SUMMARY
Cell fate transitions are essential for specification of stem cells and their niches, but the precise timing and sequence of molecular events during embryonic development are largely unknown. Here, we identify with 3D/4D microscopy unclustered precursors of dermal condensates (DC), signaling niches for epithelial progenitors in hair placodes. With population-based and single-cell transcriptomics, we define a molecular time-lapse from pre-DC fate specification through DC niche formation and establish the developmental trajectory as the DC lineage emerges from fibroblasts. Co-expression of downregulated fibroblast and upregulated DC genes in niche precursors reveals a transitory molecular state following a proliferation shutdown. Waves of transcription factor and signaling molecule expression then coincide with DC formation. Finally, ablation of epidermal Wnt signaling and placode-derived FGF20 demonstrates their requirement for pre-DC specification. These findings uncover a progenitor-dependent niche precursor fate and the transitory molecular events controlling niche formation and function.
Keywords: stem cell niche, dermal condensate precursors, dermal papilla, hair follicle morphogenesis, hair follicle formation, cell fate specification, placode progenitors, single cell
Graphical Abstract

eTOC/In Brief
Mok et al. identify niche precursors during progenitor/niche formation in mouse embryonic hair follicles. Population-based and single-cell transcriptomics show that unclustered dermal condensate precursors are at a transitional fibroblast-to-DC state marked by successive waves of transcription factor/signaling molecule expression. Niche fate acquisition is not “pre-programmed” and requires placode progenitor-derived FGF20.
INTRODUCTION
Many adult tissues are maintained during homeostasis or repaired after injury by resident stem cells as a resource of reserve cells with multilineage potential. Stem cell functions are controlled both intrinsically and by a large variety of long-range neuronal and hormonal inputs and short range signals from the microenvironment or niche (Ge and Fuchs, 2018; Heitman et al., 2018; Rezza et al., 2014). In many cases, signals are derived from specialized niche cells that control stem cell quiescence for long periods of time, regulate stem cell self-renewal, or guide their differentiation into one or more lineages of the respective tissue. Neighboring niche cell types and their signals have been identified in several tissues such as bone marrow (Wei and Frenette, 2018), intestine (Shoshkes-Carmel et al., 2018), brain (Falk and Götz, 2017), lung (Lee et al., 2017) and skin (Ge and Fuchs, 2018; Sennett and Rendl, 2012). But a detailed understanding of the molecular controls of niche fate specification during embryonic development remains limited. Equally unclear is the precise timing with which niche cells emerge relative to stem/progenitor cells and their mutual impact on each other.
In hair follicles, rapidly dividing progenitors in the proximal bulb region receive signals from the centrally-located dermal papilla (DP) niche, which controls hair follicle progenitor proliferation, migration and differentiation into several lineages of the hair shaft and its channel during continuous hair growth (Clavel et al., 2012; Sennett and Rendl, 2012). During the hair cycle after bulb degeneration and rest, the DP niche sends activating signals to the stem cells in the hair germ to regenerate a new hair bulb ( Ge and Fuchs, 2018). Many studies have investigated the developmental processes and morphogenetic events during embryonic hair follicle formation, including the emergence of stem cell and niche compartments (; Sennett and Rendl, 2012). Around embryonic day (E) 12.5, secreted epidermal Wnts activate broad dermal Wnt/β-catenin signaling (Chen et al., 2012). This is upstream of yet unidentified dermal signals that at ~E13.5 induce the fate specification of hair follicle progenitors in patterned pre-placodes (Figure 1A, hair follicle stage 0) (Paus et al., 1999) that can be molecularly identified by Dkk4 and Edar expression (Sennett and Rendl, 2012). Progenitor cell migration then forms the physically identifiable placode (Pc), (Figure 1A, stage 1) (Ahtiainen et al., 2014). Following this, Pc progenitors signal back to the dermis for formation of dermal condensates (DC) (Figure 1A, stage 1, DC1), specialized cell clusters derived from fibroblasts (Fb), which act as signaling niches for Pc progenitors to regulate continued hair follicle development (Sennett and Rendl, 2012; Sennett et al., 2015). Pc progenitors also give rise to suprabasal Sox9+ precursors of the future adult hair follicle stem cells in the bulge after completion of hair follicle morphogenesis (Ouspenskaia et al., 2016). Continued signal exchange between the DC and Pc leads to progenitor proliferation and downgrowth of the polarized hair germ (Figure 1A, stage 2, DC2), with the DC at the leading edge that, after an elongated hair peg stage, becomes engulfed by the progenitors to form the DP niche of the hair-shaft producing bulb (Grisanti et al., 2013; Sennett and Rendl, 2012). Several key signaling pathways have been identified for progenitor and niche formation (Sennett and Rendl, 2012). Wnt/β-catenin signaling is essential and most upstream for Pc (DasGupta and Fuchs, 1999; Huelsken et al., 2001) and condensate (Tsai et al., 2014) formation. Eda signaling is then required for maintaining Wnt signaling and Pc stabilization (Zhang et al., 2009). FGF20, another Wnt target, is a key Pc-derived signal required for DC niche formation (Huh et al., 2013) in a process involving cell aggregation (Biggs et al., 2018).
Figure 1. Identification of Unclustered Precursors of Dermal Condensates.
(A) Schematic depicting early key stages of hair follicle morphogenesis. Initiation of hair follicle placodes (Pc) precedes formation of the clustered dermal condensate (DC) niche. Whether DC fate specification occurs before aggregation is unclear.
(B) Immunofluorescence for Pc marker EDAR on a sagittal section of E15.0 Sox2GFP back skin. GFP high marks the DC of stage 1 (DC1, empty arrow) and stage 2 hair follicles (DC2, filled arrow). Note simultaneous detection of hair follicle stages 0, 1 and 2 at E15.0. Dotted line demarcates basement membrane. Red speckles in dermis are non-specific. DAPI marks all nuclei.
(C) Immunofluorescence whole mount staining for GFP (Sox2GFP reporter), Schwann cell marker ITGA6 and DC marker FOXD1 on E15.0 Sox2GFP back skin. Note parallel hair follicle stages in the low magnification confocal scan (top view). GFP high is in DC1 and DC2. DC2 shows anterior-posterior polarized downgrowth. GFP low marks a group of unclustered, FOXD1+ DC precursors (pre-DC, arrowhead). GFP low also marks the ITGA6+ Schwann cell network (asterisks).
(D) Magnification of inset from (C) for pre-DC (arrowhead) and DC1 (open arrow). Pre-DC are unclustered. DC1 shows clustered nuclei.
(E) Immunofluorescence whole mount staining for EDAR and SOX2 on E15.0 PdgfraH2BGFP back skin. Nuclear H2BGFP is in mesenchymal cells. Z-sections from a 3D-confocal scan are shown at the epidermal and dermal level. SOX2+ unclustered pre-DC and DC1 cells are underneath EDAR+ placodes and are Fb-type, expressing mesenchymal H2BGFP. Z values are section # of total optical sections in scan from epidermis into dermis. Section intervals = 1 µm.
(F) 3D reconstructed images of pre-DC and DC1 generated by Imaris using the confocal scans in (E).
(G) Quantification of cell density of pre-DC, DC1 and DC2. Average distances of nearest 5 neighboring nuclei for each pre-DC, DC1 and DC2 were measured in 3D reconstructions in (F) and Figure S1E. n = 3 for pre-DC, DC1 or DC2. Data are mean ± SD from two embryos. *p < 0.01.
The morphological detection of the DC after Pc initiation (Chen et al., 2012), combined with discovery of DC markers (Chen et al., 2012; Sennett and Rendl, 2012; Huh et al., 2013) and the definition of a DC gene expression signature (Sennett et al., 2015) has led to the current model of niche fate specification in response to progenitor signal(s) that coincides with condensate formation. However, the identification of unclustered dermal cells with localized high Wnt activity beneath Pcs (Zhang et al., 2009), and low-level expression of DC genes in immunophenotypically non-DC dermal cells (Sennett et al., 2015) raise the possibility of DC fate acquisition prior to cluster formation. Contextualizing these early events in hair follicle formation is further complicated by the rapidity with which morphogenesis occurs; Wnt signaling activity appears virtually simultaneously in Pc and DC (DasGupta and Fuchs, 1999; Tsai et al., 2014; Zhang et al., 2009), rendering parsing out the precise order and timing difficult. Recent work also demonstrated direct mesenchymal self-organization ability in the absence of an epidermal pre-pattern (Glover et al., 2017) further calling into question the origin and timing of niche fate specification (Figure 1A): 1) is the niche fate specified before niche cluster formation and signaling function and 2) how is the timing of niche fate acquisition related to progenitor fate and signaling?
Here, by combining 3D and 4D time-lapse live imaging with fluorescence-activated cell sorting, bulk and single-cell transcriptomics, proliferation kinetics, and mutant mouse models, we identify fated unclustered precursors of the DC niche prior to signaling center formation. Based on a molecular time-lapse of dynamically changing niche gene signatures and ordering the cell fates in developmental pseudotime we define the cell fate trajectory from Fb through DC niche precursors that are fated to become the clustered niche. In this process, DC precursors (pre-DC) undergo a transitional molecular state that is consolidated towards the DC niche biological fate through waves of transcription factor and signaling molecule upregulation. Finally, with epidermal Wnt signaling ablation, which prevents Pc formation, and with deletion of Pc-derived FGF20, which precludes DC aggregation, we demonstrate that early niche fate and emergence of unclustered DC precursors depends on pre-existing Pc progenitors and their signals.
RESULTS
Identification of Unclustered Precursors of Dermal Condensates
Previous studies have demonstrated that cell aggregation is the cellular mechanism inciting DC formation (Biggs et al., 2018; Glover et al., 2017), but when the DC niche fate is acquired and how this timing relates to placode progenitor specification remains unknown. To identify potential DC-fated precursors before cluster formation and niche establishment, we followed up on our previous work where we isolated with fluorescence-activated cell sorting (FACS) the DC as GFP+ cells from embryonic Sox2GFP knock-in reporter skin (Figure S1A) (Sennett et al., 2015). Both Sox2 promoter-driven GFP (Figures 1B and S1A) and Sox2 mRNA (Figure S1B) are highly expressed in the established DC (Driskell et al., 2009; Clavel et al., 2012), where condensed cells are visible underneath flat and barely invaginated EDAR+ hair placodes of stage 1 hair follicles (Figure 1B, open arrow) (Paus et al., 1999; Sennett et al., 2015). GFP levels remain high after initial Pc downgrowth and polarization of stage 2 hair follicle germs (Figure 1B, arrow). We named these two DC stages with high Sox2 expression DC1 and DC2 accordingly. We also observed by flow cytometry a low-level GFP population that was a mix of Schwann and DC-like cells (Figure S1A, S1B and S1C) (Sennett et al., 2015), but the difficulty of detecting low Sox2GFP levels in sagittal skin sections and absence of know-how to separately sort DC-like cells precluded further resolving this population (Figure 1B).
We surmised that low Sox2GFP-expressing DC-like cells could represent a precursor state that ramps up Sox2 expression towards a DC fate. To detect GFP low cells with high sensitivity we performed multicolor immunofluorescence in whole mount skin of E15.0 Sox2GFP embryos. In the top view, the anti-GFP signal was strong in DC1 and DC2 (Figure 1C, arrows). Also, low GFP levels were now detectable in Schwann cells (Figure 1C, asterisks), highlighted by ITGA6, forming a GFP low/ITGA6+ network within the developing skin (Figure 1C, asterisks). Importantly, we also found few loosely arranged GFP low/ITGA6− non-Schwann cells in the dermis in a location fitting a hair follicle pattern (Figure 1C, arrowhead). Co-labeling with previously identified DC marker FOXD1 (Figure S1C) (Sennett et al., 2015) labeled DC1, DC2 and these GFP low cells, suggesting that they might be DC precursors (pre-DC) before DC niche formation. High magnification images suggested that potential pre-DC cells are still unclustered, unlike the clearly visible cell aggregations of DC1 stage hair follicles (Figure 1D).
To demonstrate that potential pre-DC cells are associated with hair follicles we next performed whole mount immunofluorescence for SOX2 and EDAR on E15.0 PdgfraH2BGFP reporter back skin. Pdgfra is highly expressed in Fb-type mesenchymal cells (Figure S1C) (Hamilton et al., 2003), which are marked by nuclear GFP. In optical slices of 3D confocal scans, SOX2+ pre-DC cells at the dermal level were underneath EDAR+ Pc at the epidermal level and appeared to be unclustered compared to aggregated DC1 and DC2 (Figures 1E and S1D). Overlap of SOX2 with nuclear H2BGFP demonstrated the Fb-type of potential pre-DC cells. 3D reconstructions further illustrated the unclustered state of pre-DC cells (Figure S1E, Supplemental Movies 1–3). Z-projections demonstrated the close proximity of pre-DC to the overlying Pc (Figure 1F). Finally, to confirm that pre-DC cells are truly unclustered we measured, in 3D, the distance of each SOX2 and H2BGFP double-labeled nucleus to its five nearest neighbors for pre-DC, DC1 and DC2 (Figure 1G). Cells in DC2 clusters were significantly closer to each other compared to all others. Pre-DC cells, however, were significantly further apart compared to those of DC1 and DC2, confirming their loose initial arrangement. As other H2BGFP+ Fb were interspersed between pre-DC cells (Figure 1E), it suggests that early fate acquisition does not have to be synchronized among direct neighbors. In summary, our data thus far identified the existence of uncondensed potential DC precursor cells suggesting that the DC fate is specified prior to DC formation and function.
DC Precursor Cells Are in a Transitional Transcriptional State Towards a DC Niche Fate
After identifying potential DC precursors in stage 0 hair follicles with SOX2 protein, GFP reporter, and the DC signature gene FOXD1, we next sought to isolate this population for exploring the molecular controls of DC fate acquisition and its niche functions. Furthermore, the simultaneous presence of pre-DC, DC1 and DC2 from stage 0, stage 1 and 2 hair follicles, respectively, in E15.0 back skin provided us with an excellent opportunity to compare multiple parallel developmental stages in isolated cells. To separately purify Sox2GFP high DC1 and DC2 cells we utilized the previously identified DC marker CXCR4 (Sennett et al., 2014) that is highly expressed in DC2, has lower levels in DC1, and is absent in pre-DC (Figure 2A and 2B). To separately isolate pre-DC and Schwann cells, which are both Sox2GFP low and CXCR4−, we labeled pre-DC for mesenchymal marker PDGFRA and Schwann cells for ITGA6 to isolate them as Sox2GFPlow/CXCR4−/PDGFRA+/ITGA6− and Sox2GFPlow/CXCR4−/PDGFRA−/ITGA6+ populations, respectively. Additionally, we sorted dermal Fb as PDGFRA+/Sox2GFP- cells and PDGFRA−/Sox2GFP- cells as a negative (Neg) population containing all other remaining skin cells (Figure 2B). To confirm Fb and DC enrichment we performed qRT-PCR of known marker genes (Figure 2C). As expected, Lum was highly enriched in Fb, while Cxcr4 was absent in pre-DC and detected at increasing levels in DC1 and DC2; Sox2 expression was low in both pre-DC and Schwann cells and high in DC1 and DC2. Importantly, Schwann cell marker Fabp7 showed strong enrichment in Schwann cells and not in pre-DC (Figure S2A), and all other Fb/DC markers were also absent (Figure 2C). Finally, Foxd1 was highly expressed at all three DC stages, and not in Fb or Schwann cells, confirming successful isolation of pre-DC cells in agreement with our 3D imaging analysis.
Figure 2. DC Precursor Cells Are in a Transitional Transcriptional State Towards a DC Niche Fate.
(A) Immunofluorescence for DC marker CXCR4 and Pc marker EDAR in E15.0 back skin. CXCR4 is expressed at higher levels in DC2 (filled arrow) compared to DC1 (open arrow). Dotted line marks basement membrane. DAPI marks all nuclei.
(B) FACS isolation of pre-DC, DC1, DC2, Fb, Schwann cells (Sch), and a population of negative cells (Neg) from live, EpCAM− cells.
(C) qRT-PCR verification of marker genes in isolated Fb, pre-DC, DC1, DC2 and Schwann cells.
(D) Venn diagram of gene signatures in Fb, pre-DC and two DC stages.
(E) All genes are enriched compared to Sch and Neg. Overlaps represent commonly enriched genes in corresponding populations compared to all others. Blue outlines increased overlap between Fb/pre-DC, compared to Fb/DC1 or Fb/DC2. Red outlines overlap of pre-DC with mature DC stages. Transitional transcriptional state of pre-DC. Left: Z-score normalized expression of Fb/pre-DC overlap genes (blue) in Fb, pre-DC, DC1, and DC2 reveal progressive downregulation of Fb genes. Right: Z-score normalized expression of overlap of pre-DC/DC1/DC2 (red) and 58 established DC signature genes shows increased DC gene expression (Sennett et al., 2015). Pre-DC has intermediate levels for both Fb and DC genes.
(F and G) FPKM barplots of representative transitional genes.
(H) Experimental strategy for combined single-cell sorting and single-cell transcriptome analysis by SORT-seq. Fb, pre-DC, DC1 and DC2 were FACS-purified as in (A). Each index-sorted cell is connected to its transcriptome through the plate position.
(I) tSNE map of all sorted cells color-coded by cell type.
(J) Expression of representative DC signature genes projected onto the tSNE map from (I). Transcript counts are in linear scale.
(K) Expression of representative Fb signature genes projected onto the tSNE map from (I).
(L) Expression of representative transitional Fb and DC genes from the Fb/pre-DC and pre-DC/DC1/DC2 overlaps projected onto the tSNE map from (I).
We then proceeded with population-based transcriptome analysis for gene signature discovery by bulk RNA-sequencing (RNA-seq), as previously described (Sennett et al., 2015). We first globally compared the transcriptomes by principal component analysis of the top variably expressed genes (Figure S2B) and hierarchical clustering (Figure S2C), confirming the similarities between DC1 and DC2 and establishing multiple co-regulated gene groups within all six populations. Interestingly, pre-DC was positioned between Fb and DC1 suggesting pre-DC dually share characteristics of both dermal Fb and clustered DC. We next calculated significantly differentially expressed genes (DEGs) in each population by ANOVA (false discovery rate<0.05) and compared the resulting total of 6,454 genes. We established gene signatures using significant DEGs and selecting for genes with FPKM>1 and an expression fold change>2, resulting in 1,890 signature genes for all six cell types. To focus on the potential pre-DC and their relationship to Fb and the DC niche, we analyzed gene expression relationships in a four-way Venn diagram analysis of all significantly increased genes in Fb, pre-DC, DC1, and DC2 compared to Sch and Neg populations (Figure 2D and Table S1). A total of 1,157 genes were enriched in the four mesenchymal populations, including 692 population-unique signature genes. However, we also found several interesting population overlaps. As the DP and its embryonic DC predecessor are Fb-derived mesenchymal cells (Driskell et al., 2013), it was revealing that the pre-DC overlap with Fb (28 genes) was larger than both the unique overlaps of Fb with DC1 (1 gene) and with DC2 (5 genes) (Figure 2D, blue), suggesting that pre-DC shares transcriptional similarity with Fbs, fitting of a Fb-to-DC trajectory. Indeed, exploring gene expression shared between Fb and pre-DC demonstrated a transition of high-to-low expression from Fb to DC with intermediate levels in pre-DC (Figure 2E). Several genes included extracellular matrix components such as Ccbe1 and Eln (Figure 2F). Conversely, differential expression analysis confirmed a high overlap of 119 genes between pre-DC, DC1, and DC2 (Figure 2D, red), which included 58 previously described DC signature genes (Sennett et al., 2015). Also here, pre-DC expressed many of these shared DC signature genes at intermediate levels between Fb and DC1/DC2, suggestive of its developmental precursor status to morphologically identifiable DC (Figures 2E and 2G).
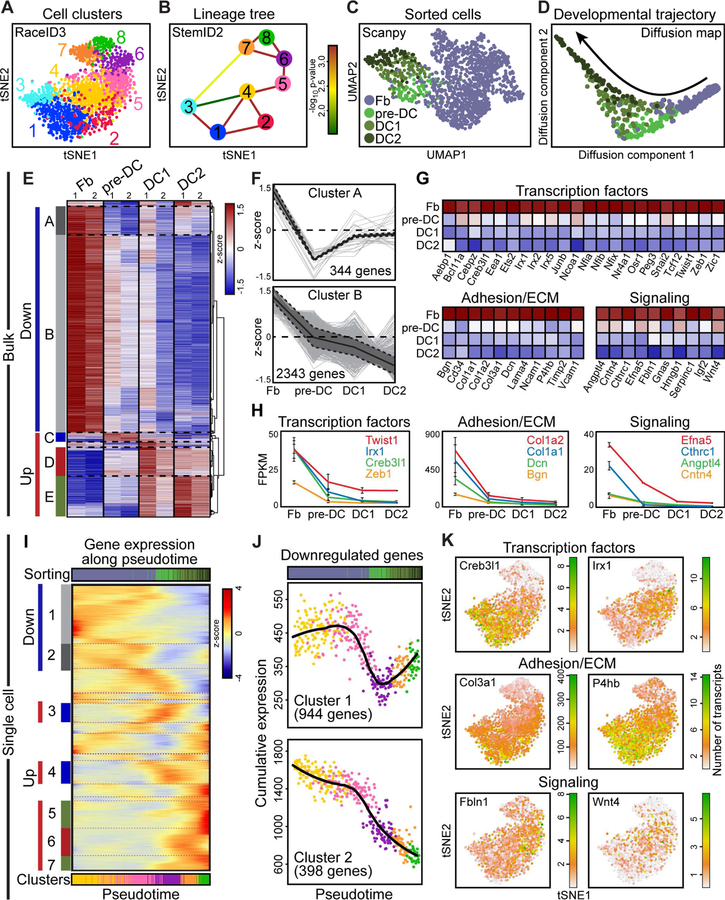
We next complemented population-based bulk RNA-sequencing with single-cell transcriptome analyses to allow for exploration of minute cell-to-cell transcriptional changes that define cell heterogeneity and differentiation (Grün et al., 201 6). Importantly, it also enables a high-resolution definition of lineage relationships along a developmental trajectory (Herman et al., 2018). To corroborate the developmental link of pre-DC between Fb and DC1/DC2, we used SORT-Seq (Muraro et al., 2016) to index-sort DC cells with our established strategy and obtain single-cell transcriptomes that are linked to the isolated cell identities (Figure 2H). We sequenced 1,371 Fb, 149 pre-DC, 152 DC1, and 151 DC2. Of those, 1,076 Fb, 107 pre-DC, 118 DC1, and 82 DC2 were used for analysis based on a minimum requirement of 6,000 transcripts/cell and established quality controls to exclude poorly sequenced cells. Based on population frequencies observed by flow cytometry it would require ~15,000 total unenriched cells to obtain similar transcriptome information of our sorted pre-DC cells (150x enrichment). We then applied the Rare Cell Type Identification 3 (RaceID3) clustering algorithm that was previously used to identify rare intestinal cell types and lineage relationships between stem cells and differentiated cells (Grün et al., 2016; Herman et al., 2018). RaceID3 clustered gene expression similarities were then visualized using t-distributed stochastic neighbor embedding (tSNE). Superimposing the Fb and DC cell identities onto the tSNE plot revealed clear distinctions between all isolated populations confirming our cell isolation strategy (Figure 2I). At the same time, it highlighted similarities between all DC stages and placed pre-DC between Fbs and DC1/DC2, similarly suggesting a lineage trajectory from Fb to DC through an intermediate pre-DC fate transition. As expected, all sorted DC populations expressed established DC signature genes, including Fgf10, Sox2, Foxd1, and Tbx18 (Grisanti et al., 2013; Sennett et al., 2015) (Figure 2J), and sorted Fb expressed established markers Col1a1, Lrig1, Dcn, and Lum (Driskell et al., 2013) (Figure 2K). Also here, expression patterns revealed gradual downregulation of Fb genes and upregulation of DC genes (Figure 2L). Altogether, these bulk and single-cell transcriptome data from purified embryonic skin cells indicate that unclustered pre-DC precursors are at a transcriptionally transitional state from Fb towards a permanent DC niche fate.
DC Precursors Are Developmental Intermediates Between Fibroblasts and the Mature DC Niche
As the identified pre-DC cells express genes characteristic of both Fbs and DC, we hypothesized that they are precursors of the mature DC niche at an intermediate developmental state between Fb and DC. To explore this lineage trajectory, we utilized RaceID3 to identify distinct cell clusters with similar transcriptome as a basis for calculating lineage relationships between related cell types (Figure 3A). Clusters 6, 7 and 8 were primarily comprised of sorted pre-DC, DC1 and DC2 cells, respectively (Figures 2H, 3A and S3A), again validating our sorting strategy. We then applied the Stem Cell Identification (StemID2) algorithm (Grün et al., 2016) to order RaceID3-identified clusters based on degree of transcriptional similarity (Figure 3B) inferring differentiation trajectories from single-cell RNA-sequencing data (Grün et al., 2016). Amongst DC clusters (clusters 6, 7, 8), pre-DC-enriched cluster 6 was closest to the Fb clusters. Amongst sorted Fb, cluster 5 was the sole calculated link to pre-DC cluster 6, suggesting that pre-DC cells emerged from closely related Fb (Figure 3B). It also demonstrated that pre-DC are the link for transition into DC1/DC2. To independently confirm the developmental trajectory, we utilized the Scanpy toolkit for Louvain clustering and diffusion maps (Wolf et al., 2018). Louvain clustering with the same 1,383 cells used for RaceID3 clustering, showed again clear distinctions between all sorted populations, placing pre-DC between Fb and DC1 (Figures 3C and S3B). Importantly, applying diffusion pseudotime, a method for ordering cells along lineage trajectories (Haghverdi et al., 2016), clearly demonstrated the lineage path from nearest Fb (Louvain cluster 5) through pre-DC towards DC1 and DC2 (Figures 3D, S3C).
Figure 3. DC Precursors are Developmental Intermediates Between Fibroblasts and The Mature DC Niche.
(A) tSNE map from Figure 2I color-coded according to RaceID3 k-medoids clustering.
(B) StemID2 lineage tree of RaceID3 k-medoids clustering output. Color of the line connecting clusters indicates p-value of link.
(C) UMAP plot of Louvain clustering (Scanpy) of Fb, pre-DC, DC1, DC2. Sorted cell identities are color-coded by cell type.
(D) Diffusion map highlighting the developmental trajectory from Fb through pre-DC to DC2 and color-coded by cell type.
(E) Trend analysis by Spearman rank clustering of differentially expressed genes from Fb to pre-DC, DC1 and DC2 in bulk RNA-seq.
(F) Expression trajectories of downregulated genes from Fb to pre-DC. Solid black lines are average z-score flanked by dotted black lines representing SD.
(G) Heatmaps of relative expression of downregulated transcription factors, adhesion/extracellular matrix, and signaling genes across the DC developmental trajectory.
(H) Absolute FPKM expression levels of representative genes from each category in (E).
(I) Self organizing map of co-regulated gene clusters across the plotted pseudotime of ordered individual cells based on the gradual transition of their transcriptomes. Bottom bar represents ordered cells color-coded by RaceID3 cluster in (A). Top bar highlights ordered cells based on sorted identities in (C).
(J) Cumulative expression plots of downregulated genes along pseudotime. Each dot represents the cumulative expression of all genes per cell in a given cluster from (I). The black line is the running average across pseudotime.
(K) Expression of representative transcription factors, adhesion/ECM molecules, and signaling factors from gene clusters 1 and 2 projected onto the tSNE map from Figure 2I.
Having established by single-cell RNA-seq analyses the differentiation trajectory from Fb to DC2, we next re-examined our bulk RNA-sequencing for gene expression trends. For this we performed hierarchical clustering noting four major trends: genes with highest expression in Fb (down A and B), and genes with highest expression in pre-DC (up C), DC1 (up D) and DC2 (up E) (Figure 3E and Table S2). 2,687 genes were downregulated from Fbs through DC2 (Figure 3F). Closer examination of these genes revealed major downregulation of Fb transcription factors, adhesion/ECM molecules, and signaling factors (Figures 3G and 3H) suggesting their involvement in rapid and sustained loss of Fb function as the DC fate is acquired and throughout DC differentiation.
Finally, we utilized our single-cell transcriptome data to define the pseudotemporal ordering of Fb and DC populations along the differentiation trajectory and explore patterns of gene regulation during niche fate acquisition. For this, we devised a self-organizing map using the Fb clusters closest to DC and the DC-enriched clusters (Figure 3I). We excluded Dlk1-expressing Fb clusters 1–3 that likely represented lower dermal Fb unrelated to DC (Figure S3D) (Driskell et al., 2013), and included Fb clusters 4 and 5 as both were closest to pre-DC-enriched cluster 6 (Figure S3E). Pseudotime gene expression analysis revealed refined trends of genes with highest expression in Fb (down clusters 1, 2), pre-DC (up clusters 3, 4), DC1 (up cluster 6), and DC2 (up clusters 5, 7) (Figure 3I and Table S3). Amongst 2,687 genes downregulated from Fb through DC2 in population-based RNA-seq (Figure 3F) and the 1,342 genes identified in single-cell transcriptomes (Figure 3J), 951 genes were in common (35% of bulk-identified targets, 70% of single cell-identified targets) (Figure S3F). Also here, these included several transcription factors (Lambert et al., 2018), adhesion and ECM components, and signaling molecules (Ramilowski et al., 2015) necessary for Fb function (Figure 3K). Creb3l1 is a secreted factor essential for collagen metabolism (Keller et al., 2018), while Irx1 and Twist1 are transcription factors that control expression of matrix-stiffening components (García-Palmero et al., 2016), and Zeb1 is known for promoting expression of vimentin (Liu et al., 2008). Additionally, expression of numerous collagens such as Col1a1, Col1a2, and Col3a1and other ECM components such as Dcn and Bgn, and P4hb were decreased between Fb and DC populations. Finally, we identified signaling molecules that are uniquely expressed in Fb, such as Efna5, Cntn4, Fbln1, and Wnt4 that are all downregulated on the path to DC lineage differentiation. Altogether these data demonstrate that Fb quickly halt transcription of Fb-associated genes as they transition through the pre-DC intermediate into the definitive DC niche fate.
Loss of Proliferation is Concomitant with Acquisition of DC Fate
It has been well established that niche cells in mature dermal papillae are quiescent and rarely divide (Tobin et al., 2003). Also the aggregated DC, as the embryonic DP precursor (Grisanti et al., 2013), was recently shown to be non-proliferative (Biggs et al., 2018). Therefore, we next explored in the unclustered DC precursors the precise timing of cell-cycle exit with respect to niche fate acquisition. As predicted, many cell cycle genes involved in molecular control of cell proliferation were downregulated from Fb to DC2 (Figure 4A). Interestingly, gene set enrichment analysis revealed a significant correlation of proliferation-associated genes in Fb compared to pre-DC, suggesting that exit of proliferation could be as early as during pre-DC fate acquisition (Figure 4B). Indeed, in pseudotime-ordered cells rapid declining expression of pro-proliferative genes such as Rbbp7, Mki67, and Pcna coincided with sharp upregulation of cell-cycle inhibitors, such as Cdkn1a and Btg1 (Zhu et al., 2013). Importantly, both events occurred prior to emergence of pre-DC and remained sustained through DC2 (Figures 4C and 4D), suggesting that cell cycle exit is a feature of pre-DC fate acquisition.
Figure 4. Loss of Proliferation is Concomitant with Acquisition of DC Fate.
(A) Hierarchical clustering of cell proliferation gene expression in bulk-isolated Fb, pre-DC, DC1, and DC2.
(B) Cell cycle genes are shut down from Fb to pre-DC through DC2. Gene Set Enrichment Analysis for cell proliferation genes shows positive correlation for Fb and negative correlation for pre-DC.
(C) Normalized running means of expressed transcript numbers in pseudotime ordered cells for pro-proliferative (Rbbp7, Mki67, Pcna) and anti-proliferative (Cdkn1a, Btg1) genes.
(D) Expression of pro-proliferative and anti-proliferative genes projected onto the tSNE map from Figure 2I.
(E) EdU pulse-chase assay for proliferation rates in Fb, pre-DC, DC1 and DC2. EdU was injected 6h, 12h, or 24h prior to harvest and analysis of E15.0 Sox2GFP skin by immunofluorescence for GFP, ITGA6 and EDAR. Representative z-sections from confocal scans are shown for interfollicular Fb and pre-DC, DC1, DC2. Examples of EdU+ proliferating cells are highlighted by empty arrowheads.
(F) Quantification of EdU+ Fb, pre-DC, DC1 and DC2. Note drastic drop of proliferation rate at 6h time point from Fb to DC precursors, sustained through DC2. n = 3 interfollicular Fb areas, pre-DC, DC1 and DC2. Data are mean ± SD from two embryos. *p < 0.05, **p < 0.01.
(G) Representative stills from time-lapse live imaging in Tbx18H2BGFP embryonic skin for detection of cell divisions. Pre-DC was identified as H2BGFP high cells after transitioning from Fbs and backtracked in time for 6 hours. Fbs were H2BGFP low. Note absence of cell divisions in pre-DC through 6 hours before fate acquisition. Two examples for mitotic cell divisions in Fb are pseudo-colored in yellow or blue.
(H) Quantification of mitotic cells during 6-hour tracking. n = 2. Data are mean ± SD from two embryos. **p < 0.01.
See also Figure S4.
To independently test when proliferation exit occurs relative to DC fate acquisition, we performed EdU proliferation assays to mark proliferating cells during S-phase of the cell cycle (Figures 4E and S4A). EdU injection 6 hours prior to E15.0 analysis showed that 33% of Fb were proliferating (Figure 4F). By contrast, < 3% of DC1 and DC2 were going through S-phase suggesting that the vast majority of maturing DC cells had exited the cell cycle. Notably, only 3% of pre-DC incorporated EdU suggesting that already during early fate acquisition precursors shut down proliferation. Analyzing 12- and 24-hour chases showed increased EdU incorporation in pre-DC and DC1 fated cells, likely reflecting their developmental history from proliferating Fb at the time of EdU injection. Interestingly, with the 12-hour EdU chase DC2 had significantly lower proliferating cells than in pre-DC and DC1, and as low as 6h levels. Given that DC1 develops into DC2 during the 12-hour chase, these results suggest that DC1 progression to DC2 does not require cell proliferation.
Finally, to confirm that exit of proliferation is already occurring during DC fate acquisition before pre-DC detection, we tracked with 4D time-lapse live imaging the cellular processes up to pre-DC fate specification (Figure 4G). We live imaged embryonic ex vivo back skins from Tbx18H2BGFP mice that express high nuclear GFP levels in the DC (Grisanti et al., 2013) (Figure S4B). Importantly, pre-DC cells also already expressed high nuclear GFP, while Fb exhibited low GFP levels (Figure 4G). Live-tracking cell divisions during a 6-hour period prior to pre-DC fate acquisition detected actively proliferating interfollicular Fb (Figure 4G and 4H). By contrast, no pre-DC cells were dividing during the same time span, indicating that proliferation exit is occurring during DC fate acquisition in the Fb-to-pre-DC transition.
Three Distinct Waves of Transcription Factor and Signaling Molecule Upregulation Occur Along the DC Niche Developmental Trajectory
Besides proliferation shutdown and gradual loss of Fb gene expression signature, gradual upregulation of the DC niche signature occurs during DC fate acquisition (Figures 2E, 2G, 2L, 3E and 3I). We next explored the precise timing of molecular fate acquisition and consolidation from Fb to pre-DC, through DC1 and DC2. We discovered three distinct waves of gene upregulation for pre-DC fate acquisition, DC1 clustering, and hair germ downgrowth stages at DC2, in both bulk (Figure 5A) and single-cell (Figure 5C and Table S3) transcriptional analyses. The first wave was evident in gene clusters whose transient expression peaked early at pre-DC and then dropped, which likely includes factors necessary for fate acquisition and other early functions. Two additional waves of co-regulated gene clusters had sustained expression through DC2, but differed only in when their expression peaked: mid genes were enriched in genes achieving maximal expression at DC1, and late genes reached maximal expression in DC2, at the end of the pseudotime. Of the three upregulated clusters, the mid cluster contained the most heterogeneity in expression dynamics, and included genes with gradually ramped up expression beginning in pre-DC that is sustained throughout, and genes with more delayed upregulation patterns.
Figure 5. Three Distinct Waves of Transcription Factor and Signaling Molecule Upregulation Occur Along the DC Niche Developmental Trajectory.
(A) Bulk RNA-seq expression trajectories of up-regulated genes from Fb to DC2 comprising Spearman rank clustered groups C (early, pre-DC peak), D (mid, DC1 plateau), and E (late, max at DC2) from Figure 3E.
(B) Heatmap of relative expression (z-score) of transcription factors in groups C, D, and E (blue, red, green bars) from Figure 3E.
(C) Cumulative expression plots of early, mid and late upregulated genes along pseudotime from Figure 3I. Each dot represents the cumulative expression of all genes per cell in a given cluster. The black line is the running average across pseudotime.
(D) Left panel: Expression of early upregulated transcription factors projected onto the tSNE map from Figure 2I. Right panel: Parallel in situ hybridization for Twist2 and immunofluorescence for anti-GFP staining. Note Twist2 expression in pre-DC, which is downregulated in DC1. DAPI marks all nuclei.
(E) Expression of mid and late upregulated transcription factors projected onto the tSNE map from Figure 2I.
(F) Normalized running means of expressed transcript numbers in pseudotime ordered cells. Transcription factors are upregulated in three waves during specification of DC precursors towards the mature DC niche fate.
(G) Cumulative percent of sum of upregulated expression of 51 transcription factors in early, mid, and late waves. Means and range of cumulative percent in each transcription factor wave are represented by thin solid and thick faded lines, respectively.
(H) Heatmap of relative expression (z-score) of signaling-related genes in groups C, D, and E (blue, red, green bars).
(I) Expression of representative upregulated signaling molecules projected onto the tSNE map from Figure 2I.
(J) Normalized running means of expressed transcript numbers in pseudotime ordered cells. Signaling molecules are upregulated in three waves early during specification of DC precursors and consolidation of DC niche fate.
(K) Cumulative percent of sum of upregulated expression of 43 signaling molecules in early, mid, and late waves. Means and range of cumulative percent in each transcription factor wave are represented by thin solid and thick faded lines, respectively.
All three waves contained many transcription factors that could represent potential master regulators for DC specification and development (Figure 5B–5G and Table S4). Transcription factors expressed in the early wave included known key fate regulators such as Twist2 (Dermo1) (Li et al., 1995), the Wnt signaling transcription factor Lef1, and the TGF-β effector Smad3 (Figures 5B, 5D, 5F and S5A). Twist2 knock-out animals have sparse hair, among many other abnormalities (Šošić et al., 2003). Similarly, Lef1 knock-out results in abrogation of whiskers and back skin hair follicles (van Genderen et al., 1994). We confirmed Twist2 mRNA expression in pre-DC by in-situ hybridization, which is later shut down at the DC1 stage (Figure 5D). Many more transcription factors were in the mid gene cluster (Figures 5E, 5F and S5B). Mid-expressing transcription factors include Trps1, Tshz1, Foxd1, Prdm1, and Sox18. Mutations in Trps1 are associated with Ambras Syndrome, a hypertrichosis disorder (Fantauzzo et al., 2008). Mutations in Sox18 have been shown to impact differentiation of adult dermal papillae (Villani et al., 2017). Finally, we resolved many transcription factors whose expression peaked in DC2, including Notch target genes Hey1 and Hes5, as well as Glis2, Foxp1, and Alx4 (Figures 5E, 5F and S5C). Foxp1 has noted functions in maintaining hair follicle stem cell quiescence (Leishman et al., 2013). In total, we discovered 11 early-wave, 16 mid-wave, and 24 late-wave transcription factors (Figure 5G) that followed these temporally-restricted upregulation patterns along pseudotime, suggesting that a coordinated array of molecular changes is required for pre-DC to mature through DC1 and DC2.
As the dermal papilla acts as a major signaling niche for epithelial progenitors during postnatal hair growth and for stem cells in cycling adult hair follicles, we finally explored the expression and upregulation of signaling molecules during DC fate specification and niche formation. Also here, we discovered a distinct pattern of signaling waves (Figures 5H–5K and Table S4). Signaling factors expressed in the short time window around pre-DC include the Wnt inhibitor Dkk1, Cyr61, and Fst (Figures 5I, 5J and S5D). Cyr61 is dynamically expressed in wound healing to reduce fibrosis (Jun and Lau, 2010). Fst knock-out is associated with abrogated hair follicle development (Nakamura et al., 2003). The mid-acting signaling molecules included well known regulators Fgf10, the TGF-β modulator Ltbp1, and Sema6a (Figures 5I, 5J and S5E); late-acting signaling molecules included key ligands such as Igfbp4, Inhba, and Rspo3 (Figure 5I, 5J and S5F) – many of which continue to be expressed in dermal papillae (Rezza et al., 2016). The global display, as cumulative percent of total, corroborates a resolution of 8 early, 11 mid, and 24 late upregulated signaling factors (Figure 5K). Overall these data suggest that DC development, from fate acquisition in a subset of Fb through aggregated DC1 and DC2, is accompanied by three distinct waves of transcription factor and signaling molecule expression that in concert are likely regulating the specification of DC fate and initiation of niche function.
DC Cell Fate Specification Requires Signals from Pre-existing Placodes
Our data suggest that the transition to pre-DC fate from Fb and to niche formation is coordinated by waves of transcription factor and signaling molecule upregulation. Resolving whether this is a pre-programmed cell-autonomous process or whether it relies on intercompartmental signals from pre-existing epithelial placode progenitors directly establishes the developmental hierarchy of progenitor and niche fate specification. Wnt/β-catenin signaling in the epidermis is essential for Pc formation and hair follicle morphogenesis, including the formation of the mature DC (Huelsken et al., 2001; Zhang et al., 2009).We next asked whether the early pre-DC fate, which developmentally precedes DC1 and DC2, is established in the absence of Pc progenitors. To this end, we blocked Pc formation by conditional Wnt/β-catenin signaling ablation in embryonic epidermis of K14-Cre;β-cateninfl/fl mice. In E14.5 control skin we confirmed the developmental stages of SOX2+ ITGA6− DCs underneath EDAR+ Pcs, including pre-DC (Figures 6A and S6A). Additional double immunofluorescence with FOXD1 confirmed pre-DC cell identity (Figure S6B). In β-catenin-ablated skin, as predicted, EDAR+ Pc and SOX2+ ITGA6− mature DCs were absent (Figures 6B, S6A). Intriguingly, in the absence of Pc we also failed to detect groups of unclustered pre-DC, by either SOX2+/ITGA6− (Figure 6B, S6A and S6C) or SOX2+/FOXD1+/ITGA6− (Figure S6B) immunofluorescence, indicating that fate acquisition of pre-DC cells was entirely abolished. These data demonstrate that DC cell fate specification from Fb requires the presence of placode progenitors that likely provide essential Pc-derived signals.
Figure 6. DC Cell Fate Specification Requires Pre-existing Placodes and Progenitor-Derived Fgf20.
(A, B) Ablation of Pc formation in β-catenin conditional knock out (cKO) epidermis. Immunofluorescence whole mount staining for EDAR, ITGA6 and SOX2. Z-sections from a 3D-confocal scan are shown at the epidermal and dermal level. Z values are section # of total optical sections in scan from epidermis into dermis. Section intervals = 1 µm. (A) Pre-DC (arrowhead) are specified underneath EDAR+ placodes in E14.5 β-cateninfl/fl control embryos. (B) Pre-DC fail to become specified in K14-Cre;β-cateninfl/fl cKO skins.
(C, D) Ablation of DC fate specification in Fgf20 knock out mice. Immunofluorescence whole mount staining for GFP, ITGA6 and SOX2. (C) Pre-DC (arrowhead) are specified underneath GFP+ placodes in E14.5 Fgf20Cre-GFP/+ control embryos. GFP is expressed in Pc progenitors. (D) Pre-DC fail to become specified in Fgf20Cre-GFP/LacZ KO knockout skins. Note Fgf20 promoter-driven GFP is still expressed in KO Pcs.
See also Figure S6.
Placode Progenitor-Derived FGF20 Is Required for DC Cell Fate Specification
A placode-derived signal potentially important for specifying the DC precursor fate is FGF20, which is both a well-known downstream target of Wnt/β-catenin signaling and also required for formation of the mature DC (Huh et al., 2013). Recent studies have further demonstrated that FGF signaling is required for cell migration to form mature DC clusters, however it is unclear whether it regulates DC fate acquisition itself (Biggs et al., 2018; Glover et al., 2017). Having discovered unclustered DC precursors prior to formation of mature DC, we next asked whether Fgf20 is required for induction of the pre-DC fate. To this end, we generated Fgf20-ablated embryos by crossing viable and fertile Fgf20Cre-GFP/Cre-GFP knockout mice with Fgf20LacZ/+ mice (Huh et al., 2012, 2015). In E14.5 heterozygous Fgf20Cre-GFP/+ control skins, we observed normal hair follicle development with multiple DC stages underneath GFP+ Pc (Figures 6C and S6D). In Fgf20 knockout skins of Fgf20Cre-GFP/LacZ embryos aggregated mature DCs were absent underneath GFP+ Pc, as described previously (Figures 6D and S6D) (Huh et al., 2013). Intriguingly, here like in the Wnt/β-catenin signaling ablation above, SOX2+/ITGA6− or SOX2+/FOXD1+ pre-DC cells were also entirely absent (Figures 6D, S6D, S6E and S6F), suggesting that FGF20 is a key Pc-derived signal essential for DC fate specification, prior to cell aggregation.
DISCUSSION
The timing of Fb-to-DC niche transformation has been traditionally described by the emergence of a characteristic tightly clustered DC beneath a thickened epidermal placode (Millar, 2002). At the earliest identified stage of bona fide HF morphogenesis, distinct Pcs develop before morphological DC clusters form. Recently, two studies have demonstrated that the structured DC forms through directed migration of dermal Fb (Biggs et al., 2018; Glover et al., 2017), but the precise timing and specific molecular drivers of DC fate-acquisition in relation to the physical aggregation process and to the timing of Pc progenitor specification remained unexplored.
Here we demonstrate that de novo expression of the definitive (clustered) DC markers, Sox2 (Clavel et al., 2012; Driskell et al., 2009) and Foxd1 (Sennett et al., 2015), first appears before any appreciable DC clustering. Taking advantage of Sox2 and other DC molecular markers, both pre- and post-clustering, we were now able to define the molecular landscape of the earliest fate-acquired DC precursors and their most closely related Fb, further refining our previous molecular characterization of the established DC niche (Sennett et al., 2015). Reinforcing our novel sorting strategy for population-based RNA sequencing analyses of distinct unclustered DC precursors and clustered DC stages, unbiased lineage prediction from single-cell transcriptomes further reconstructed the DC differentiation path of the bulk approach. Importantly, it also provided a highly resolved account of the molecular transitional states and developmental trajectory from non-committed Fb to DC precursors through DC1 and DC2.
Complementing the model of DC niche formation through directed migration from ex vivo live imaging (Biggs et al., 2018; Glover et al., 2017), we found that while Fb surrounding the DC can have robust proliferative activity, the earliest fate-acquired, Sox2+ DC precursors have significantly diminished proliferation indicating that the mature DC is a product of continual Fb recruitment (Figure 7A). We interestingly observed an early steady decline in the expression of proliferation-related genes in Fb most closely-related to pre-DC in pseudotime. This preemptive suppression of proliferation occurs prior to upregulation of early DC-fate genes and is particularly intriguing since the establishment of asymmetrically positioned DC is required in maintaining cell-fate asymmetry in the epithelium giving rise to both hair follicle stem cells and progenitors during early down growth of developing follicles (Cetera et al., 2018). Therefore, by extension, it can be posited that tight regulation of DC size by proliferation suppression is vital to maintain its asymmetric positioning for proper morphogenesis and hair follicle stem cell establishment. The potential necessity of cell-cycle exit for acquisition of DC fate, the cell intrinsic and/or extrinsic mechanisms to achieve cell cycle arrest, and the precise molecular controls that prevent over-recruitment through migration remain to be studied in more detail.
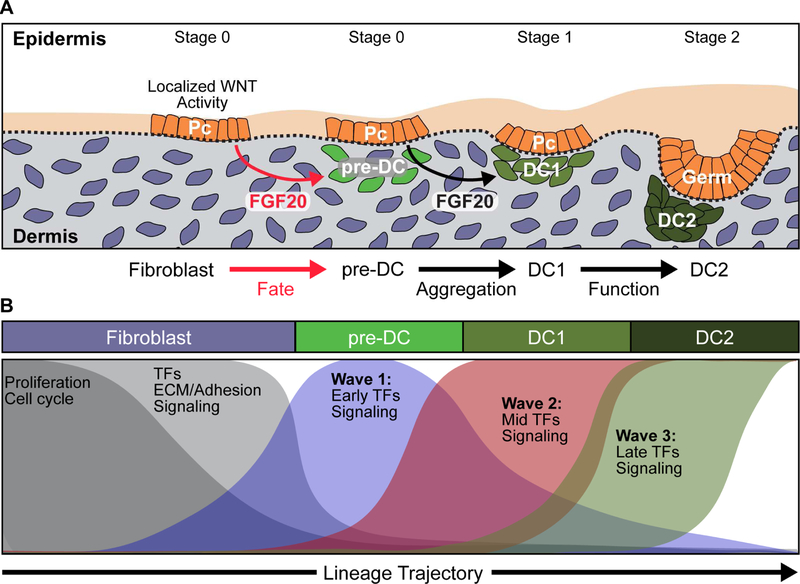
Figure 7. Model of DC Fate Specification and Maturation During Hair Follicle Formation.
(A) Schematic depicting the developmental hierarchy and timing of Pc and DC niche fate specification and maturation. Unknown dermal signals induce Pc progenitors. Local Wnt signaling activity upregulates FGF20. Pc-derived FGF20 is first required for fate specification of DC precursors that are at a transitional molecular state towards the DC niche biological fate. FGF20 then regulates cell aggregation to form the mature DC niche.
(B) Key steps of the DC niche fate trajectory during the Fb-to-DC transition. Exit from the cell cycle and progressive loss of Fb fate coincide with progressive acquisition of the DC niche fate through three upregulation waves of DC transcription factors and signaling molecules.
From both our bulk and single-cell transcriptome analyses, we identified transcription factors enriched in pre-DC including Twist2, Lef1 and Smad3 indicating an immediate shift in the gene regulatory network during early niche fate acquisition, potentially acting as trailblazers for downstream upregulation of mid- and late-expressed transcription factors and genes for DC formation and function such as those encoding cell-adhesion and signaling proteins (Figure 7B). The reconstructed lineage order in pseudotime also revealed a subset of Fb closely related to pre-DC raising the possibility of a yet earlier precursor stage to the DC niche fate. While pinpointing their precise spatial location poses a considerable challenge because of the paucity of distinctly expressed marker genes, clustering analyses place these cells broadly among upper dermal Fb which harbor multipotent dermal Fb progenitors (Driskell et al., 2013).
Epithelial-mesenchymal interdependence during early HF morphogenesis has been shown by multiple compartment-specific disruptions affecting the development of its counterpart (Chen et al., 2012; Tsai et al., 2014; Zhang et al., 2009). By preventing Pc formation through epidermal β-catenin ablation (Huelsken et al., 2001), we determined from the absence of pre-DC, early DC fate acquisition is not an autonomous pre-program of the upper dermis, but rather requires local directive signals from spatially patterned Pc. Genetic ablation of Fgf20 has previously demonstrated its requirement for DC formation by hair follicle stage 1 through cell migration (Biggs et al., 2018; Huh et al., 2013). Here we provide evidence that FGF20 directly promotes the early DC precursor fate before DC cluster formation. Similarly, recent identification of an Fgf20-expressing olfactory epithelial stem cell niche revealed a Wnt-regulated FGF20 requirement for formation of the underlying mesenchymal condensations that form nasal turbinates (Yang et al., 2018). Taken together these data indicate that the progenitors in nascent hair follicle pre-Pc signal to underlying Fb at least in part through FGF20 to induce the early DC fate prior to cluster-forming migration, and unequivocally demonstrate that progenitor fate precedes establishment of its supportive niche.
We now propose an updated model of progenitor and niche specification during early hair follicle morphogenesis (Figure 7A and 7B): Wnt-dependent Pc progenitors signal to underlying dermal Fb via the FGF/FGFR signaling axis initiating a cascade of dynamic transcriptional waves that regulate DC niche fate specification. Through multifaceted signaling interactions between Pc, pre-DC and unspecified interfollicular dermal Fb, fate specified DC precursor cells centrally migrate to form the clustered DC (DC1). Continued pre-DC fate acquisition and migration incorporates additional cells to the DC cluster, while in the earliest established DC the new transcriptional regulatory network – set up by the first transcriptional wave – begins to upregulate expression of the molecular machinery crucial for signaling functions from the DC niche to the progenitors above.
STAR METHODS
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were housed at the Center for Comparative Medicine and Surgery (CCMS) at Icahn School of Medicine at Mount Sinai (ISMMS) and all performed animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at ISMMS. Timed matings were set up to obtain embryos at the ages indicated in figure legends (embryonic day 14.0 to 15.0), sex not known. Embryo ages were determined by designating the morning of vaginal plug detection as E0.5. Sox2GFP (Ellis et al., 2004), Tbx18H2BGFP (Grisanti et al., 2013) and PdgfraH2BGFP (Hamilton et al., 2003) fluorescent reporter mice were generated as previously described. Fgf20Cre-GFP and Fgf20LacZ mice for Fgf20 knockout were generated as previously described (Huh et al., 2012; Huh et al., 2015). β-catenin epidermal-specific cKO were generated by mating β-cateninfl/fl (Brault et al., 2001) with K14-Cre (Dassule et al., 2000) mice.
EdU incorporation
For EdU-uptake proliferation assays, pregnant females mated with Sox2GFP males were injected intraperitoneally with a single dose of EdU/PBS (500 µg/g body weight) 6, 12 or 24 hours prior to sacrifice. EdU detection was performed with the Click-It EdU AlexaFluor 647 Imaging Kit (Life Technologies) according to manufacturer’s instructions.
Immunofluorescence staining
Tissue sections were embedded in OCT (Tissue Tek) and cut at a thickness of 10 µm with a cryostat (Leica). Whole-mounted embryo skins or sections from E15.0 embryos were fixed in 4% PFA and washed with PBS. Whole-mount samples were additionally permeabilized for 2 hr in 0.3% Triton X-100 in PBS. Tissues were blocked in PBS-Triton with BSA/NDS for 2 hour at room temperature. Primary antibody labelling against EDAR (goat, 1:100, R&D), SOX2 (Goat, 1:100, Stemgent), FOXD1 (Goat, 1:100, Santa Cruz), GFP (chicken, 1:800, Abcam), and CXCR4 (Rat, 1:100, BD Pharmagen) were carried out overnight at 4ºC. After PBS washes, samples were incubated with Rhodamine Red-X-, AlexaFluor 488-, AlexaFluor 555-, or AlexaFluor 647-conjugated donkey anti-goat, rabbit, chicken or rat secondary antibodies (Jackson Immunoresearch, Invitrogen). Samples were again PBS washed, counterstained with 4’6’-diamidino-2-phenilindole (DAPI) and mounted onto slides with antifade mounting medium.
Image acquisition and processing
Whole mount immunofluorescence and stained tissue sections were imaged using Leica SP5 DMI confocal and Leica DM5500 widefield microscopes, respectively, both equipped with Leica LASAF software. For 3D whole mount imaging, z-stacks of up to 60 planes with 1 µm vertical intervals were acquired with Leica 40X, 63X or 100X oil-immersion lenses. 3D stack images from confocal microscopy were processed and analyzed with ImageJ/FIJI (NIH) or Imaris 3/4D image visualization and analysis software (Bitplane).
Single-molecule mRNA fluorescence in situ hybridization
Single-molecule mRNA fluorescence in situ hybridization (FISH) was performed on whole mount skin from E15.0 embryos using RNAscope (Advanced Cell Diagnostics) according to the manufacturer’s instructions. Briefly, back skins were fixed in 4% PFA, dehydrated through a graded methanol (MeOH) series (50%, 70% and 100% MeOH/H2O), and were stored at −20°C overnight in 100% MeOH. The following day, skins were rehydrated and pretreated with hydrogen peroxide and protease III (2.5 HD Reagent Kit-RED), and hybridized with the Twist2 probe (ACD #489121) overnight at 40 C. The following day, skins were washed, postfixed in 4% PFA for 15 minutes at room temperature, followed by signal amplification by provided hybridizing horseradish peroxidase-labeled probes and fast red signal detection, as described in the manufacturer’s kit instructions. Following the RNAscope protocol, slides were processed for co-immunofluorescence and imaged as described above. The red chromogenic substrate was visualized as autofluorescence in the red fluorescent channel.
E15.0 cell isolation and sorting
To generate single-cell suspensions from E15.0 Sox2GFP dorsal skins, tissues were collected and processed as previously described (Sennett et al., 2015). Briefly, back skins were harvested by microdissection and were pooled for FACS for bulk RNA-sequencing while skin from a single embryo was taken for single cell sorting prior to single-cell RNA-sequencing. Skins were then digested in a dispase (Invitrogen)/collagenase (0.03%, Worthington) solution with 20 U/µl of DNase (Roche) at 4ºC overnight followed the next day by 30 minutes incubation at 37ºC. Cells were then filtered through 40µm-pore nylon cell strainers, followed by centrifugation at 350xg for 10 minutes. Resuspended cell pellets were stained with primary antibodies against EpCAM (BrV605-conjugated rat Ab, 1:100, Biolegend), CXCR4 (PE-conjugated rat Ab, 1:100, Biolegend), ITAG6 (BrV421-conjugated rat Ab, 1:100, Biolegend), PDGFRA (biotinylated rat Ab, 1:50, Affymetrix). This was followed by secondary labeling with Streptavidin-APC (1:200, Biolegend). Dead cells were identified by DAPI uptake. Cells were sorted on a FACSAria II fluorescence cytometry sorter (BD Biosciences) for both bulk and single cell RNA-sequencing. For single cell RNA-sequencing, live fibroblasts, pre-DC, DC1, and DC2 were index sorted into 384-well hard shell plates (Biorad) pre-loaded with 5 µl of vapor-lock (QIAGEN) containing a 100–200 nl mix of RT primers, dNTPs, and synthetic mRNA spike-ins. After sorting, plates were immediately quick spun and frozen to −80°C.
Real-Time qRT-PCR
Total RNA from the FACS-sorted cells was purified with the Absolutely RNA Nanoprep Kit (Agilent) and quantified with the NanoDrop spectrophotometer (Thermo Scientific). Reverse transcription was performed with the SuperScript III First-Strand Synthesis System using oligo(dT) primers (Invitrogen). Real-time qRT-PCR was performed with a LightCycler 480 (Roche) instrument using LightCycler DNA Master SYBR Green I reagents (Roche). Fold changes between samples were calculated based on the 2−∆∆Ct method and normalized to Gapdh.
cDNA library preparation for bulk RNA-sequencing
Total RNA from the FACS-sorted cells was purified with the Absolutely RNA Nanoprep Kit (Agilent). RNA concentration and quality were measured by the Agilent Bio-analyzer. Samples with RIN (RNA integrity number) scores of 9.6 or higher were further processed. 5 µl of RNA from each sample was used for reverse transcription, followed by amplification using the RNA Ovation RNA-Seq System V2 (NuGEN). Using the Ovation Ultralow DR Library System (NuGEN), cDNA libraries were generated from 100 ng amplified cDNA with a unique barcoded adaptor for each sample. Library concentration and quality were quantified by Qbit (Invitrogen) and the Agilent Bioanalyzer. Samples were then sequenced on the Illumina HiSeq 200 platform using a 50-nt single-read setting.
cDNA library preparation for single-cell RNA-sequencing
Single-cell RNA-sequencing was performed using the SORT-seq protocol (Muraro et al., 2016). Briefly, after sorting cells were lysed at 65°C for 5 minutes. RT and second strand mixes were dispensed by the Nanodrop II liquid handling platform for cDNA library preparation (GC Biotech). The aqueous phase was separated from the oil phase after pooling all cells from one plate into one library, followed by IVT transcription. For each 384 well plate, 384 primers (1 library of 384 cells) was used for mRNA reverse transcription, conversion to double-stranded cDNA, and in vitro transcription. Each primer consists of a 24 bp polyT stretch, a 4 bp random molecular barcode (UMI), a cell-specific 8 bp barcode, the 5’ Illumina TruSeq small RNA kit adapter, and a T7 promoter. Single-cell double-stranded cDNAs were pooled and in-vitro transcribed for linear amplification following the CEL-Seq 2 protocol. Illumina sequencing libraries were prepared using TruSeq small RNA primers (Illumina) and sequenced paired-end at a read length of 75 bp using the Illumina NextSeq.
4D time-lapse live imaging
Tbx18H2BGFP E14.0 embryonic dorsal skin was harvested by microdissection in ice-cold PBS, sandwiched between a 8 µm Nuclepore filter (Whatman) and the bottom of a 35 mm Lumox culture dish (Greiner) such that the epidermal side of the skin was in contact with the Lumox membrane, i.e. the bottom of the dish. The skin-membrane sandwich was stabilized with Matrigel (Corning). Explants were cultured in DMEM (phenol red free, Invitrogen) containing 10% fetal bovine serum, 1% HEPES, and 1% penicillin-streptomycin. After overnight culture, 3D Z-stack images of up to 20 planes with 2 µm vertical intervals were acquired at 10 or 25 minute intervals during a total of 16–20 hours using a Zeiss LSM880 equipped with a 20X Plan Apo 0.8 NA air objective or a Zeiss Axio Observer Z1 Yokogawa spinning disk equipped with a 20x Plan Apo 0.4 NA air objective. During imaging sessions, explants were maintained in a live-cell chamber at 37ºC with 5% CO2 and humidity control.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of DC clustering state
3D reconstructed images for nearest neighbor measurements of pre-DC, DC1, DC1 and interfollicular dermal fibroblasts were generated in Imaris. Automated segmentation of each cell was then performed by placing a point sphere at the geometric center of each nucleus from which each point’s discrete coordinates were used to quantify the absolute distances between nuclei. Multiple hypothesis testing of the data from the average distance between 5 nearest neighbors among cells from pre-DC, DC1, DC2 and interfollicular fibroblasts were performed using Anova: single factor.
Quantification of proliferation by EdU-uptake
Percent EdU+ among pre-DC, DC1, DC2 and interfollicular dermal cells was manually counted from acquired 3D confocal scans in ImageJ/FIJI. Sox2GFP+/ITGA6− cells were considered as DC cells whereas GFP−/ITGA6− cells were counted as interfollicular dermal cells. For each injection time point skins from 2 embryos were analyzed, from which 3 DCs/regions of each morphological stage were quantified (i.e. 3 follicles x 4 stages per embryo per time point). Statistical analysis was performed using one-tailed Student’s t-test.
Quantification of proliferation by cell division during time-lapse live imaging
To measure proliferation with time-lapse live imaging, 4D images acquired from explants were processed and analyzed with Imaris. 15,625um2 areas (350×350pixels) containing pre-DC or interfollicular dermis were obtained by cropping from larger 20X fields. Areas containing pre-DC were identified by locating DCs at the final time point and back tracing in time through the clustering process over a 6-hour period whereas the areas containing interfollicular dermis were defined as not containing any DC over the entire imaging session. Total cell numbers and mitotic events were quantified manually. Hypothesis testing was performed using Student’s t-test.
Quantification of number of DCs per field of view
SOX2+ pre-DC were quantified in whole mount stained back skins of β-cateninfl/fl wild type and K14-Cre;β-cateninfl/fl conditional knockout, as well as in Fgf20Cre-GFP/+ heterozygous and Fgf20Cre-GFP/LacZ knockout. For each genotype 2 embryos were analyzed. Pre-DC were counted in 4 field of views of 10x widefield live scans (601,267 μm2) in each embryonic skin. Statistical analysis was performed using one-tailed Student’s t-test.
Bulk RNA-sequencing data analysis
All raw RNA sequencing reads were mapped to the mouse genome (mm10) with TopHat v2.0.3 (Trapnell et al., 2009) coupled with the Bowtie2 (Langmead and Salzberg, 2012) and aligned with default parameters. Transcriptomes were assembled and fragments per kilobase per million reads (FPKM) for each gene were computed with Cufflinks v2.1.1 (Trapnell et al., 2010) with default parameters. Differentially expressed genes (DEGs) were identified using Cuffdiff (with default parameters except for the library normalization method was upper quartile normalization, where FPKMs were scaled via the ratio of the 75 quartile fragment counts to the average 75 quartile value across all libraries) and ANOVA with the Benjamini-Hochberg correction for multiple hypothesis testing with significance cut off FDR <0.05. The Fisher exact test was used for enrichment analysis with the same multiple hypotheses testing correction procedure and cut off. Principle component analyses (PCA) were performed for samples using BioJupies (Torre et al., 2018). Population signature genes were defined by DEGs with a FPKM≥1, and a fold enrichment≥2 compared to all other populations. Signature genes of multiple population overlaps featured in the Venn diagram analysis were defined similarly as individual-population signature genes by which each overlapping population must individually meet minimal expression level and fold-enrichment constraints.
Selection of genes used in developmental trajectory analyses were obtained by excluding genes with too low expression across all stages (FPKMany population < 1) and genes with similar expression levels across each developmental stage (FPKMmax < 2*FPKMmin). The remaining genes were log2 transformed and z-score standardized to obtain 0-mean and unit standard deviation. Gene clustering was performed with Morpheus (Broad Institute: https://software.broadinstitute.org/morpheus) using Spearman rank correlation with average linkage. Gene ontology and KEGG pathway enrichment analyses were carried out using Enrichr (Chen et al., 2013). Gene Set Enrichment Analysis (Subramanian et al., 2005) was performed using GenePattern (Reich et al., 2006). Curated lists were used to identify transcription factors (Lambert et al., 2018) and signaling molecules (Ramilowski et al., 2015).
Single-cell RNA-sequencing data analysis
Analysis of single-cell RNA-seq data was performed independently in an R environment (R 3.5.1, RStudio 1.1.453) and a Python environment (Python 3.6.5, Jupyter 1.0.0). Cells with more than 6000 transcripts/cell were included in the analysis. Additionally, mitochondrial genes, ERCC spike-ins, and non-expressed genes were excluded from downstream analysis. The remaining 1383 cells and 18483 genes were analyzed independently using the RaceID3 package (R-based) (Grün et al., 2016; Herman et al. , 2018) or the Scanpy package (Python-based) (Wolf et al., 2018).
For RaceID3 analyses, normalization was performed by simple rescaling, which is done by dividing the transcript counts per cell by the total number of transcripts, and multiplying this by the minimum total number of transcripts across cells. Principal components with overrepresentation of genes related to GO annotations “cell proliferation” and “cell cycle” were removed using the CCcorrect function. Evaluation of transcriptome similarities by 1-Pearson’s correlation coefficient followed by k-medoids clustering was used to cluster cells and t-distributed stochastic neighbor embedding (tSNE) (Van Der Maaten and Hinton, 2008) was used for visualization.
Using the Scanpy module, highly variable genes were extracted, data was log-normalized, and principal components were computed. Principal components showing significant overrepresentation of genes linked to the GO annotations “cell proliferation” and “cell cycle” in the top or bottom 1% quantile of loadings were removed in a fashion similar to the CCcorrect function in RaceID3. Cells were clustered using the Louvain algorithm (Blondel et al., 2008), and visualized using UMAP (McInnes and Healy, 2018).
Pseudotime analysis was performed on a subset of cells: clusters 4, 5, 6, 7, and 8 in RaceID3 analysis and clusters 1, 2, 3, and 5 in Scanpy analysis which comprise dermal condensate cells and fibroblast precursors of the latter. Cell order along pseudotime was inferred using StemID2 (Grün et al., 2016; Herman et al., 2018) or diffusi on pseudotime (Haghverdi et al., 2016). Curated lists were used to identify transcription factors (Lambert et al., 2018) and signaling molecules (Ramilowski et al., 2015).
For easier reproducibility, the complete code is available online (https://github.com/rendllab and https://github.com/kasperlab).
Cumulative percent was calculated as the running sum of FPKM values divided by the total FPKM sum. Cumulative percent plots for early, mid, and late expressed genes were generated by calculating the mean cumulative percent values for each position (cell) in the SOM order.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti GFP | Abcam | Cat# ab13970; RRID:AB_300798 |
| Goat polyclonal anti EDAR | R&D Systems | Cat# AF745; RRID:AB_355565 |
| Rat anti ITGA6 (Clone GoH3) | Biolegend | Cat# 313602; RRID:AB_345296 |
| BV421-conjugated, rat monoclonal anti ITGA6 (Clone GoH3) | Biolegend | Cat# 313623; RRID:AB_2562243 |
| Goat polyclonal anti FOXD1 (C-19) | Santa Cruz Biotechnology | Cat# sc-47585; RRID:AB_2105295 |
| Rat monoclonal anti CXCR4 (Clone 2B11) | BD Pharmingen | Cat# 551852; RRID:AB_394273 |
| PE-conjugated, rat monoclonal anti CXCR4 (Clone L276F12) | Biolegend | Cat# 146506; RRID:AB_2562783 |
| Rabbit polyclonal anti SOX2 | Stemgent | Cat# 09–0024; RRID:AB_2195775 |
| Goat polyclonal anti SOX2 (D-17) | Santa Cruz Biotechnology | Cat# sc-17319; RRID:AB_661259 |
| Rat monoclonal anti PDGFRA (Clone APA5) | eBioscience | Cat# 16–1401-82; RRID:AB_529483 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| EdU | Invitrogen | Cat# A10044 |
| Critical Commercial Assays | ||
| Absolutely RNA Nanoprep Kit | Agilent | Cat# 400753 |
| Ovation RNA-Seq System V2 | NuGEN | Cat# 0511-A01 |
| MinElute Reaction Cleanup Kit | Qiagen | Cat# 28204 |
| Superscript III | Invitrogen | Cat# 18080093 |
| LightCycler 480 SYBR Green I Master | Roche | Cat# 04707516001 |
| RNAscope 2.5 HD Reagent Kit-RED | Advanced Cell Diagnostics | Cat# 322350 |
| RNAscope Probe - Mm-Twist2 | Advanced Cell Diagnostics | Cat# 489121 |
| Click-iT Plus EdU Alexa Fluor 647 Imaging Kit | Invitrogen | Cat# C10640 |
| Thermo Scientific reagents for CEL-Seq2, Primers for CEL-Seq2 | Muraro et al., 2016 | N/A |
| Deposited Data | ||
| Raw and Analyzed Data (Bulk and Single-Cell) | This paper | GEO:GSE122026 |
| E14.5 DC and Fb signature genes | Sennett et al 2015 | GEO:GSE70288 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Sox2GFP: B6;129S-Sox2tm2Hoch/J | The Jackson Laboratory | Cat# 017592 |
| Mouse: PdgfraH2BGFP: B6.129S4-Pdgfratm11(EGFP)Sor/J | The Jackson Laboratory | Cat# 007669 |
| Mouse: Tbx18H2BGFP | Grisanti et al., 2013 | N/A |
| Mouse: K14-Cre: B6N.Cg-Tg(KRT14-cre)1Amc/J | The Jackson Laboratory | Cat# 018964 |
| Mouse: β-cateninfl/fl: B6.129-Ctnnb1tm2Kem/KnwJ | The Jackson Laboratory | Cat# 004152 |
| Mouse: Fgf20Cre-GFP: Fgf20tm1.1Dor | Huh et al., 2015 | N/A |
| Mouse: Fgf20LacZ: Fgf20tm2.1(cre/EGFP)Dor | Huh et al., 2012 | N/A |
| Mouse: CD1 IGS | Charles River | Strain code: 022 |
| Oligonucleotides | ||
| Primer: Lum Forward: GTTGGGAATGTGGGGGATGT Reverse: TCTCAGGATGAAAAAGGCCTGC | This paper | |
| Primer: Sox2 Forward: GTTGGGAATGTGGGGGATGT Reverse: TCTCAGGATGAAAAAGGCCTGC | This paper | |
| Primer: Foxd1 Forward: GTTGGGAATGTGGGGGATGT Reverse: TCTCAGGATGAAAAAGGCCTGC | This paper | |
| Primer: Cxcr4 Forward: GTTGGGAATGTGGGGGATGT Reverse: TCTCAGGATGAAAAAGGCCTGC | This paper | |
| Primer: Fabp7 Forward: GTTGGGAATGTGGGGGATGT Reverse: TCTCAGGATGAAAAAGGCCTGC | This paper | |
| Software and Algorithms | ||
| Imaris | Bitplane | Version 9.2.1 |
| ImageJ | NIH | https//imagej.nih.gov/ij/index.html |
| FlowJo | FlowJo | Version 7.6.5 |
| TopHat v2.0.3 | Trapnell et al., 2009 | Version 2.0.3 |
| Bowtie2 | Langmead and Salzberg, 2012 | Version 2.0.0 |
| Cufflinks v2.1.1 | Trapnell et al., 2010 | Version 2.1.1 |
| Genepattern | Reich et al., 2006 | http://software.broadinstitute.org/cancer/software/genepattern |
| Morpheus | https://software.broadinstitute.org/morpheus | N/A |
| RaceID3 and StemID2 algorithm | Grun et al., 2016 | N/A |
| R | https://www.r-project.org/ | Version 3.5.1 |
| Rstudio | https://www.rstudio.com/products/rstudio/download/ | Version 1.1.453 |
| BioJupies | Torre et al., 2018 | https://amp.pharm.mssm.edu/biojupies/ |
| Python | https://www.python.org/downloads/ | Version 3.6.5 |
| Jupyter | http://jupyter.org/ | Version 1.0.0 |
| Scanpy | Wolf et al., 2018 | Version 1.2.2 |
| Other | ||
| 384-well hard shell plates for sorting and SORT-Seq | Biorad | HSP3801 |
| Nanodrop II Liquid Handling Platform | Innovadyne | N/A |
| FACS Aria II | BD Biosciences | N/A |
Highlights.
Precursors of the hair follicle niche are specified before niche cluster formation
Bulk/single-cell transcriptomics defines niche fate at molecular transitional state
Successive transcription factor and signaling waves mark niche fate acquisition
Niche fate acquisition is not “pre-programmed” and requires FGF20 from progenitors
ACKNOWLEDGEMENTS
We thank Elena Ezhkova (ISMMS) for sharing mice and reagents. Many thanks to the personnel at the ISMMS Flow Cytometry and Microscopy CoRE and to Venu Pothula for technical assistance with single cell sorting. Thanks go to the personnel at the Genome Technology Center at NYU, and to Judith Vivié for single-cell RNA sequencing at the Hubrecht Institute (Netherlands). Additionally, we thank Marja Mikkola (U. Helsinki), Peggy Myung (Yale) and Martina Rangl (Cornell) for helpful discussions and valuable comments. K.W.M was supported by The Science Appearance Career Development Award fellowship from the Dermatology Foundation. N.H. was supported by grants T32GM007280 from NIH/NIGMS and F30AR070639 from NIH/NIAMS. A.M. was supported by NIH grants U54-HL127624 (LINCS-DCIC) and U24-CA224260 (IDG-KMC). D.M.O. was supported by NIH grant HL111190 and R21DC017042. M.R. was supported by grants from NIH/NIAMS (R01AR071047; R01AR063151) and New York State Department of Health (NYSTEM-C029574), and a fellowship from the Irma T. Hirschl Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AND SOFTWARE AVAILABILITY
Software
RaceID3 and StemID2 were run on RStudio version 1.1.453. Scanpy was run in Python version 3.6.5 in a Jupyter Notebook version 1.0.0. The scripts run are available at https://github.com/rendllab and https://github.com/kasperlab.
Data Resources
The accession number for the sequencing data reported in this paper is NCBI GEO:GSE122026 (bulk and single-cell RNA-sequencing.)
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Ahtiainen L, Lefebvre S, Lindfors PH, Renvoisé E, Shirokova V, Vartiainen MK, Thesleff I, and Mikkola ML (2014). Directional Cell Migration, but Not Proliferation, Drives Hair Placode Morphogenesis. Dev. Cell 28, 588–602. [DOI] [PubMed] [Google Scholar]
- Biggs LC, Mäkelä OJM, Myllymäki S-M, DasRoy R, Närhi K, Pispa J, Mustonen T, and Mikkola ML (2018). Hair follicle dermal condensation forms via Fgf20 primed cell cycle exit, cell motility, and aggregation. Elife 7, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume JL, Lambiotte R, and Lefebvre E (2008). Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp 2008, 1–12. [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, and Kemler R (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264. [DOI] [PubMed] [Google Scholar]
- Cetera M, Leybova L, Joyce B, and Devenport D (2018). Counter-rotational cell flows drive morphological and cell fate asymmetries in mammalian hair follicles. Nat. Cell Biol 20, 541– 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Jarrell A, Guo C, Lang R, and Atit R (2012). Dermal -catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development 139, 1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel C, Grisanti L, Zemla R, Rezza A, Barros R, Sennett R, Mazloom AR, Chung C-Y, Cai X, Cai C-L, et al. (2012). Sox2 in the dermal papilla niche controls hair growth by fine-tuning BMP signaling in differentiating hair shaft progenitors. Dev. Cell 23, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R, and Fuchs E (1999). Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568. [DOI] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, and McMahon AP (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775–4785. [DOI] [PubMed] [Google Scholar]
- Driskell RR, Giangreco A, Jensen KB, Mulder KW, and Watt FM (2009). Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 136, 2815– 2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, et al. (2013). Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, and Pevny L (2004). SOX2, a Persistent Marker for Multipotential Neural Stem Cells Derived from Embryonic Stem Cells, the Embryo or the Adult. Dev Neurosci 26, 148–165. [DOI] [PubMed] [Google Scholar]
- Falk S, and Götz M (2017). Glial control of neu rogenesis. Curr. Opin. Neurobiol 47, 188–195. [DOI] [PubMed] [Google Scholar]
- Fantauzzo KA, Tadin-Strapps M, You Y, Mentzer SE, Baumeister FAM, Cianfarani S, Van Maldergem L, Warburton D, Sundberg JP, and Christiano AM (2008). A position effect on TRPS1 is associated with Ambras syndrome in humans and the Koala phenotype in mice. Hum. Mol. Genet 17, 3539–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Palmero I, Torres S, Bartolomé RA, Peláez-García A, Larriba MJ, Lopez-Lucendo M, Peña C, Escudero-Paniagua B, Muñoz A, and Casal JI (2016). Twist1-induced activation of human fibroblasts promotes matrix stiffness by upregulating palladin and collagen α1(VI). Oncogene 35, 5224–5236. [DOI] [PubMed] [Google Scholar]
- Ge Y, and Fuchs E (2018). Stretching the limits : from in wound healing and cancer. Nat. Publ. Gr [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Genderen C, Okamura RM, Fariñas I, Quo RG, Parslow TG, Bruhn L, and Grosschedl R (1994). Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev 8, 2691–2703. [DOI] [PubMed] [Google Scholar]
- Glover JD, Wells KL, Matthäus F, Painter KJ, Ho W, Riddell J, Johansson JA, Ford MJ, Jahoda CAB, Klika V, et al. (2017). Hierarchical patterning modes orchestrate hair follicle morphogenesis [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisanti L, Clavel C, Cai X, Rezza A, Tsai SY, Sennett R, Mumau M, Cai CL, and Rendl M (2013). Tbx18 targets dermal condensates for labeling, isolation, and gene ablation during embryonic hair follicle formation. J. Invest. Dermatol 133, 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Muraro MJ, Boisset J-C, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, et al. (2016). De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell 19, 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Büttner M, Wolf FA, Buettner F ., and Theis FJ (2016). Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, and Soriano P (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol 23, 4013– 4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman N, Saxena N, and Rendl M (2018). Advancing insights into stem cell niche complexities with next-generation technologies. Curr. Opin. Cell Biol 55, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JS, Sagar, and Grün D (2018). FateID in fers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods 15, 379–386. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Li L, and Fuchs E (2014b). Emerging interactions between skin stem cells and their niches. Nat. Med 20, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, and Birchmeier W (2001). β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 105, 533–545. [DOI] [PubMed] [Google Scholar]
- Huh S-H, Närhi K, Lindfors PH, Häärä Otsa, Yang Lu, Ornitz DM, and Mikkola ML (2013). Fgf20 governs formation of primary and secondary dermal condensations in developing hair follicles. Genes Dev 27, 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Jones J, Warchol ME, and Ornitz DM (2012). Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Warchol ME, and Ornitz DM (2015). Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. Elife 2015, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J Il, and Lau LF (2010). The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat. Cell Biol 12, 676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RB, Tran TT, Pyott SM, Pepin MG, Savairayan R, McGillivray G, Nickerson DA, Bamshad MJ, and Byers PH (2018). Monoallelic and Biallelic CREB3L1 Variant Causes Mild and Severe Osteogenesis Imperfecta, Respectively. Genet Med 5, 355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, and Weirauch MT (2018). The Human Transcription Factors. Cell 172, 650– 665. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, et al. (2017). Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 170, 1149–1163.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman E, Howard JM, Garcia GE, Miao Q, Ku AT, Dekker JD, Tucker H, and Nguyen H (2013). Foxp1 maintains hair follicle stem cell quiescence through regulation of Fgf18. Development 140, 3809–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cserjesi P, and Olson EN (1995). Dermo-1: a novel twist related bHLH protein expressed in the developing dermis. Dev. Biol 172, 280–292. [DOI] [PubMed] [Google Scholar]
- Liu Y, El-Naggar S, Darling DS, Higashi Y, and Dean DC (2008). Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 135, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten LJP, and Hinton GE (2008). Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- McInnes L, and Healy J (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. J. Open Source Softw 3, 861. [Google Scholar]
- Millar SE (2002). Molecular mechanisms regulating hair follicle development. J. Invest. Dermatol 118, 216–225. [DOI] [PubMed] [Google Scholar]
- Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJP, et al. (2016). A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst 3, 385–394.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Matzuk MM, Gerstmayer B, Bosio A, Lauster R, Miyachi Y, Werner S, and Paus R (2003). Control of pelage hair follicle development and cycling by complex interactions between follistatin and activin. FASEB J 17, 497–499. [DOI] [PubMed] [Google Scholar]
- Ouspenskaia T, Matos I, Mertz AF, Fiore VF, and Fuchs E (2016). WNT-SHH Antagonism Specifies and Expands Stem Cells prior to Niche Formation. Cell 164, 156–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R, Müller-Röver S, Van Der Veen C, Maurer M, Eichmüller S, Ling G, Hofmann U, Foitzik K, Mecklenburg L, and Handjiski B (1999). A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J. Invest. Dermatol 113, 523–532. [DOI] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, et al. (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, and Mesirov JP (2006). GenePattern 2.0 [2]. Nat. Genet 38, 500–501. [DOI] [PubMed] [Google Scholar]
- Rezza A, Sennett R, and Rendl M (2014). Adult stem cell niches: cellular and molecular components. Curr. Top. Dev. Biol 107, 333–372. [DOI] [PubMed] [Google Scholar]
- Rezza A, Wang Z, Sennett R, Qiao W, Wang D, Heitman N, Mok KW, Clavel C, Yi R, Zandstra P, et al. (2016). Signaling Networks among Stem Cell Precursors, Transit-Amplifying Progenitors, and their Niche in Developing Hair Follicles. Cell Rep 14, 3001–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennett R, and Rendl M (2012). Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin. Cell Dev. Biol 23, 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennett R, Rezza A, Dauber KL, Clavel C, and Rendl M (2014). Cxcr4 is transiently expressed in both epithelial and mesenchymal compartments of nascent hair follicles but is not required for follicle formation. Exp. Dermatol 23, 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennett R, Wang Z, Rezza A, Grisanti L, Roitershtein N, Sicchio C, Mok KW, Heitman NJ, Clavel C, Ma’ayan A, et al. (2015). An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Dev. Cell 34, 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massasa EE, Itzkovitz S, and Kaestner KH (2018). Nature 557, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šošić D, Richardson J. a, Yu K, Ornitz DM, and Olson EN (2003). Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell 112, 169–180. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DJ, Gunin A, Magerl M, Handijski B, and Paus R (2003). Plasticity and cytokinetic dynamics of the hair follicle mesenchyme: Implications for hair growth control. J. Invest. Dermatol 120, 895–904. [DOI] [PubMed] [Google Scholar]
- Torre D, Lachmann A, and Ma’ayan A (2018). BioJupies: Automated Generation of Interactive Notebooks for RNA-seq Data Analysis in the Cloud. BioRxiv 352476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S-Y, Sennett R, Rezza A, Clavel C, Grisanti L, Zemla R, Najam S, and Rendl M (2014). Wnt/β-catenin signaling in dermal condensates is required for hair follicle formation. Dev. Biol 385, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani R, Hodgson S, Legrand J, Greaney J, Wong HY, Pichol-Thievend C, Adolphe C, Wainwight B, Francois M, and Khosrotehrani K (2017). Dominant-negative Sox18 function inhibits dermal papilla maturation and differentiation in all murine hair types. Development 144, 1887–1895. [DOI] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol 19, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LM, Huh S-H, and Ornitz DM (2018). FGF20-Expressing, Wnt-Responsive Olfactory Epithelial Progenitors Regulate Underlying Turbinate Growth to Optimize Surface Area. Dev. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, et al. (2009). Reciprocal Requirements for EDA/EDAR/NF-κB and Wnt/β-Catenin Signaling Pathways in Hair Follicle Induction. Dev. Cell 17, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R, Zou S-T, Wan J-M, Li W, Li X-L, and Zhu W (2013). BTG1 inhibits breast cancer cell growth through induction of cell cycle arrest and apoptosis. Oncol. Rep 30, 2137– 2144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.