SUMMARY
The molecular chaperone Hsp90 stabilizes and activates client proteins. Co-chaperones and post-translational modifications tightly regulate Hsp90 function and consequently lead to activation of clients. However, it is unclear whether this process occurs abruptly or gradually in the cellular context. We show that casein kinase-2 phosphorylation of the co-chaperone folliculin-interacting protein 1 (FNIP1) on priming serine-938 and subsequent relay phosphorylation on serine-939, 941, 946, and 948 promotes its gradual interaction with Hsp90. This leads to incremental inhibition of Hsp90 ATPase activity and gradual activation of both kinase and non-kinase clients. We further demonstrate that serine/threonine protein phosphatase 5 (PP5) dephosphorylates FNIP1, allowing the addition of O-GlcNAc (O-linked N-acetylglucosamine) to the priming serine-938. This process antagonizes phosphorylation of FNIP1, preventing its interaction with Hsp90, and consequently promotes FNIP1 lysine-1119 ubiquitination and proteasomal degradation. These findings provide a mechanism for gradual activation of the client proteins through intricate crosstalk of post-translational modifications of the co-chaperone FNIP1.
In Brief
Sager et al. show that casein-kinase-2-mediated sequential phosphorylation of the co-chaperone FNIP1 leads to incremental inhibition of Hsp90 ATPase activity and gradual activation of both kinase and non-kinase clients. O-GlcNAcylation antagonizes phosphorylation of FNIP1, preventing its interaction with Hsp90, and consequently promotes FNIP1 ubiquitination and proteasomal degradation.
INTRODUCTION
The molecular chaperone heat shock protein 90 (Hsp90) is essential in eukaryotes and is involved in looking after ~200 target proteins, also referred to as client proteins (Picard, 2002; Schopf et al., 2017; Verba and Agard, 2017). Hsp90 exists as a dimer, and each protomer consists of (1) an N-terminal domain, where the nucleotide-binding pocket resides; (2) a middle-domain; and (3) a C-terminal domain, which is constitutively dimerized with the other protomer. The N-M domains of Hsp90 are connected by a charged linker (Mayer, 2010; Prodromou, 2016; Röhl et al., 2013). Hsp90 chaperone function is linked to its ability to bind and hydrolyze ATP (Panaretou et al., 1998; Zierer et al., 2016), which allows for a series of conformational changes known as the chaperone cycle, wherein an Hsp90 dimer progresses from an N-terminally “open” to a “closed” conformation. Co-chaperones and post-translational modifications (PTMs) regulate the Hsp90 chaperone cycle and, consequently, the chaperoning and activity of the client proteins (Sager et al., 2018b; Sahasrabudhe et al., 2017).
As the name suggests, folliculin-interacting protein 1 (FNIP1) was initially identified as a binding partner of the tumor suppressor folliculin (FLCN) (Baba et al., 2006). Our subsequent work revealed FNIP1 as a co-chaperone of Hsp90 that acts as a scaffold to load FLCN onto Hsp90 (Sager et al., 2018a; Woodford et al., 2016). FNIP1 is also involved in chaperoning of both kinase and non-kinase clients. FNIP1 does not have any known functional domains; however, based on amino acid sequence alignments, conserved regions were identified and named as A–D. The C-terminal domain of FNIP1 (amino acids 929–1,166 or fragment D) preferentially interacts with the middle domain of Hsp90. This fragment and the full-length FNIP1 are potent inhibitors of Hsp90 ATPase activity (Woodford et al., 2016). Small-molecule inhibitors that target the nucleotide-binding pocket of the N-terminal domain of Hsp90 also inhibit its ATPase activity and lead to degradation of the client proteins (Neckers et al., 2018). However, FNIP1-mediated inhibition of Hsp90 ATPase activity appears to decelerate the chaperone cycle, not inhibit it completely, as overexpression of FNIP1 stabilizes and activates client proteins. This can also be reversed by the co-chaperone Aha1, which is the activator of the Hsp90 ATPase function and competes with FNIP1 for binding to Hsp90 (Woodford et al., 2016).
Recombinant FNIP1 devoid of any PTMs binds to and forms a complex with Hsp90 in vitro. However, in the cellular context, it is unclear whether PTMs of FNIP1 play a role in its ability to bind to Hsp90 and influence the activity of the client proteins. In this study, we used the FNIP1-D fragment (Hsp90-binding region of FNIP1) and identified a series of serine residues (938, 939, 941, 946, and 948) that were phosphorylated by casein-kinase-2 (CK2) both in vitro and in vivo. This led to stepwise binding of FNIP1 to Hsp90, incremental inhibition of Hsp90 ATPase activity, and gradual activation of both kinase and non-kinase clients. We further demonstrated that protein phosphatase 5 (PP5) dephosphorylates FNIP1, allowing for subsequent O-GlcNAcylation (addition of O-linked N-acetylglucosamine) of the priming serine-938. This antagonized phosphorylation of FNIP1 and disrupted its interaction with Hsp90 as well as promoted ubiquitination of FNIP1 lysine-1119 and eventual degradation of FNIP1 in the proteasome. These data provide a mechanism for gradual activation of the kinase and non-kinase client proteins through intricate crosstalk of PTMs of the co-chaperone FNIP1.
RESULTS
CK2 Sequentially Phosphorylates FNIP1 Co-chaperone
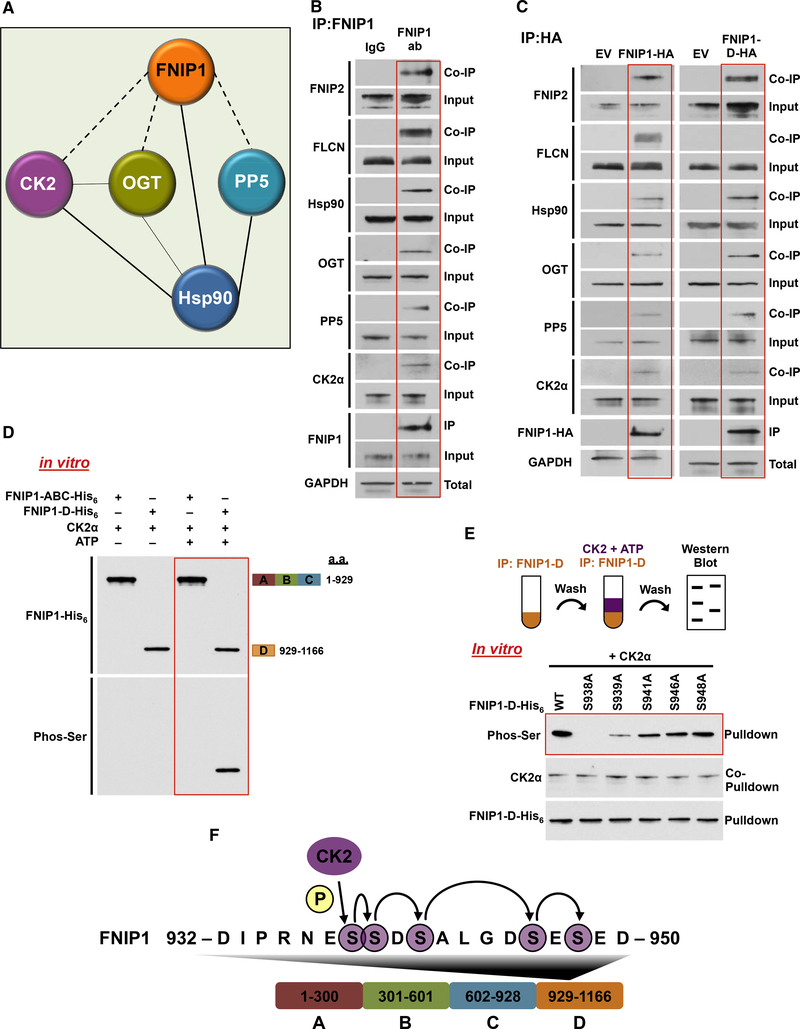
FNIP1 is a co-chaperone of Hsp90 (Woodford et al., 2016). To gain further insight into the binding partners and their involvement in regulation of FNIP1, we immunoprecipitated endogenous FNIP1 from HEK293 cells followed by mass spectrometry (MS) analysis. We identified Hsp90, CK2, O-linked N-acetylglucosamine (GlcNAc) transferase (OGT), and serine/threonineprotein phosphatase 5 (PP5) to interact with FNIP1 (Figure 1A; Table S1). We confirmed these data by immunoprecipitation of FNIP1 from HEK293 cells followed by immunoblotting (Figure 1B). FLCN and FNIP2 proteins are known binding partners of FNIP1 (Woodford et al., 2016), and they were used as positive controls. FNIP1 does not have any known functional domains; however, our previous work has shown that its C-terminal region (amino acids 929–1,166 or fragment D) binds to Hsp90 and inhibits its ATPase activity (Woodford et al., 2016). Transient expression and immunoprecipitation of FNIP1-D-HA from HEK293 showed that the FNIP1-D fragment, like full-length FNIP1 hemagglutinin (HA), also binds to Hsp90, CK2, OGT, and PP5 (Figure 1C). We next showed that CK2 is capable of phosphorylating FNIP1 on the FNIP1-D fragment on serine, but not threonine, residues in vitro (Figures 1D and S1A).
Figure 1. Sequential Phosphorylation of the FNIP1 Co-chaperone by CK2 Kinase.
(A) Endogenous FNIP1 protein was isolated from HEK293 cells, and the profile of interacting proteins was determined by MALDI-TOF.
(B) FNIP1 was immunoprecipitated (IP) from HEK293 cell lysates using anti-FNIP1 or immunoglobulin G (IgG) (control) and immunoblotted with indicated antibodies to confirm protein interactions.
(C) FNIP1-HA, FNIP1-D-HA, and empty vector (EV) were transiently expressed and isolated by IP from HEK293 cells. Indicated coIP proteins were immunoblotted with indicated antibodies to confirm protein interactions.
(D) Indicated FNIP1-His6 fragments were used as substrates of CK2α in an in vitro kinase assay. Phosphorylation of serine residues was assessed by immunoblotting using a pan-anti-phosphoserine antibody.
(E) FNIP1-D-His6 and the indicated non-phosphomutants were bacterially expressed and purified. These proteins were used in an in vitro kinase assay with CK2α kinase. Serine phosphorylation was detected by immunoblotting using a pan-anti-phosphoserine antibody.
(F) Schematic representation of the relay phosphorylation of serine residues in the FNIP1-D fragment.
We examined the sequence of the FNIP1-D fragment and found that S946 was the only serine to fit the canonical CK2 consensus sequence (Cesaro and Pinna, 2015). This serine, however, was present in a stretch of residues that included a number of other serine residues as well as multiple aspartic and glutamic acids. CK2 is well known to be capable of multisite phosphorylation, with non-canonical consensus sequences recognizing acidic residues or phosphorylated serine residues in close proximity to the serine of interest (Cesaro and Pinna, 2015). In fact, FNIP1-S938 was identified as a possible CK2 phosphorylation site in a systematic investigation for these non-canonical hierarchical consensus sequences (St-Denis et al., 2015). We therefore made non-phosphorylatable alanine mutants of this series of serine residues (S938, S939, S941, S946, and S948) and bacterially expressed and purified these mutants as well as the wild-type FNIP1-D fragment. We performed an in vitro kinase assay using CK2 and ATP followed by immunoblotting with pan-phosphoserine antibody, which showed a gradual reduction of serine phosphorylation from S948A to S938A (Figures 1E and S1B). Interestingly, this was not due to alteration of FNIP1-D binding to CK2. Our data here therefore suggest that CK2 phosphorylates these serine sites in the FNIP1-D fragment in a relay manner (Figure 1F).
PP5 Relay Dephosphorylation of FNIP1 Disrupts Its Interaction with Hsp90
PP5 is a serine/threonine-protein phosphatase and also a cochaperone of Hsp90 (Schopf et al., 2017). Since PP5 interacts with FNIP1, we decided to check its ability to dephosphorylate FNIP1. Expression and purification of the FNIP1-D fragment as well as the non-phosphorylating alanine mutants (S938A, S939A, S941A, S946A, and S948A) from bacteria followed by in vitro phosphorylation with CK2 again confirmed serine phosphorylation of FNIP1-D in a relay manner (Figures 2A and S2A). Addition of PP5 to these reactions led to dephosphorylation of wild-type FNIP1-D, but not its non-phosphorylatable alanine mutants, even though PP5 interacted with all of the mutants (Figures 2A and S2B). We repeated this experiment in HEK293 cells by transiently expressing wild-type FNIP1-D-HA or its individual non-phosphorylatable alanine mutants (S938A, S939A, S941A, S946A, and S948A) followed by immunoprecipitation and incubation with recombinant and active PP5-glutathione S-transferase (GST). Immunoblotting of these samples produced similar results as our in vitro experiments, showing serine dephosphorylation of only wild-type FNIP1-D-HA (Figure 2B). We also saw that phosphorylation of FNIP1-D promotes its interaction with Hsp90; this interaction occurs gradually based on the phosphorylation status of the serine series, and the FNIP1-D-S938A mutation blocks both its phosphorylation and binding to Hsp90 (Figures 2B and S2C). Our findings suggest that PP5 dephosphorylates FNIP1-D in a relay manner by initially removing phosphate from the S948 residue. This is supported by the fact that PP5 completely dephosphorylates all the modified serine residues on wild-type FNIP1-D both in vitro and in vivo (i.e., no serine phosphorylation signal for wild-type FNIP1-D-HA), and the common feature among all of the phosphomutants is the lack of phosphorylation on S948. Therefore, initial dephosphorylation of S948 on FNIP1-D is essential for subsequent dephosphorylation of the other serine sites (Figure 2C).
Figure 2. PP5 Relay Dephosphorylation of FNIP1 and Its Disruption from Hsp90.
(A) Recombinant FNIP1-D-His6 and indicated non-phosphomutants were phosphorylated in vitro by CK2α and then incubated with or without recombinant PP5-GST. Phosphorylation of serine residues was examined by immunoblotting using a pan-anti-phosphoserine antibody.
(B) HEK293 cells were transiently co-transfected with FNIP1-D-HA and the indicated non-phosphomutants, IP, and then incubated with recombinant PP5-GST. Samples were immunoblotted for serine phosphorylation and interaction with Hsp90. LE, long exposure; SE, short exposure.
(C) Schematic representation of PP5 mediated dephosphorylation of FNIP1-D serine residues.
Sequential Phosphorylation of FNIP1 Incrementally Inhibits Hsp90 ATPase Activity
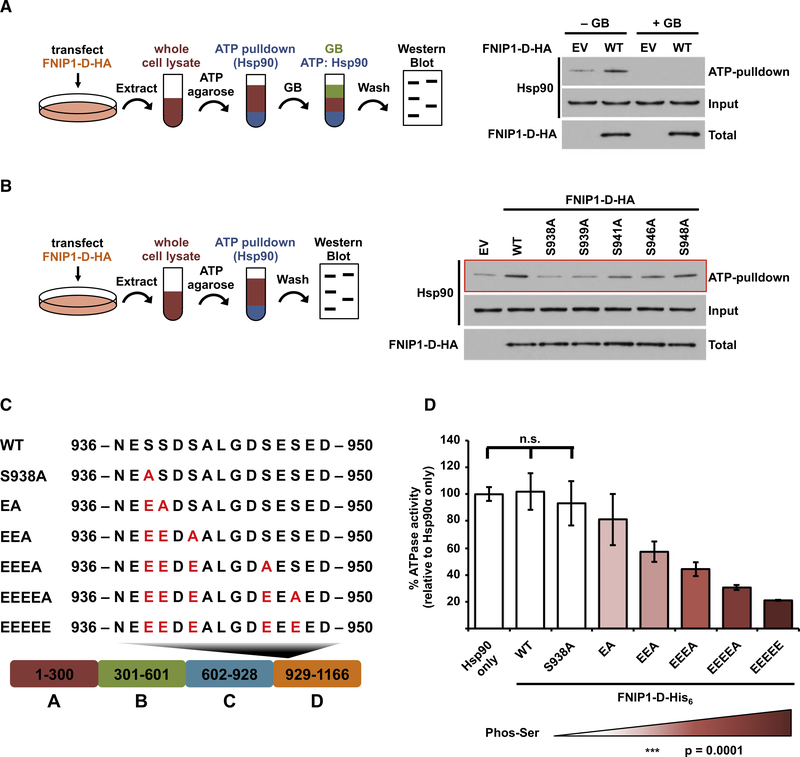
The ability of Hsp90 to bind and hydrolyze ATP is coupled to its chaperone function (Panaretou et al., 1998; Zierer et al., 2016). Our data show that CK2 mediated relay phosphorylation of FNIP1 impacts its binding to Hsp90; however, the consequence of this interaction toward Hsp90 ATP binding and hydrolysis remains unknown. We have previously shown that overexpression of FNIP1 in HEK293 cells increases Hsp90 binding to ATP agarose (Woodford et al., 2016). Here, we showed that overexpression of FNIP1-D-HA in HEK293 cells is sufficient to enhance Hsp90 binding to ATP agarose (Figure 3A). Transient expression of non-phospho-FNIP1-D mutants (S938A, S939A, S941A, S946A, and S948A) impacted Hsp90 binding to ATP in a similar manner as their interaction with Hsp90 (Figure 3B). We have previously shown that FNIP1-D purified from HEK293 cells inhibits Hsp90 at a 0.4:1 molar ratio (Woodford et al., 2016). Since FNIP1-D from HEK293 cells is post-translationally modified, we sought to dissect the impact of the individual serine phosphorylation states on ATPase activity of Hsp90-FLAG (Figure S3A).
Figure 3. FNIP1-D Serine Phosphorylation Gradually Inhibits Hsp90 ATPase Activity.
(A) FNIP1-D-HA (wild-type [WT]) was overexpressed in HEK293 cells, and the cell lysates were challenged with ATP-agarose for 1 h at 4°C, washed, and then incubated with or without 10 μM of the Hsp90 inhibitor ganetespib (GB) for an additional 1 h at 4°C. ATP-agarose was washed, and ATP-bound Hsp90 was analyzed by immunoblotting. EV was used as control.
(B) Non-phosphorylatable FNIP1-D-HA (WT) and its mutants were transiently expressed in HEK293 cells, and cell lysates were incubated with ATP-agarose. Hsp90 binding to ATP was examined by immunoblotting.
(C) Schematic representation of creating FNIP1-D phosphomimetic mutants for expression and purification from bacteria. EV was used as control.
(D) Inhibitory effect of the recombinant FNIP1-D-His6 phosphomimetic mutants on ATPase activity of Hsp90α-FLAG isolated from PC3 cells was measured in vitro. A Student’s t test was performed to assess statistical significance (n.s., not significant).
While we avoided use of phosphomimetic mutations for cell-based assays in this study, in order to capture the different intermediate phosphorylation states on bacterially expressed FNIP1-D for in vitro experiments, we constructed a series of sequential phosphomimetic mutants (Figures 3C and S3B). We next measured Hsp90 ATPase activity in the presence of bacterially expressed and purified wild-type FNIP1-D-His6 (devoid of any PTMs) and showed that it can inhibit Hsp90 ATPase activity in a dose-dependent manner (Figures S3C and S3D). Since a 0.4:1 molar ratio of FNIP1-D-His6 to Hsp90 does not inhibit Hsp90 ATPase activity, we used the same amounts of the phosphomimetic serine FNIP1-D mutants and showed that FNIP1-D-His6-S938E/S939A (FNIP1-D EA) slightly inhibited (by 19%) the ATPase activity of Hsp90-FLAG (Figure 3D).
Earlier, we showed that phosphorylation of S938 leads to relay phosphorylation of S939, S941, S946, and S948. Here, our data show that the FNIP1-D phosphomimetic S938E/S939E/S941E/ S946E/S948E (ie phosphomimetic of all serine residues in the series) significantly inhibits the ATPase activity of Hsp90 by 79% (Figures 3D and S3E). Our intermediate phosphomimetic serine FNIP1-D mutants led to gradual inhibition of Hsp90 ATPase activity similar to the pattern seen with Hsp90 binding and ATP binding. Taken together, our data presented here provide strong evidence that relay serine phosphorylation of FNIP1-D causes an incremental binding to Hsp90 and, consequently, gradually inhibits its ATPase activity.
Stepwise Phosphorylation of FNIP1 Gradually Activates Both Kinase and Non-kinase Clients
We previously reported that Lst4 is not an ortholog of FNIP1 in the budding yeast Saccharomyces cerevisiae (Woodford et al., 2016). Additionally, overexpression of FNIP1 under the GAL1 promoter caused lethality in yeast with Hsp90α as the sole copy of Hsp90 (Woodford et al., 2016). Here, we expressed FNIP1-FLAG under a significantly weaker and constitutive promoter, ADH1 (alcohol dehydrogenase 1), and showed that yeast cells were viable (Figures S4A and S4B). We next confirmed that FNIP1-FLAG under either the GAL1 or ADH1 promoter is capable of interacting with Hsp90α (Figure S4C). Like in HEK293 cells, FNIP1-FLAG is also subject to serine phosphorylation in yeast, and expression of the non-phosphomutants showed a gradual reduction in serine phosphorylation of FNIP1-S948A to S938A mutants (Figure S4D). These mutants also bound in a similar manner to Hsp90α (Figure S4D). The baker’s yeast S. cerevisiae has been proven to be a valuable tool to examine the effect of Hsp90 on chaperoning of its client proteins (Echtenkamp and Freeman, 2012). Here, we deciphered the impact of relay phosphorylation of FNIP1 on chaperoning of kinase and non-kinase clients in yeast. We used three well-established Hsp90 kinase clients: mitogen-activated protein (MAP) kinase Mpk1/Slt2, Ste11, and v-Src.
MAP kinase Mpk1/Slt2 is an endogenous kinase client of yeast Hsp90, which mediates a cell wall integrity pathway (Millson et al., 2005). We used the downstream reporter gene RLM1-LacZ to monitor Mpk1/Slt2 activity in yeast expressing empty vector (EV), wild-type FNIP1-FLAG, or its non-phosphomutants. Caffeine activates the RLM1-LacZ reporter (Truman et al., 2006), and we found that reporter activity was significantly increased in FNIP1-FLAG-expressing cells, and this effect was abrogated with the FNIP1-FLAG-S938A mutant (Figure 4A). Expression of individual non-phospho-FNIP1-FLAG mutants S939A, S941A, S946A, and S948A led to gradual activation of the reporter RLM1-LacZ and, hence, activation of the cell wall integrity pathway (Figure 4A). Another endogenous yeast kinase client is Ste11 (a Raf-1 ortholog) (Flom et al., 2008; Louvion et al., 1998), and a constitutively active variant, Ste11ΔN, expressed under the GAL1 promoter can readily be detected in yeast cells (Figure 4B). However, Ste11ΔN protein was moderately increased in cells expressing FNIP1-FLAG, whereas FNIP1-FLAG-S938A-expressing cells exhibited Ste11ΔN expression similar to that seen in the empty-vector-expressing cells. We observed a gradual increase in Ste11ΔN protein levels in cells expressing non-phospho-FNIP1-FLAG-S938A to S948A mutants correlating with the degree of FNIP1 serine phosphorylation (Figure 4B).
Figure 4. Relay Serine Phosphorylation of FNIP1 Gradually Activates Both Kinase and Non-kinase Clients.
(A) RLM1-LacZ activity was measured in yeast expressing WT hHsp90α and WT FNIP1-FLAG and its non-phosphomutants. EV was used as a control. Cells were grown to mid-log phase and stressed with 8 mM caffeine for 4 h. Data are presented as mean ± SD derived from three independent experiments. A Student’s t test was performed to assess statistical significance (**p < 0.01). FNIP1-FLAG and Hsp90α were visualized by immunoblotting.
(B) Yeast with WT hHsp90α, expressing WT FNIP1-FLAG, and its non-phosphomutants were also co-expressing Ste11ΔN-cMyc under the GAL1 promoter. EV was used as a control. Cells were grown on glucose (−) or galactose (+) media, and Ste11ΔN-cMyc protein expression was examined by immunoblotting.
(C) GAL1-v-SRC was transformed into yeast cells with WT hHsp90α, containing WT FNIP1-FLAG and its non-phosphomutants. EV was used as a control. Cells were grown on glucose (−) or galactose (+) media. v-Src and total phosphotyrosine were detected by immunoblotting.
(D) GR-lacZ activity was examined in cells expressing WT hHsp90α and WT FNIP1-FLAG and its non-phosphomutants. EV was used as a control. Data are expressed as a percentage of the activity observed in WT cells and presented as the mean ± SD from values obtained in three independent experiments. A Student’s t test was performed to assess statistical significance (***p < 0.001). FNIP1-FLAG and Hsp90α were visualized by immunoblotting.
(E) The above yeast strains expressing HSE-LacZ were heat shocked at 39°C for 40 min. The heat shock response was measured in three independent experiments. All data represent mean ± SD. EV was used as a control. A Student’s t test was performed to assess statistical significance (****p < 0.0001). FNIP1-FLAG and Hsp90α were examined by immunoblotting.
(F) CFTR-HA was expressed in the above yeast strains and treated with cycloheximide (CHX; 50 mM) and harvested at the indicated time points. CFTR-HA FNIP1-FLAG and Hsp90α were analyzed by immunoblotting.
(G) Quantification and representation of kinase and non-kinase clients stability and/or activity obtained from data in (A)–(F). Results are presented as a heatmap.p values are for Pearson correlation of the trend between number of phospho-sites in each FNIP1 construct and client stability or activity.
We next used another well-established Hsp90 kinase client, v-Src (Nathan and Lindquist, 1995). v-SRC expression under the GAL1 promoter and v-Src-mediated tyrosine phosphorylation of proteins were detectable in yeast (Figure 4C). v-Src expression and activity were dramatically increased in yeast cells expressing FNIP1-FLAG (Figure 4C). Expression of FNIP1-FLAG-S938A reduced this effect to the same levels seen in yeast cells with EV (Figure 4C). Expression of the FNIP1-FLAG phosphomutants S939A, S941A, S946A, and S948A gradually increased v-Src protein levels and activity (Figure 4C).
We next explored the impact of FNIP1 and its phosphorylation in chaperoning of a set of well-established non-kinase clients: glucocorticoid receptor (GR) (Garabedian and Yamamoto, 1992; Picard et al., 1990; Pratt et al., 2004), heat shock transcription factor (HSF1), and cystic fibrosis transmembrane conductance regulator (CFTR) protein. We obtained data similar to the results obtained for kinase clients. More specifically, FNIP1-FLAG expression increased GR activity in yeast, whereas the S938A mutant had no effect compared to EV (Figure 4D). The expression of individual non-phospho-S939A, S941A, S946A, and S948A FNIP1-FLAG mutants led to a gradual increase of GR activity (Figure 4D). We observed a similar pattern with the heat shock element (HSE)-LacZ activity in yeast expressing wild-type FNIP1-FLAG and the phosphomutants (Figure 4E). Lastly, we assessed the impact of FNIP1 and its phosphomutants on the stability of CFTR protein, a client of Hsp90 (Loo et al., 1998; Youker et al., 2004). We expressed HA-tagged CFTR under a constitutive promoter, PGK1 (phosphoglycerate kinase), in wild-type yeast with Hsp90α expressing FNIP1-FLAG or its phosphomutants. The rate of CFTR protein degradation was determined by cycloheximide (CHX) chase analysis. Our data showed that overexpression of FNIP1-FLAG significantly improved CFTR stability compared to the EV-expressing wild-type yeast with Hsp90α (Figure 4F). The non-phosphomutant FNIP1-FLAG-S938A expressing cells had similar CFTR stability to the wild-type yeast with Hsp90α, and this was improved in mutants S939A to S948A (Figure 4F). Our data obtained for the clients were quantified and summarized in a heatmap (Figures 4G and S4E–S4H). FNIP1 expression in yeast appears to activate the kinase and nonkinase clients in a manner that is dependent on FNIP1 phosphorylation. This effect is completely abrogated with expression of FNIP1-FLAG-S938A mutant and is gradually restored from S939A to S948A as phosphorylation increases. These data also mirror the pattern of FNIP1-FLAG phosphomutants binding to Hsp90 (Figure S4D).
O-GlcNAcylation of the Priming S938 Antagonizes FNIP1 Phosphorylation
Our proteomic analysis as well as immunoprecipitation experiments identified OGT as a binding partner for FNIP1 and FNIP1-D (Figures 1A–1C). OGT modifies serine and threonine residues by the addition of GlcNAc (Zachara et al., 2015). To determine if OGT post-translationally modifies FNIP1-D, we bacterially expressed and purified FNIP1-D-His6 and its non-phosphomutants followed by the addition of active OGT and its substrate, UDP-GlcNAc. Our in vitro data showed that OGT-FLAG can modify FNIP1-D and forms a complex with the recombinant FNIP1-D-His6 and its non-phosphomutants; however, O-GlcNAcylation of FNIP1-D was completely abrogated in the S938A mutant (Figures 5A and S5A). This suggests that S938, which is the priming serine required for relay phosphorylation, can also be O-GlcNAcylated. We also obtained similar data in HEK293 cells and demonstrated that S938 is the only site in the FNIP1-D fragment that is subject to O-GlcNAcylation (Figure 5B). FNIP1-D phosphomutants are binding with the same affinity of wildtype FNIP1-D to OGT (Figure 5B). The sequential increase of FNIP1-D phosphorylation seems to go hand in hand with an equally increased affinity for Hsp90 (Figures 5B and S5B). Interestingly, overexpression of OGT-V5 in HEK293 cells completely abrogates serine phosphorylation of FNIP1-D, presumably by increasing O-GlcNAcylation of the S938 site and blocking the priming residue for the relay phosphorylation as well as by affecting CK2 activity (Tarrant et al., 2012) (Figures 5B and 5C).
Figure 5. O-GlcNAcylation Antagonizes Phosphorylation of FNIP1.
(A) In vitro O-GlcNAcylation of recombinant FNIP1-D-His6 and its non-phosphomutants. O-GlcNAcylation and interaction of FNIP1-D-His6 and its mutants with OGT-FLAG were assessed by immunoblotting.
(B) FNIP1-D-HA and its non-phosphomutants were co-expressed with either EV or OGT-V5 in HEK293 cells. Serine phosphorylation and O-GlcNAcylation of FNIP-1-D-HA and its mutants as well as their interaction with Hsp90 and OGT were assessed by IP and immunoblotting.
(C) Schematic representation of O-GlcNAcylation of FNIP1.
FNIP1 O-GlcNAcylation Is a Prerequisite for K1119 Ubiquitination and Proteasomal Degradation
To gain further insight into regulation of FNIP1 by O-GlcNAcylation, we first examined the cellular distribution of FNIP1-FLAG and FNIP1-FLAG-S938A in HEK293 cells and showed that PTM of S938A does not affect its cellular localization (Figures 6A and S6A). OGT is an essential gene in mammalian cells (Groves and Zachara, 2017). Therefore, to examine the effects of loss of OGT on FNIP1 stability, we transiently expressed FNIP1-D-HA and FNIP1-D-HA-S938A mutant in inducible OGT knockout mouse embryonic fibroblasts (OGT(f/y) Cre-ERT2-GFP). In the presence of 4-hydroxytamoxifen (4-OHT), ERT2-Cre chimera dimerizes and translocates to the nucleus, where recombination of the loxP sites results in an Ogt null allele (Ogt is X linked). Treating these cells with 0.5μM 4-OHT for 48 h led to loss of OGT (Figure S6B). Cells were transfected with FNIP1-D-HA wild-type (WT) and S938A 24 h after 4-OHT induction and then extracted 24 h after transfection. Interestingly, OGT knockout caused an increase in the levels of WT FNIP1-D-HA, but not the S938A mutant, suggesting that O-GlcNAcylation may affect FNIP1 stability.
Figure 6. O-GlcNAcylation Leads to Ubiquitination of K1119-FNIP1 and Proteasomal Degradation.
(A) HEK293 cells were transiently transfected with FNIP1-FLAG and S938A mutant. DAPI was used for nuclear staining, and cells were analyzed by immunofluorescence microscopy.
(B) FNIP1-D-FLAG and S938A mutant were transiently expressed alone or co-expressed with OGT-V5 or CK2α-HA in HEK293 cells. 100 μM of the OGA inhibitor PUGNAc was used to treat the cells only expressing FNIP1-D-FLAG and S938A mutant overnight. FNIP1-D-FLAG and its mutant were IP, and their serine phosphorylation and O-GlcNAcylation were detected by immunoblotting.
(C) Cells in (B) were used in chase experiment by treating them with 50 μM CHX followed by protein extraction at the indicated time points. The half-life of FNIP1-D-FLAG and S938A mutant was analyzed by immunoblotting.
(D) Ubiquitination of samples in (B) detected by immunoblotting.
(E) FNIP1-D-FLAG and K1119R mutant were transiently expressed alone or co-expressed with OGT-V5 in HEK293 cells. PUGNAc was used in cells only expressing FNIP1-D-FLAG and K1119R mutant. FNIP1-D-FLAG and its mutant were IP, and ubiquitination was detected by immunoblotting.
(F) Samples in (E) were treated with 50 μM CHX followed by protein extraction at the indicated time points. The half-life of FNIP1-D-FLAG and K1119R mutant was examined by immunoblotting.
(G) Schematic representation of O-GlcNAcylation of FNIP1-S938 followed by ubiquitination of K1119.
To verify this, we used 100 μM PUGNAc (an inhibitor of O-GlcNAcase [OGA]) to treat HEK293 cells overnight, which caused a shift in equilibrium from O-phosphorylation to O-GlcNAcylation (Figure 6B). Additionally, we co-expressed FNIP1-D-FLAG or S938A mutant with OGT-V5 to increase O-GlcNAcylation or CK2-HA to enhance serine phosphorylation of FNIP1-D in HEK293 cells (Figure 6B). We used these conditions to determine if O-GlcNAcylation or phosphorylation affects FNIP1-D stability. Treating these cells with 50 μM CHX was sufficient to conduct chase experiments. Our data showed that O-GlcNAcylation of FNIP1-D leads to its destabilization, whereas CK2-mediated phosphorylation protects FNIP1-D from degradation (Figure 6C). Consistent with these data, we further demonstrated that overexpression of OGT-V5 or inhibition of OGA with PUGNAc caused ubiquitination of FNIP1-D, whereas increased CK2-mediated phosphorylation or non-phosphorylatable S938A mutation completely protected FNIP1-D from ubiquitination (Figure 6D).
There are eight lysine residues (K982, K1055. K1071, K1074, K1094, K1117, K1119, and K1134) within the FNIP1-D fragment. We therefore mutated each lysine to an arginine residue in order to identify the ubiquitination site. We first showed that only the FNIP1-D-K1119R mutant is more stable than the other lysine mutants as well as the WT FNIP1-D fragment (Figure S6C). Further experiments also revealed that FNIP1-D-HA-K1119R is unable to be ubiquitinated even with overexpression of OGT-V5 or PUGNAc-mediated inhibition of OGA in HEK293 cells, therefore providing further evidence for ubiquitination of K1119 in the FNIP1-D fragment (Figure 6E). We repeated our chase experiments with FNIP1-D-HA and FNIP1-D-HA-K1119R in HEK293 cells either co-expressing OGT-V5 or treated with the OGA inhibitor PUGNAc. As expected, hyper-O-GlcNAcylation led to destabilization of WT FNIP1-D-HA (Figure 6F). However, this effect was abrogated with the FNIP1-D-HA-K1119R mutant (Figure 6F). Our data suggest that O-GlcNAcylation of FNIP1-D-S938 is a prerequisite for ubiquitination of K1119 and degradation of FNIP1-D (Figure 6G). Phosphorylation of FNIP1-D-S938 can block O-GlcNAcylation, ubiquitination, and degradation of FNIP1-D.
DISCUSSION
Research during the past few years has provided a clear picture on how co-chaperones and PTMs can be integrated in the functional cycle of Hsp90 and consequently the chaperoning and activity of the client proteins (Bachman et al., 2018; Sahasrabudhe et al., 2017; Verba et al., 2016). However, it is unclear whether optimal activity of the clients is achieved in a stepwise, controlled manner. Our data presented here provide a mechanism where CK2-mediated relay phosphorylation of the co-chaperone FNIP1 on S938, S939, S941, S946 and S948 leads to its interaction with Hsp90 and inhibition of Hsp90 ATPase activity incrementally, based on FNIP1 phosphorylation state. However, unlike small molecules that inhibit Hsp90 function and lead to client degradation (Neckers et al., 2018), FNIP1 appears to decelerate the ATPase activity and gradually activate the client proteins. The mechanism of FNIP1 deceleration of Hsp90 ATPase activity is likely very different from that of small-molecule inhibitors. Our data for the well-established non-kinase clients (HSF1, GR, and CFTR) are in agreement with our previous studies as well as other groups’ findings that slowing of the chaperone cycle through the actions of co-chaperones or PTMs benefits stability and activity of this class of Hsp90 clients (Mollapour et al., 2014; Sahasrabudhe et al., 2017; Wang et al., 2006). It is noteworthy that some of our in vivo experiments rely on overexpression of FNIP1 and its phosphomutants, which may derail the dynamics of the regulatory co-chaperone network. However, the use of appropriate controls and the constitutive ADH1 promoter in our functional yeast studies supports the idea that these effects are a result of phosphorylation of FNIP1.
What is puzzling to us, however, is how deceleration of the chaperone cycle leads to activation of kinases. This is not in agreement with previous studies with a hyperactive yeast Hsp90-T22I, which was shown to cause hyperactivation of the kinase clients Slt2/Mpk1 (Truman et al., 2006). Although FNIP1 binding decelerates Hsp90 ATPase activity in vitro, this appears to be a surrogate for the aggregate effect of the action of FNIP1 on Hsp90. The ATPase activity likely does not correlate directly to the activation of Hsp90 kinase clients, as there is a nuanced series of intermediate states that affect client protein maturation and cannot be captured by a single metric. While we can readily observe the effect of FNIP1 on Hsp90 function cumulatively through ATPase activity, FNIP1 seems to be affecting the chaperone cycle in other ways. FNIP1 prefers binding to the Hsp90 “closed” conformation, and its collective actions appear to be favorable for both kinase and non-kinase client activation.
We demonstrate here that degree of phosphorylation of FNIP1 allows titration of the activation of the client proteins. Taking advantage of phosphomutants, we were able to capture and functionally decipher the effect of the intermediate phosphorylation states of FNIP1 and consequent gradual activation of the clients. We currently lack the phospho-specific antibodies to phosphorylated FNIP1 that would be needed to determine the abundance of intermediate phosphorylation states of FNIP1 at steady state and dissect the role each site plays in client activation. Our findings presented here, however, provide a mechanistic understanding of how the chaperone machinery may respond to the needs of the cell. Control over the degree of FNIP1 phosphorylation can allow titration of client activation depending on cellular needs.
Despite the vast amount of knowledge gained from our data, there are, however, two broad and overarching questions that remain to be answered: how does FNIP1 decelerate Hsp90 ATPase activity at the atomic level, and how does the FNIP1 interaction with Hsp90 translate to the activation of client proteins? Our previous work provided some evidence that FNIP1 binds to the middle domain of Hsp90 and perhaps masks the catalytic residue within this domain. Additional work such as structural analysis would help identify contact sites between FNIP1 and Hsp90 and whether phosphorylated serine residues create a contact surface between FNIP1-D and Hsp90. Additionally, it is unclear how exactly FNIP1 exerts this regulation over client maturation and activation. For example, does FNIP1 directly play an important role on the Hsp90-client axis, or does it function through its effect on the dynamics of other cochaperones?
The second part of our study focused on regulation and stabilization of FNIP1. Our interactome data identified both OGT and PP5 as binding proteins of FNIP1. We further demonstrated that PP5 dephosphorylates FNIP1 in a reverse pattern to its phosphorylation. In other words, PP5 dephosphorylates S948, which is essential for subsequent dephosphorylation of the S946, S941, S939, and S938. We further showed that OGT-mediated O-GlcNAcylation of FNIP1-S938 antagonizes its phosphorylation and consequently leads to ubiquitination of FNIP1-K1119 and ultimately its proteasomal degradation (Figure 7). These additional modifications provide further control over the stability and activity of FNIP1 and therefore its impact on the chaperoning of clients.
Figure 7. Post-translational Regulation of the Co-chaperone FNIP1.
CK2-mediated relay phosphorylation of FNIP1 promotes its interaction with the molecular chaperone Hsp90. This leads to gradual activation and enhanced activity and stability of the kinase and non-kinase clients. PP5-mediated dephosphorylation of FNIP1 enables O-linked N-acetylglucosamine (GlcNAc) transferase (OGT) to O-GlcNAcylate S938 on FNIP1. This consequently causes a ubiquitination of K1119 and subsequent degradation of FNIP1 in the proteasome.
Although in this study, we are focusing on PTMs of FNIP1 and the impact on its regulation as well as chaperoning of the client proteins, it is worth mentioning that OGT is an Hsp90 client. Previous work has demonstrated O-GlcNAcylation of Hsp90 (Fierro-Monti et al., 2013; Frank et al., 2014; Zhang et al., 2012) as well as CK2, which leads to its decreased activity and degradation of this kinase (Tarrant et al., 2012). Here, we also confirmed OGT to be an Hsp90 client, and overexpression of FNIP1 caused a slight upregulation of OGT (Figures S7A and S7B). However, surprisingly, total O-GlcNAcylation of protein lysate was unaffected (Figure S7B). We speculate that the hyperactive FNIP1 co-chaperone may increase OGT client activity, which in turn leads to O-GlcNAcylation and degradation of FNIP1, serving as a negative feedback loop for FNIP1 regulation. Additionally, CK2 did not appear to be an Hsp90 client, since inhibition of Hsp90 did not lead to degradation of this kinase (Figure S7A). Ultimately, our study raises new questions with regards to relay phosphorylation of co-chaperones. How exactly is FNIP1 co-chaperone affecting the activation of Hsp90 client proteins? Does a similar relay mechanism exist in other co-chaperones of Hsp90 or indeed Hsp90 itself? How do they impact the chaperoning of client proteins? This concept warrants further investigation.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact Mehdi Mollapour (mollapom@upstate.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Plasmids
For mammalian expression pcDNA5-FNIP1-HA and pcDNA5-FNIP1-D-HA constructs were cloned previously (Woodford et al., 2016). FNIP1 and FNIP1-D were subcloned with a FLAG tag into pcDNA3 (see Table S2). pZW6-CK2α-HA was purchased from Addgene (Plasmid #27086). OGT (Human, Isoform 1, 1036 amino acids) was purchased from Harvard Plasmids (HsCD00042534) and cloned into pcDNA 3.1 pDestNV5 (ThermoFisher Scientific). OGT gene was subcloned into pcDNA3 plasmid with a FLAG tag (see Table S2). The EVs containing these tags were used and also referred to as controls. For bacterial expression, FNIP1-ABC and FNIP1-D were subcloned into pRSETA containing an N-terminal His6 tag (ThermoFisher Scientific). Codon-optimized FNIP1-D was synthesized and subcloned into pET41 containing a C-terminal His6 tag. For yeast expression, FNIP1 was subcloned into p424ADH (ATCC® 87373). pYES2-FNIP1-FLAG was cloned previously (Woodford et al., 2016). pHCA/rGR (Garabedian and Yamamoto, 1992), constitutively expressing glucocoticoid receptor (GR) under control of the Alcohol dehydrogenase promoter (ADH1), the GR reporter vector pΔS26X, a URA3 vector which expresses β-galactosidase (encoded by lacZ) as a reporter gene under control of a promoter bearing 3 × GR response elements (Schena et al., 1989). 2xRLM1-LacZ reporter plasmid (Truman et al., 2006), 4XHSE-LacZ-pUp41a (Truman et al., 2006), pSM1152 (PPGK-CFTR-HA) plasmid (Youker et al., 2004), and YpRS426-GAL1-v-Src plasmid (Murphy et al., 1993) were all reported previously. STE11ΔN-cMYC-pYES2 was created using primers in (Table S2). Point mutations were made using site-directed mutagenesis (see Table S2) and confirmed by DNA sequencing.
Yeast Strains
The yeast strain pp30 (MATa, trp1–289, leu2–3,112, his3–200, ura3–52, ade2–101, lys2–801, hsc82KANMX4, hsp82KANMX4) expressing Hsp90α-Ycplac111 as the sole Hsp90 was used in this study. These yeast strains were reported previously (Mollapour et al., 2011).
Mammalian Cell Culture
Cultured human embryonic kidney (HEK293) cells were grown in Dulbecco’s Modified Eagle Medium (DMEM, Sigma-Aldrich), PC3 cells were grown in RPMI-1640 medium (Sigma-Adrich), and MEF-OGT(f/y)-Cre-ERT2-GFP cells were grown in low-glucose DMEM (Sigma-Aldrich). All media was supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich). HEK293 and PC3 cells were acquired from (American Type Culture Collection, ATCC). MEF-OGT cells were obtained from Dr. Natasha Zachara at the CardioPEG CoreC4 at Johns Hopkins University School of Medicine (Kazemi et al., 2010; O’Donnell et al., 2004). All cell lines were maintained in a CellQ incubator (Panasonic Healthcare) at 37°C in an atmosphere containing 5% CO2.
METHOD DETAILS
Yeast Growth Media
Yeast cells were grown on YPDA (2% (w/v) Bacto peptone, 1% yeast extract, 2% glucose, 20 mg/liter adenine), YPGal (2% (w/v) Bacto peptone, 1% yeast extract, 2% galactose, 20 mg/liter adenine) and YPRaf (2% (w/v) Bacto peptone, 1% yeast extract, 2% raffinose, 20 mg/liter adenine). Selective growth was on dropout 2% glucose (DO) medium with appropriate amino acids (Adams et al., 1997). Medium pH was adjusted to 6.8 with NaOH before autoclaving.
Bacterial Expression and Protein Purification
FNIP1-ABC-His6, FNIP1-D-His6, and FNIP1-D phosphomutants were expressed and purified from E. coli strain BL21 (DE3). Transformed cells were grown at 37°C in LB with 50 mg/L ampicillin until OD600 = 0.6. The cultures were then cooled to 30°C, induced with 1mM IPTG, and grown to OD600 = 1.2. Cells were harvested by centrifugation and lysed by sonication in fresh lysis buffer without detergent (20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM MgCl2, protease inhibitor cocktail (Roche) and PhosSTOP (Roche)). After sonication Triton X-100 was added to a final concentration of 1% prior to pelleting insoluble debris by centrifugation. Supernatant was collected and FNIP1 fragment expression was assessed by immunoblotting. Isolation of FNIP1-His6 fragments was accomplished by two sequential Ni-NTA agarose (QIAGEN) pulldowns. Lysate was incubated with Ni-NTA agarose (QIAGEN) for 2hr at 4°C. Proteins bound to Ni-NTA agarose were washed three times with lysis buffer (as above) followed by two washes with 50mM imidazole in lysis buffer. They were then eluted in 500mM imidazole in lysis buffer and concentrated in 10K Amicon® Ultra Centrifugal Filters (Millipore). Concentrations were determined using the Micro BCA Protein Assay Kit (Thermo Scientific) per manual protocol. Purified protein was run on an SDS-PAGE gel and Coomassie stained to confirm purity prior to use in assays.
Protein Extraction, IP, ATP-pulldown and Immunoblotting
Protein extraction from both yeast and mammalian cells was carried out using methods previously described (Mollapour et al., 2010). For immunoprecipitation, mammalian cell lysates were incubated with anti-FLAG or anti-HA antibody conjugated agarose beads (Sigma) for 2 hr at 4°C, or with anti-FNIP1 antibody for 1 hr followed by protein G agarose for 2 hr at 4°C. For ATP pulldown, lysates were incubated with ATP conjugated agarose for 1 hr at 4°C. Immunopellets were washed 4 times with fresh lysis buffer (20mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM MgCl2, 0.1% NP40, protease inhibitor cocktail (Roche), and PhosSTOP (Roche)) and eluted in 5x Laemmli buffer. Precipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Co-immunoprecipitated proteins were detected by immunoblotting with indicated dilutions of antibodies recognizing 1:8000 FLAG, 1:10000 6x-His (ThermoFisher Scientific), 1:8000 Hsp90–835-16F1, 1:10,000 GAPDH (ENZO Life Sciences), 1:4000 FLCN, 1:1000 PP5, 1:2000 OGT, 1:2000 CK2α, 1:2000 p-Akt (S473), 1:4000 Akt, 1:2000 GR, 1:1000 O-GlcNAc, 1:2000 HA, 1:1000 myc, 1:2000 V5 (Cell Signaling), 1:10,000 GST (GE Healthcare), 1:1000 phospho-serine, 1:1000 phospho-threonine (Simga-Aldrich), 1:1000 phospho-tyrosine, 1:1000 v-src (Millipore), 1:1000 ubiquitin (Santa Cruz Biotechnology), 1:2000 FNIP1, 1:2000 FNIP2 (NCI), 1:1000 FNIP1 (antibodies-online.com). Secondary antibodies raised against mouse, rabbit, and rat (Cell Signaling) and goat (Santa Cruz Biotechnology) were used at 1:4000 dilution (See Key Resources Table).
Protein Purification from Mammalian Cells
Protein extraction was carried out using methods previously described (Mollapour et al., 2010). Purification was accomplished by two sequential immunoprecipitations. Hsp90α-FLAG for ATPase measurements was expressed in PC3 cells and OGT-FLAG for in vitro GlcNAcylation was expressed in HEK293 cells. Cell lysate was incubated with anti-FLAG antibody conjugated agarose beads (Sigma) for 2hr at 4°C. Immunopellets were washed four times with fresh high salt lysis buffer (20mM Tris-HCl (pH 7.4), 500 mM NaCl, 1 mM MgCl2, 1% NP40, protease inhibitor cocktail (Roche), and PhosSTOP (Roche)) and subsequently competed off with 3x FLAG-peptide (Sigma-Aldrich) in detergent-free fresh lysis buffer (20mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM MgCl2, protease inhibitor cocktail (Roche), and PhosSTOP (Roche)) at 4°C for 1hr two times. Protein was concentrated in 50K Amicon® Ultra Centrifugal Filters (Millipore). A second round of immunoprecipitation, competition and concentration was then performed. Concentrations were determined using the Micro BCA Protein Assay Kit (Thermo Scientific) per manual protocol. Purified protein was run on an SDS-PAGE gel and Coomassie stained to confirm purity prior to use in assays.
Protein Extraction From Yeast
Yeast cells were collected from liquid culture by centrifugation and resuspended in 500μl of protein extraction buffer (20mM Tris-HCl (pH 7.4), 500 mM NaCl, 1 mM MgCl2, protease inhibitor cocktail (Roche), and PhosSTOP (Roche)) and two pellet volumes of acid washed glass beads. To lyse the cells, tubes were agitated using a bead beater (mini-Beadbeater 8, Biospec Products, USA) for 30 s at maximum speed and then 30 s on ice. This procedure was repeated 6X followed by (10,000Xg; 5 s) to pellet the beads and unbroken cells. The supernatant was transferred to a new microfuge tube and centrifuged (10,000Xg; 10 minutes) to pellet insoluble aggregates. Supernatant was then transferred to a fresh microfuge.
Assays for Hsp90 Client Activity in Yeast
Ste11ΔN and v-Src induction and activation were analyzed as described previously (Mollapour et al., 2010). Expressed Ste11ΔNcMyc, v-Src protein were detected with cMyc mouse antibody (Cell signaling Technology), EC10 mouse antibody (Millipore) respectively. v-Src activity with 4G10 mouse anti-phosphotyrosine (Millipore). β-galactosidase assay for measuring GR-LacZ expression (Garabedian and Yamamoto, 1992), HSE-LacZ expression (Hjorth-Sørensen et al., 2001), and RLM1-LacZ (Truman et al., 2006) were described previously (Mollapour et al., 2014).
β-Galactosidase Assay
Yeast cells were grown overnight to exponential phase with a cell density of 2–3 × 106 cells per ml in 50ml of the same medium at 30°C. Then, dexamethasone (DEX) was added to a final concentration of 30 μM, followed by incubation at 30°C for 2.5hr to activate the GR-LacZ. HSE-LacZ expressing yeast cells were heat shock at 39°C for 40 min. Cells were collected by centrifugation (2000 × g; 5 minutes), washed once with dH2O, and frozen at −80°C. β-Galactosidase activity was measured as previously described (Mollapour et al., 2011). Cell lysate (10 μl) was mixed with equal volume of 2XZ-buffer(60mM Na2HPO4, 60mM NaH2PO4, 5mM KCl, 0.5mM MgSO4, pH adjusted to 7.0). The mixture was added to 700 μL of 2mg/ml ONPG solution (O-Nitrophenyl-β-D-galactopyranoside dissolved in 1X Z buffer) prewarmed at 30°C. The reaction was stopped by adding 500 μL of 1M Na2CO3. The optical density at 420nm (OD420) of each reaction mixture was determined. The protein concentration of the lysate was determined by the BioRad assay. The β-Galactosidase activity was calculated using the following formula: Enzyme Activity = 1000 × OD420/minute/[10 μL × protein concentration(μg/μl)]
Hsp90 ATPase Activity Measurement
Hsp90α-FLAG was isolated from PC3 cells and FNIP1-D-His6 WT and phosphomutants were isolated from bacteria as described above. ATPase activity of Hsp90α and its inhibition by FNIP1-D were measured using the PiPer Phosphate Assay Kit (Life Technologies) as described in the manufacturer’s protocol. Standard curve with linear fit line was created from 0–20μM Pi final concentration reactions. 750ng of Hsp90α and aforementioned amounts of FNIP1-D were added to each reaction with 1mM ATP as substrate. Reactions were run in triplicate and incubated at 37°C for over 1 hour. 10μM GB (Synta) was used in drug reactions. ATP turnover was calculated as mmol Pi per mol Hsp90α per minute, and relative ATPase activity was calculated from those values, with the value of Hsp90α alone representing 100% activity.
In vitro FNIP1 Phosphorylation and Dephosphorylation
FNIP1-ABC, FNIP1-D fragment and its phospho mutants were isolated by incubating 2mg of protein extracts with 50 μl of Ni-NTA agarose (QIAGEN) for 2hr. The Ni-NTA agrose beads was washed with 30μM imidazole and then incubated with 50ng of baculovirus expressed and purified active CK2α-GST (SignalChem) for in vitro kinase assay. The assay was carried out in 50mM Tris-HCl (pH7.5), 10mM MgCl2 and 0.2mM ATP, at 28°C for 15 min. The reaction was either quenched by addition of an equal volume of protein loading buffer, or washed 3X with 100mMNaCl, 50mMTris,pH7.5 followed by resuspension in a buffer containing 100mM NaCl, 50mM Tris pH7.5, 2 m M DTT, and 1mM MnCl2. PP5 mediated dephosphorylation was performed as previously described (Dushukyan et al., 2017). The reaction was started by adding 0.25 μM PP5-GST and the samples were incubated at 30°C for 15 min. Samples were quenched by addition of an equal volume of protein loading buffer and followed by SDS/PAGE analysis.
In vitro FNIP1 O-GlcNAcylation
O-GlcNAcylation was performed in vitro using bacterially expressed and purified FNIP1-D-His6 and phosphomutants S938A, S939A, S941A, S946A and S948A. These proteins were bound to Ni-NTA agarose and then incubated with 10ng of OGT-FLAG purified from HEK293 cells and 0.2mM UDP-GlcNAc in 25μl reactions (50mM Tris-HCl, 12.5mM MgCl2, and 1mM DTT, pH7.5) for 15 min at 30°C. The reaction was stopped by addition of protein loading buffer and SDS/PAGE analysis.
Mass Spectrometry Analysis
Visible bands were excised from the gel manually and cut into small pieces approximately 1 mm × 1 mm. These gel pieces were destained using 1:1 30 mM potassium ferricyanide: 100 mM sodium thiosulfate for 10 minutes. The destained gel pieces were then washed with 25 mM ammonium bicarbonate and acetonitrile alternatively for 5 minutes each wash. This cycle of 5 minute 25 mM ammonium bicarbonate wash followed by 5 minute acetonitrile wash was repeated 3 times. To further prepare the gel pieces for digestion, the gel pieces were then dehydrated in 100% acetonitrile. After removing all acetonitrile, 25 μl of porcine trypsin (Promega) dissolved in 25 mM ammonium bicarbonate at a concentration of 4 μg/ml was added to the gel pieces. The gel pieces were then kept at room temperature overnight (approximately 12–16 hours). Following digestion, the supernatant was transferred to a second tube, and acetonitrile was added to the gel pieces to complete the extraction of digested peptides. This extract was added to the first supernatant and this combined solution, containing the extracted peptides was frozen and lyophilized. The peptides were resuspended in 5 μl of 100:99:1 acetonitrile: water: trifluoroacetic acid immediately prior to spotting on the MALDI target. For MALDI analysis, the matrix solution consisted of alpha-cyano-4-hydroxycinnamic acid (Aldrich Chemical Co. Milwaukee, WI) saturating a solution of 1:1:0.01 acetonitrile: 25 mM ammonium citrate: trifuoroacetic acid. Approximately 0.15 μl of peptide solution was spotted on the MALDI target immediately followed by 0.15 μl of the matrix solution. This combined solution was allowed to dry at room temperature. MALDI MS and MS/MS data was then acquired using the ABSCIEX TOF/TOF® 5800 Mass Spectrometer. Resultant peptide mass fingerprint and peptide sequence data was submitted to the UniProt database using the Mascot search engine to which relevance is calculated and scores are displayed. Data are presented in (Table S1) and (Figure 1A).
Immunofluorescence Staining and Microscopy
HEK293 cells were plated overnight on glass coverslips (#1) and then transiently transfected with pcDNA3-FNIP1-FLAG or pcDNA3-FNIP1-FLAG-S938A overnight. Cells were fixed in 4% paraformaldehyde for 20 minutes at room temperature, washed 3x with fresh PBS, and then permeabilized with 0.1% Triton X-100 at room temperature for 4 minutes followed 3 washes with fresh PBS. Cells were blocked in PBSAT (1% BSA, 0.5% Triton X-100 in PBS) for 30 minutes at room temperature. Cells were then incubated with primary mouse anti-FLAG (ThermoFisher Scientific) 1:250 in PBSAT for 2 hours at room temperature and then washed 3x with fresh PBSAT. Coverslips were then incubated with secondary anti-mouse Alexa Fluor® 594 (ThermoFisher Scientific) 1:900 in PBSAT at room temperature for 1 hour and then washed 3x with PBSAT followed by 2 washes with PBS. Coverslips were then mounted onto glass slides using ProLong® Gold antifade mounting media with DAPI (4′,6-diamidino-2-phenylindole) in fluorescence microscopy. (ThermoFisher Scientific). Images were obtained using a Zeiss LSM780 confocal microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistics were performed using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla, California, USA, https://www.graphpad.com). Statistical significance was ascertained between individual samples using a parametric unpaired t test. Trends were calculated through Pearson correlation coefficient testing, using the total number of phospho-sites present in each molecule as the X variable. Significance was denoted as asterisks in each figure: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Error bars represent the standard deviation (SD) for three independent experiments, unless it is indicated.
DATA AND SOFTWARE AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PRIDE: PXD012229.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-FLAG tag | Thermo Scientific | Cat# PA1-984B; RRID:AB_347227 |
| Mouse anti-FLAG tag | Thermo Scientific | Cat# F3165 |
| Mouse anti-6x-His epitope tag (HIS.H8) | Thermo Scientific | Cat# MA1-21315; RRID:AB_557403 |
| Rat anti-Hsp90 (16F1) | Enzo Life Sciences | Cat# ADI-SPA-835; RRID:AB_11181205 |
| Mouse anti-GAPDH (1D4) | Enzo Life Sciences | Cat# ADI-CSA-335; RRID:AB_10617247 |
| Rabbit anti-FLCN (D14G9) | Cell Signaling Technology | Cat# 3697; RRID:AB_2231646 |
| Rabbit anti-PP5 | Cell Signaling Technology | Cat# 2289; RRID:AB_2168757 |
| Rabbit anti-OGT | Cell Signaling Technology | Cat# 24083; RRID:AB_2716710 |
| Rabbit anti-CK2α | Cell Signaling Technology | Cat# 2656; RRID:AB_2236816 |
| Rabbit anti-phospho-Akt S473 (D9E) | Cell Signaling Technology | Cat# 2289; RRID:AB_2315049 |
| Mouse anti-Akt (2H10) | Cell Signaling Technology | Cat# 2967; RRID:AB_331160 |
| Rabbit anti-GR (D6H2L) | Cell Signaling Technology | Cat# 12041; RRID:AB_2631286 |
| Mouse anti-O-GlcNAc (CTD110.6) | Cell Signaling Technology | Cat# 9875; RRID:AB_10950973 |
| Rabbit anti-HA tag (C29F4) | Cell Signaling Technology | Cat# 3724; RRID:AB_1549585 |
| Rabbit anti-myc tag (71D10) | Cell Signaling Technology | Cat# 2278; RRID:AB_490778 |
| Rabbit anti-V5 tag | Cell Signaling Technology | Cat# 13202; RRID:AB_2687461 |
| Goat anti-GST | GE Healthcare | Cat# 27-4577-01; RRID:AB_771432 |
| Mouse anti-phospho-serine (PSR-45) | Sigma-Aldrich | Cat# P5747; RRID:AB_477376 |
| Mouse anti-phospho-threonine (PTR-8) | Sigma-Aldrich | Cat# P6623; RRID:AB_477393 |
| Mouse anti-phospho-tyrosine (4G10) | Millipore | Cat# 05–321; RRID:AB_309678 |
| Mouse anti-v-src (clone 327) | Millipore | Cat# MABS193; RRID:AB_11205595 |
| Mouse anti-ubiquitin (P4D1) | Santa Cruz Biotech | Cat# sc-8017; RRID:AB_628423 |
| Rabbit anti-FNIP1 (181) | NCI (Baba et al., 2006) | N/A |
| Rabbit anti-FNIP2 (3G) | NCI (Hasumi et al., 2008) | N/A |
| Goat anti-FNIP1 | Antibodies-online.com | Cat# ABIN238670; RRID:AB_10775640 |
| Anti-mouse secondary | Cell Signaling Technology | Cat# 7076; RRID:AB_330924 |
| Anti-rabbit secondary | Cell Signaling Technology | Cat# 7074; RRID:AB_2099233 |
| Anti-rat secondary | Cell Signaling Technology | Cat# 7077; RRID:AB_10694715 |
| Anti-goat secondary | Santa Cruz Biotech | Cat# sc-2020; RRID:AB_631728 |
| Anti-mouse Alexa Fluor secondary | ThermoFisher Scientific | Cat# A-21203; RRID:AB_2535789 |
| Bacterial and Virus Strains | ||
| BL21(DE3) | New England BioLabs | Cat# C2527I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SNX-2112 | Duke University; Dr. Timothy Haystead (Barrott and Haystead, 2013) |
CAS# 908112-43-6 |
| CHX | Research Products International | Cat# C81040; CAS# 66-81-9 |
| Caffeine | Sigma-Aldrich | Cat# C0750–500G |
| ATP | Sigma-Aldrich | Cat# FLAAS; CAS# 34369-07-8 |
| UDP-GlcNAc | Sigma-Aldrich | Cat# U4375; CAS# 91183-98-1 |
| PUGNAc | Sigma-Aldrich | Cat# A7229; CAS# 132489-69-1 |
| 4-Hydroxytamoxifen | Sigma-Aldrich | Cat# SML1666; CAS# 68392-35-8 |
| Critical Commercial Assays | ||
| Mirus TransIT-2020 | MirusBio | Cat# MIR5405 |
| PiPer Phosphate Assay | ThermoFisher Scientific | Cat# P22061 |
| Anti-FLAG M2 affinity gel | Sigma-Aldrich | Cat# A2220; RRID:AB_10063035 |
| Anti-HA agarose | ThermoFisher Scientific | Cat# 26182; RRID:AB_2532162 |
| Protein G agarose | ThermoFisher Scientific | Cat# 15-920-010 |
| ATP agarose | Novus Biologicals | Cat# 510–0002 |
| Ni-NTA Agarose | ThermoFisher Scientific | Cat# 88221 |
| ProLong Gold antifade reagent with DAPI | ThermoFisher Scientific | Cat# P36935 |
| Experimental Models: Cell Lines (Sex) | ||
| HEK293 (Female) | ATCC | Cat# CRL-1573; RRID:CVCL_0045 |
| PC3 (Male) | ATCC | Cat# CRL-1435; RRID:CVCL_0035 |
| MEFs OGT (f/y) Cre-ERT2 GFP (Male) | Dr. Natasha Zachara; CardioPEG CoreC4, Johns Hopkins University | Kazemi et al., 2010; O’Donnell et al., 2004 |
| Experimental Models: Organisms/Strains | ||
| PP30-Hsp90α | Piper et al., 2003 | n/a |
| Oligonucleotides | ||
| DNA primers | Eurofins Genomics | See Table S2 |
| Recombinant DNA | ||
| pcDNA5-FNIP1-D-HA | Baba et al., 2006 | n/a |
| pZW6-CK2α-HA | Addgene (Turowec et al., 2010) | Cat# 27086 |
| pDONR221-OGT | Harvard Plasmids | HsCD00042534 |
| pcDNA 3.1 pDestNV5 | ThermoFisher Scientific | Cat# 12290010 |
| pYES2-FNIP1-FLAG | Woodford et al., 2016 | n/a |
| p424-ADH | ATCC | Cat# 87373 |
| pRSETA | ThermoFisher Scientific | Cat# V35120 |
| pET-41a(+) | Millipore | Cat# 70556 |
| pHCA/rGR | Garabedian and Yamamoto, 1992 | n/a |
| PΔS26X | Schena et al., 1989 | n/a |
| 2xRLM1-LacZ | Truman et al., 2006 | n/a |
| 4XHSE-LacZ-pUp41a | Truman et al., 2006 | n/a |
| pSM1152 (PPGK-CFTR-HA) | Youker et al., 2004 | n/a |
| YpRS426-GAL1-v-Src | Murphy et al., 1993 | n/a |
| STE11ΔN-cMYC-pYES2 | This paper | n/a |
| Deposited Data | ||
| Mass spectrometry data | This study | PXD012229 |
| Software and Algorithms | ||
| GraphPad Prism 7.0 | GraphPad Software | https://www.graphpad.com/ |
Highlights.
Casein-kinase-2-mediated sequential phosphorylation of the co-chaperone FNIP1
FNIP1 relay phosphorylation leads to gradual activation of Hsp90 clients
Serine/threonine protein phosphatase 5 (PP5) dephosphorylates FNIP1
O-GlcNAcylation causes ubiquitination and proteasomal degradation of FNIP1
ACKNOWLEDGMENTS
We thank J. L. Brodsky for the yeast CFTR-HA plasmids and G. Yildirir for cloning OGT-V5 plasmid. This work was supported with funds from the National Institute of General Medical Sciences of the NIH under award number R01GM124256 (M.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was also partly supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E (L.S.); SUNY Upstate Medical University, The Upstate Foundation (M.M. and D.B.); the Carol M. Baldwin Breast Cancer Fund (D.B. and M.M.); and the Urology Care Foundation-American Urological Association (M.M.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2019.01.018.
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, and Stearns T (1997). Methods in Yeast Genetics (Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF 3rd, Hartley JL, et al. (2006). Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. USA 103, 15552–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman AB, Keramisanou D, Xu W, Beebe K, Moses MA, Vasantha Kumar MV, Gray G, Noor RE, van der Vaart A, Neckers L, and Gelis I (2018). Phosphorylation induced cochaperone unfolding promotes kinase recruitment and client class-specific Hsp90 phosphorylation. Nat. Commun 9, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrott JJ, and Haystead TA (2013). Hsp90, an unlikely ally in the war on cancer. FEBS J. 280, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesaro L, and Pinna LA (2015). The generation of phosphoserine stretches in phosphoproteins: mechanism and significance. Mol. Biosyst 11, 2666–2679. [DOI] [PubMed] [Google Scholar]
- Dushukyan N, Dunn DM, Sager RA, Woodford MR, Loiselle DR, Daneshvar M, Baker-Williams AJ, Chisholm JD, Truman AW, Vaughan CK, et al. (2017). Phosphorylation and ubiquitination regulate protein phosphatase 5 activity and its prosurvival role in kidney cancer. Cell Rep 21, 1883–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echtenkamp FJ, and Freeman BC (2012). Expanding the cellular molecular chaperone network through the ubiquitous cochaperones. Expanding the cellular molecular chaperone network through the ubiquitous cochaperones. Biochim. Biophys. Acta 1823, 668–673. [DOI] [PubMed] [Google Scholar]
- Fierro-Monti I, Echeverria P, Racle J, Hernandez C, Picard D, and Quadroni M (2013). Dynamic impacts of the inhibition of the molecular chaperone Hsp90 on the T-cell proteome have implications for anti-cancer therapy. PLoS ONE 8, e80425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom GA, Lemieszek M, Fortunato EA, and Johnson JL (2008). Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol. Biol. Cell 19, 5249–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LA, Sutton-McDowall ML, Brown HM, Russell DL, Gilchrist RB, and Thompson JG (2014). Hyperglycaemic conditions perturb mouse oocyte in vitro developmental competence via beta-O-linked glycosylation of heat shock protein 90. Hum. Reprod 29, 1292–1303. [DOI] [PubMed] [Google Scholar]
- Garabedian MJ, and Yamamoto KR (1992). Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol. Biol. Cell 3, 1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves JA, and Zachara NE (2017). Characterization of tools to detect and enrich human and mouse O-GlcNAcase. Glycobiology 27, 791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hong SB, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, and Schmidt LS (2008). Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene 415, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth-Sørensen B, Hoffman ER, Lissin NM, Sewell AK, and Jakobsen BK (2001). Activation of heat shock transcription actor in yeast is not influenced by the levels of expression of heat shock proteins. Mol. Microbiol 39, 914–923. [DOI] [PubMed] [Google Scholar]
- Kazemi Z, Chang H, Haserodt S, McKen C, and Zachara NE (2010). O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner. J. Biol. Chem 285, 39096–39107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo MA, Jensen TJ, Cui L, Hou Y, Chang XB, and Riordan JR (1998). Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 17, 6879–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvion JF, Abbas-Terki T, and Picard D (1998). Hsp90 is required for pheromone signaling in yeast. Mol. Biol. Cell 9, 3071–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP (2010). Gymnastics of molecular chaperones. Mol. Cell 39, 321–331. [DOI] [PubMed] [Google Scholar]
- Millson SH, Truman AW, King V, Prodromou C, Pearl LH, and Piper PW (2005). A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 4, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Donnelly AC, Beebe K, Tokita MJ, Lee MJ, Lee S, Morra G, Bourboulia D, Scroggins BT, et al. (2010). Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol. Cell 37, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Truman AW, Xu W, Vaughan CK, Beebe K, Konstantinova A, Vourganti S, Panaretou B, Piper PW, et al. (2011). Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol. Cell 41, 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Bourboulia D, Beebe K, Woodford MR, Polier S, Hoang A, Chelluri R, Li Y, Guo A, Lee MJ, et al. (2014). Asymmetric Hsp90 N domain SUMOylation recruits Aha1 and ATP-competitive inhibitors. Mol. Cell 53, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Bergman M, and Morgan DO (1993). Suppression of c-Src activity by C-terminal Src kinase involves the c-Src SH2 and SH3 domains: analysis with Saccharomyces cerevisiae. Mol. Cell. Biol 13, 5290–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, and Lindquist S (1995). Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol 15, 3917–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L, Blagg B, Haystead T, Trepel JB, Whitesell L, and Picard D (2018). Methods to validate Hsp90 inhibitor specificity, to identify off-target effects, and to rethink approaches for further clinical development. Cell Stress Chaperones 23, 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell N, Zachara NE, Hart GW, and Marth JD (2004). Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol 24, 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, and Pearl LH (1998). ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17, 4829–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D (2002). Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci 59, 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, and Yamamoto KR (1990). Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 348, 166–168. [DOI] [PubMed] [Google Scholar]
- Piper PW, Millson SH, Mollapour M, Panaretou B, Siligardi G, Pearl LH, and Prodromou C (2003). Sensitivity to Hsp90-targeting drugs can arise with mutation to the Hsp90 chaperone, cochaperones and plasma membrane ATP binding cassette transporters of yeast. Eur. J. Biochem 270, 4689–4695. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Galigniana MD, Morishima Y, and Murphy PJ (2004). Role of molecular chaperones in steroid receptor action. Essays Biochem. 40, 41–58. [DOI] [PubMed] [Google Scholar]
- Prodromou C (2016). Mechanisms of Hsp90 regulation. Biochem. J 473, 2439–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl A, Rohrberg J, and Buchner J (2013). The chaperone Hsp90: changing partners for demanding clients. Trends Biochem. Sci 38, 253–262. [DOI] [PubMed] [Google Scholar]
- Sager RA, Woodford MR, and Mollapour M (2018a). The mTOR independent function of Tsc1 and FNIPs. Trends Biochem. Sci 43, 935–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager RA, Woodford MR, Neckers L, and Mollapour M (2018b). Detecting posttranslational modifications of Hsp90. Methods Mol. Biol 1709, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabudhe P, Rohrberg J, Biebl MM, Rutz DA, and Buchner J (2017). The plasticity of the Hsp90 co-chaperone system. Mol. Cell 67, 947–961. [DOI] [PubMed] [Google Scholar]
- Schena M, Freedman LP, and Yamamoto KR (1989). Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 3, 1590–1601. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM, and Buchner J (2017). The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol 18, 345–360. [DOI] [PubMed] [Google Scholar]
- St-Denis N, Gabriel M, Turowec JP, Gloor GB, Li SS, Gingras AC, and Litchfield DW (2015). Systematic investigation of hierarchical phosphorylation by protein kinase CK2. J. Proteomics 118, 49–62. [DOI] [PubMed] [Google Scholar]
- Tarrant MK, Rho HS, Xie Z, Jiang YL, Gross C, Culhane JC, Yan G, Qian J, Ichikawa Y, Matsuoka T, et al. (2012). Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol 8, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW, Millson SH, Nuttall JM, King V, Mollapour M, Prodromou C, Pearl LH, and Piper PW (2006). Expressed in the yeast Saccharomyces cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2p (Mpk1p) cell integrity stress-activated protein kinase. Eukaryot. Cell 5, 1914–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowec JP, Duncan JS, French AC, Gyenis L, St Denis NA, Vilk G, and Litchfield DW (2010). Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: strategies to identify CK2 substrates and manipulate its activity in mammalian cells. Methods Enzymol. 484, 471–493. [DOI] [PubMed] [Google Scholar]
- Verba KA, and Agard DA (2017). How Hsp90 and Cdc37 lubricate kinase molecular switches. Trends Biochem. Sci 42, 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verba KA, Wang RY, Arakawa A, Liu Y, Shirouzu M, Yokoyama S, and Agard DA (2016). Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352, 1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44 (D1), D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, et al. (2006). Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127, 803–815. [DOI] [PubMed] [Google Scholar]
- Woodford MR, Dunn DM, Blanden AR, Capriotti D, Loiselle D, Prodromou C, Panaretou B, Hughes PF, Smith A, Ackerman W, et al. (2016). The FNIP co-chaperones decelerate the Hsp90 chaperone cycle and enhance drug binding. Nat. Commun 7, 12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, and Brodsky JL (2004). Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15, 4787–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara N, Akimoto Y, and Hart GW (2015). The O-GlcNAc modification In Essentials of Glycobiology, Third Edition, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, and Prestegard JH, et al. , eds. (Cold Spring Harbor Laboratory Press; ), pp. 239–251. [Google Scholar]
- Zhang F, Snead CM, and Catravas JD (2012). Hsp90 regulates O-linked b-N-acetylglucosamine transferase: a novel mechanism of modulation of protein O-linked β-N-acetylglucosamine modification in endothelial cells. Am. J. Physiol. Cell Physiol 302, C1786–C1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierer BK, Rübbelke M, Tippel F, Madl T, Schopf FH, Rutz DA, Richter K, Sattler M, and Buchner J (2016). Importance of cycle timing for the function of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol 23, 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PRIDE: PXD012229.