEF2C ablation in endothelial cells reduces retinal vessel loss and suppresses pathologic retinal neovascularization in oxygen-induced retinopathy. The American journal of pathology 180, 2548–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]