Abstract
Purpose
Molecular characterization of prostate cancer, including The Cancer Genome Atlas, has revealed distinct subtypes with underlying genomic alterations. One of these core subtypes, SPOP (speckle-type POZ protein) mutant prostate cancer, has previously only been identifiable via DNA sequencing, which has made the impact on prognosis and routinely used risk stratification parameters unclear.
Methods
We have developed a novel gene expression signature, classifier (Subclass Predictor Based on Transcriptional Data), and decision tree to predict the SPOP mutant subclass from RNA gene expression data and classify common prostate cancer molecular subtypes. We then validated and further interrogated the association of prostate cancer molecular subtypes with pathologic and clinical outcomes in retrospective and prospective cohorts of 8,158 patients.
Results
The subclass predictor based on transcriptional data model showed high sensitivity and specificity in multiple cohorts across both RNA sequencing and microarray gene expression platforms. We predicted approximately 8% to 9% of cases to be SPOP mutant from both retrospective and prospective cohorts. We found that the SPOP mutant subclass was associated with lower frequency of positive margins, extraprostatic extension, and seminal vesicle invasion at prostatectomy; however, SPOP mutant cancers were associated with higher pretreatment serum prostate-specific antigen (PSA). The association between SPOP mutant status and higher PSA level was validated in three independent cohorts. Despite high pretreatment PSA, the SPOP mutant subtype was associated with a favorable prognosis with improved metastasis-free survival, particularly in patients with high-risk preoperative PSA levels.
Conclusion
Using a novel gene expression model and a decision tree algorithm to define prostate cancer molecular subclasses, we found that the SPOP mutant subclass is associated with higher preoperative PSA, less adverse pathologic features, and favorable prognosis. These findings suggest a paradigm in which the interpretation of common risk stratification parameters, particularly PSA, may be influenced by the underlying molecular subtype of prostate cancer.
INTRODUCTION
Prostate cancer is a clinically and molecularly heterogeneous disease.1-4 Current risk stratification guidelines, such as those from the National Comprehensive Cancer Network,5 the American Urological Association/American Society for Therapeutic Radiology and Oncology,6 and the European Association of Urology–European Society for Radiotherapy–Oncology–International Society of Geriatric Oncology7 use clinical and pathologic parameters, including the level of prostate-specific antigen (PSA), to guide management decisions for clinically localized disease. PSA is also used in a number of other clinical scenarios in prostate cancer, including initial diagnosis, monitoring for recurrence after primary therapy, and monitoring disease burden and treatment response for metastatic disease.
The emerging next-generation DNA and RNA sequencing data point toward distinct molecular subclasses of prostate cancer,2-4,8,9 but their clinical impact remains unclear. The Cancer Genome Atlas (TCGA) study identified seven core prostate cancer subtypes, which were defined by underlying genomic alterations.4 One key molecular subclass, which represents approximately 10% of prostate cancers, harbors recurrent missense mutations in the E3 ubiquitin ligase component SPOP.2-4,10-13 To date, identification of SPOP mutations has required mutational analysis using genomic (DNA) sequencing. These cohorts typically have limited follow-up,3,4,10 thus limiting definitive conclusions about the clinical effect of the molecular subtypes. In contrast, gene expression (RNA) data are widely available from a variety of prostate cancer cohorts, often with the long follow-up necessary to define prognostic impact.14-21
Here, we reported the first development and validation of the Subclass Predictor Based on Transcriptional Data (SCaPT) model, which we used to define key prostate cancer molecular subclasses, including SPOP mutant cancer, using gene expression data. We used the SCaPT model to predict the molecular subclass from a retrospective cohort that included 1,626 patient samples and a prospective cohort that included 6,532 samples using genome-wide microarray gene expression data from a clinically available prognostic assay (Decipher; GenomeDx Biosciences, Vancouver, BC, Canada), and we explored the clinicopathologic and prognostic associations of key molecular subclasses of prostate cancer.
METHODS
Prostate Cancer Tumor Samples and Microarray Data
We used a total of 8,559 radical prostatectomy (RP) tumor expression profiles for training, testing, and validation. For training and testing, we used RNA sequencing expression and DNA mutation data from TCGA prostate cancer project (n = 333)4 and the Weill Cornell Medicine (WCM) sequencing (n = 68) cohort. For validation, expression profiles of retrospective (n = 1,626) and prospective (n = 6,532) cohorts were derived from the Decipher Genomics Resource Information Database (GRID) registry (ClinicalTrials.gov identifier: NCT02609269). The retrospective GRID cohort was pooled from seven published microarray studies: Cleveland Clinic,22 Erasmus MC,23 Johns Hopkins,15 Memorial Sloan Kettering Cancer Center,1 Mayo Clinic (Mayo I and Mayo II),20,24 and Thomas Jefferson University.21 The prospective GRID cohort was from clinical use of the Decipher test. DNA and RNA from the TCGA and WCM cohorts were extracted from fresh frozen RP tumor tissue, as previously described.4 RNA from the GRID cohorts was extracted from routine formalin-fixed, paraffin-embedded RP tumor tissues, amplified, and hybridized to Human Exon 1.0 ST microarrays (Thermo Fisher Scientific, Waltham, MA).24,25
SPOP Mutant Transcriptional Signature
We developed the SPOP mutant transcriptional signature, which includes 212 genes differentially expressed between SPOP mutant and wild-type samples from TCGA prostate cancer RNA sequencing data4 (Fig 1). Low-expressed genes (mean resem [RNA-seq by expectation-maximization] < 1) were filtered before the analysis. Specifically, we identified significantly differentially expressed genes by comparing SPOP mutant and wild-type cases as determined from DNA mutational analyses among TCGA samples that lacked ETS family gene fusions (ERG, ETV1, ETV4, and FLI1) using Wilcoxon rank-sum test,26 and controlled for false discovery using Benjamini-Hochberg adjustment (false-discovery rate ≤ 0.0001).27 By performing DESeq2 (DESeq2 v1.20.0; https://bioconductor.org/packages/release/bioc/html/DESeq2.html) on SPOP mutant and wild-type cases (the same approach used initially), we found 300 differentially expressed genes at a false discovery rate of < 0.05 via DESeq2, and 105 genes that overlapped with the 212-gene list. The overlap between two methods was significant (P < 2.2 × 10−16), which confirmed similar results when applying these methods.
Fig 1.
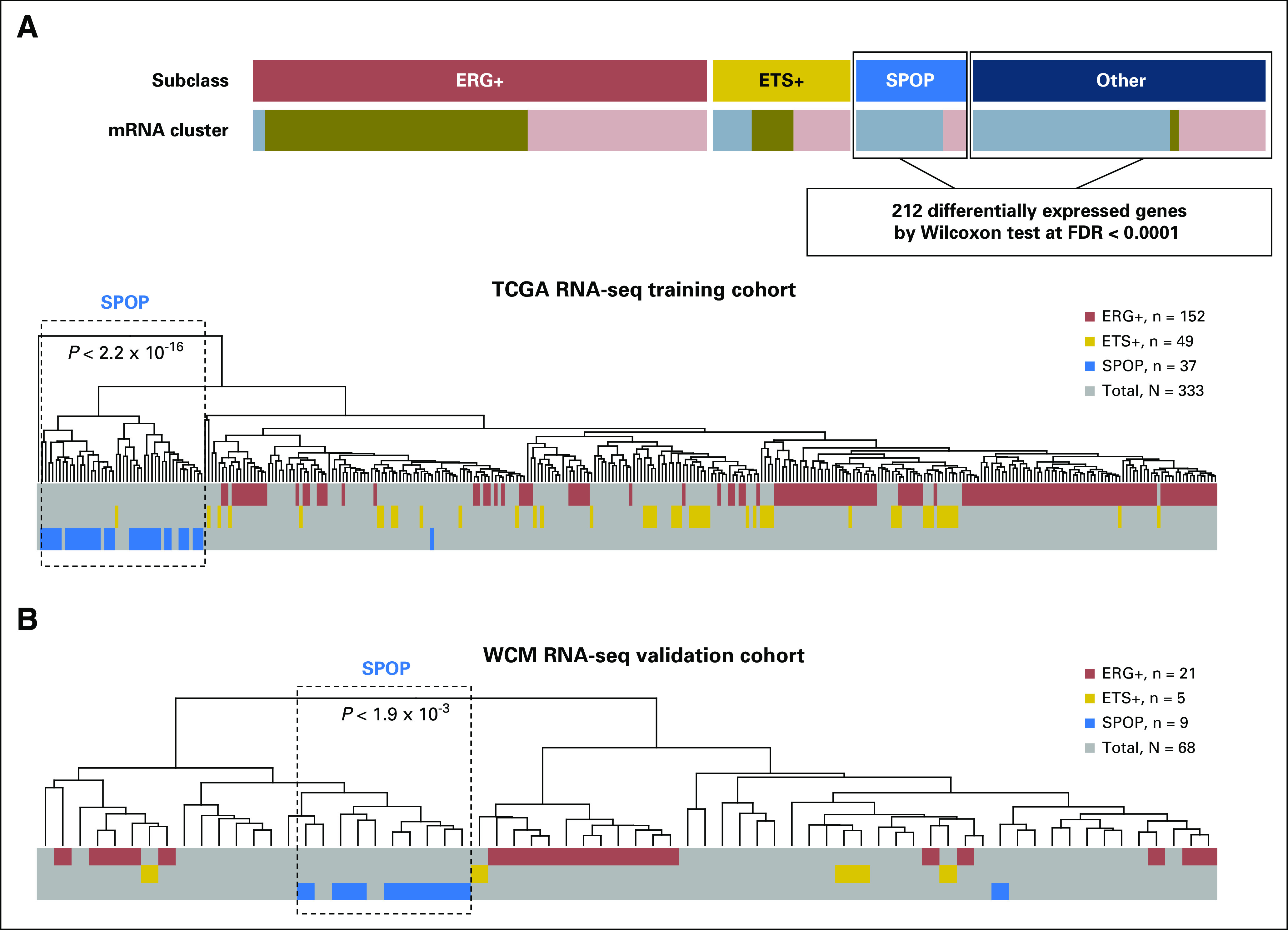
SPOP mutant transcriptional signature. (A) SPOP mutant transcriptional signature that included 212 differentially expressed genes between SPOP mutant and wild-type samples from The Cancer Genome Atlas (TCGA) non-ETS fusion RNA sequencing (RNA-seq) data. The signature was generated from the TCGA study and tested back in the TCGA training cohort. Significant enrichment of SPOP mutant cases was based on hierarchical clustering of 333 TCGA prostate cancer samples. Different colors represent molecular subclasses from genomic and transcriptomic annotations. (B) Significant enrichment of SPOP mutant case from 68 Weill Cornell Medicine (WCM) prostate cancer samples with SPOP mutant transcriptional signature on the basis of hierarchical clustering. ERG, ERG-fusion position; ETS: other ETS fusion positive; FDR, false-discovery rate.
SCaPT Development on the Basis of SPOP Mutant Transcriptional Signature and the Support Vector Machine Model
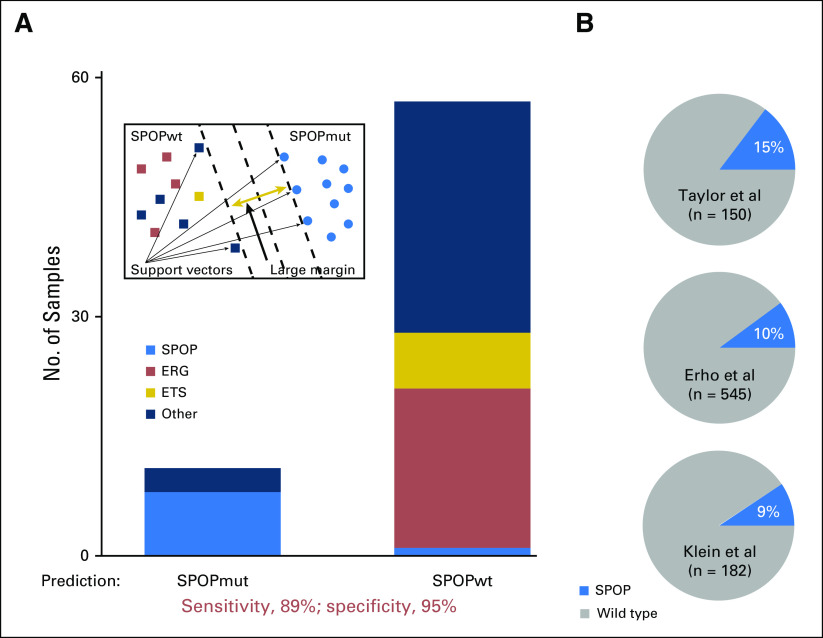
To predict tumors in the SPOP mutant subclass in the absence of DNA sequencing data—that is, microarray data sets—we developed the SCaPT model on the basis of the support vector machine (SVM) model.28-30 Given a set of training data marked with two categories, SVM builds a model that assigns testing data into one category or the other, which makes it a nonprobabilistic binary linear classifier (Fig 2A). In our SCaPT model, training data were defined as the transcriptional z-scores of SPOP mutant signature from TCGA cohort. Testing data would be the transcriptional z-scores from RNA sequencing or microarray expression data of SPOP mutant signature. By performing 10-fold cross-validation on the TCGA training data set, we found cost—cost of constraints violation—equal to 0.04 to yield the model with the highest sensitivity and specificity. In the following analysis, we used cost equal to 0.04 to predict SPOP mutant subclass.
Fig 2.
High accuracy and confidence of SPOP mutant (SPOPmut) subclass prediction on Weill Cornell Medicine (WCM) and Gene Expression Omnibus (GEO) data set by the SCaPT (SubClass Predictor Based on Transcriptional Data) model. (A) SCaPT model example and its SPOPmut prediction on WCM prostate cancer RNA sequencing (RNA-seq) data. Different colors represent molecular subclasses from genomic and transcriptomic annotations. (B) The SPOPmut prediction of the SCaPT model on three independent exon array data downloaded from the GEO database. Data from Taylor et al,1 Erho et al,24 and Kelin et al.22 wt, wild type.
Prostate Cancer Molecular Subclass Prediction by Decision Tree
In each individual study of the retrospective and prospective GRID cohorts, the SPOP mutant subclass was first predicted using the SCaPT model. Next, using a decision tree and previously developed microarray-based classifiers for the ERG-positive and ETS-positive subtypes,25 we classified the remaining cases in each cohort. Some cases with both predicted SPOP mutant and ERG-positive/ETS-positive status were classified as conflict subclass, and the rest without SPOP mutant calling and outlier expression were considered as other subclass (Data Supplement).
Statistical Analysis
Statistical analyses were performed in R (version 3.1.2; https://www.r-project.org/). All statistical tests were two sided, with the significance level set at P < .05. Univariable logistic regression analyses were performed on the combined cohort to test the statistical association between SPOP mutant status and clinical variables, including age, race, preoperative PSA, Gleason score, lymph node invasion, surgical margin status, extracapsular extension, and seminal vesicle invasion. We evaluated the associations between SPOP mutant status and patient outcomes, including biochemical recurrence, metastasis, and prostate cancer–specific mortality, on the basis of Kaplan-Meier analysis. Preoperative PSA from TCGA and Taylor cohorts were downloaded from cBioPortal (http://www.cbioportal.org/).31,32
RESULTS
Development and Validation of a Transcriptional Signature for SPOP Mutant Prostate Cancer
To build an SPOP mutant prediction model that could be applied to RNA expression data, we first developed a transcriptional signature of SPOP mutant tumors. As SPOP mutations are mutually exclusive with ERG and other ETS rearrangements,13 we excluded prostate cancer samples with ETS fusions to define SPOP mutant–specific gene expression effects (Fig 1A). Using TCGA RNA sequencing data, we identified 212 differentially expressed genes between SPOP mutant and wild-type samples (Data Supplement). Among those 212 genes, we found that upregulated genes from the SPOP mutant subgroup were enriched in transport vesicle membrane and oxidoreductase activity, but that there was no significant enriched function in downregulated genes from the SPOP mutant subgroup (Data Supplement). Applying the SPOP mutant transcriptional signature on the training data of 333 TCGA prostate cancer samples, we found significant enrichment (P < 2.2 × 10−16) of SPOP mutant cases on the basis of unsupervised clustering (Fig 1A and Data Supplement).
To test the SPOP mutant transcriptional signature, we used the WCM RNA sequencing cohort with SPOP mutant annotations based on whole-exome DNA sequencing (Data Supplement). In this independent cohort, we found significant enrichment (P < 1.9×10−3) of SPOP mutant tumors in one subcluster based on unsupervised clustering (Fig 1B and Data Supplement). We next used the SPOP mutant transcriptional signature in a locked SVM model—that is, with fixed parameters on the basis of the training step using the TCGA cohort (see Methods)—to generate scores for each sample in the WCM cohort. We found 89% sensitivity and 95% specificity of SPOP mutant prediction compared with DNA mutation annotation (Fig 2A). Finally, we applied the SCaPT model to the GRID microarray expression data, which do not have SPOP mutation annotation from DNA analysis. Between 9% and 15% of samples in the cohorts were predicted as SPOP mutant subclass, which is consistent with the known prevalence of SPOP mutations at the genomic level in previous prostate cancer studies3,4,10,12 (Fig 2B). Overall, these results demonstrated that the SCaPT model predicted SPOP mutant subclass on the basis of the transcriptional data with high accuracy and confidence.
Molecular Subtyping of 8,158 Patients Using the SCaPT and Decision Tree
We applied the SCaPT model and decision tree to 8,158 patients from retrospective and prospective GRID cohorts. Among the retrospective cohort with 1,626 RP specimens, we predicted 9% (range, 2% to 13%) of samples to be SPOP mutant subclass (Data Supplement). Previously defined expression thresholds25 classified 42% (35% to 68%) as ERG positive and 11% (8% to 13%) as non-ERG ETS positive, as well as 35% without outlier expression, which we defined as an “other” subtype (Fig 3A and Data Supplement). Approximately 2% of samples with both predicted SPOP mutant and ERG-positive/ETS-positive status were classified as conflict cases. Among the prospective cohort with 6,532 RP specimens, we predicted 8% of cases to be SPOP mutant subclass, 41% as ERG positive, 12% as ETS positive, 39% as other subtype, and 1% as conflict cases (Fig 3C). The percentage of each molecular subclass was consistent with that reported in previous prostate cancer studies,1-4,14,25 which supports the validity of our approach.
Fig 3.

The SPOP mutant prediction and its impacts on clinical and prognostic outcomes from retrospective (n = 1,626) and prospective GRID (n = 6,532) cohorts. (A) The pie chart of predicted molecular subclasses from the retrospective cohort with 1,626 samples, on the basis of the SCaPT (SubClass Predictor based on Transcriptional data) model and decision tree. Different colors represent molecular subclasses. (B) Associations between predicted SPOP mutant status and clinical variables via univariable analysis in the retrospective cohort, with SPOP wild type as reference. Box size indicates the significance from univariable analysis. (C) The pie chart of predicted molecular subclasses from the prospective GRID cohort with 6,532 samples, on the basis of the SCaPT model and decision tree. Different colors represent molecular subclasses. (D) Associations between predicted SPOP mutant status and clinical variables via univariable analysis in the prospective GRID cohort, with SPOP wild type as reference. Box size indicates the significance from univariable analysis. ERG, ERG-fusion position; ETS, other ETS fusion positive; OR, odds ratio; PSA, prostate-specific antigen.
SPOP Mutant Subclass Associated With Favorable Pathology at RP
We used binominal univariable analysis to compare clinical and pathologic characteristics between SPOP mutant and wild-type subclasses (Figs 3B and 3D). SPOP mutant subclass was less likely to harbor adverse pathologic features, such as positive surgical margins, extraprostatic extension, and seminal vesicle invasion, compared with wild-type subclass in both retrospective and prospective cohorts (Figs 3B and 3D). Surprisingly, SPOP mutant cancers were associated with higher preoperative PSA in both retrospective (odds ratio [OR], 2.00; 95% CI, 1.43 to 2.76; P < .001) and prospective cohorts (OR, 1.28; 95% CI, 1.01 to 1.60; P = .074).
In contrast, consistent with prior reports,25,33-36 tumors in the ERG-positive subclass were associated with lower preoperative PSA in both retrospective (OR, 1.51; 95% CI, 1.27 to 1.79; P < .001) and prospective cohorts (OR, 1.47; 95% CI, 1.29 to 1.68; P < .001), but were more likely to have extraprostatic extension in both retrospective (OR, 1.37; 95% CI, 1.16 to 1.62; P = .002) and prospective cohorts (OR, 1.43; 95% CI, 1.28 to 1.60; P < .001; Data Supplement). These data suggest that the SPOP mutant subclass was associated with more favorable pathologic outcomes at RP, but expresses higher levels of PSA and was associated with tumors from older men, whereas the opposite was true in the ERG-positive subclass of prostate cancer.
Consistent Association Between SPOP Mutation and Higher PSA in Multiple Cohorts
The inverse association of PSA and prognosis in specific prostate cancer subtypes has potential clinical implications. To independently validate the association of SPOP mutant status and higher preoperative PSA, we examined multiple distinct RP cohorts (GRID, TCGA, Taylor, and WCM). We observed a similar trend of higher preoperative PSA in SPOP mutant cases, and the SPOP mutant subclass was more enriched in the higher PSA subgroups (PSA > 10; Fig 4A). All cohorts demonstrated significantly higher PSA in SPOP mutant than in the ERG-positive subclass via Kolmogorov-Smirnov test. On univariable analysis across cohorts, SPOP mutant status was significantly associated with higher preoperative PSA (Fig 4B), whereas ERG fusion status was significantly associated with lower preoperative PSA (Fig 4C).
Fig 4.
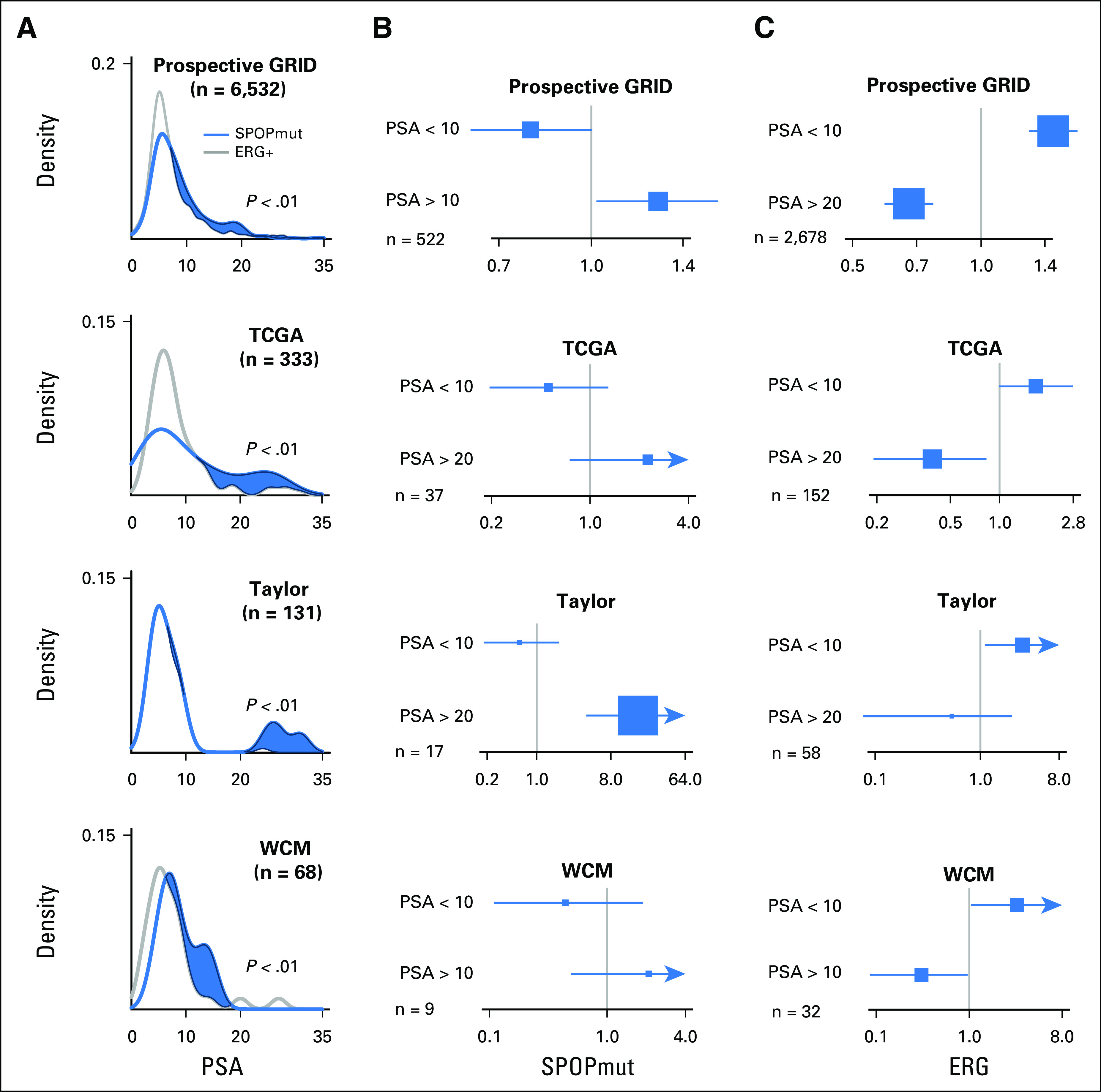
Association of SPOP mutant (SPOPmut) status and higher prostate-specific antigen (PSA) from four independent studies. (A) Enrichment of SPOPmut cases among higher PSA subgroups from prospective GRID, The Cancer Genome Atlas (TCGA), Taylor, and Weill Cornell Medicine (WCM) cohorts. P value indicates the significant difference between SPOPmut and ERG-positive cases via Kolmogorov-Smirnov test in each cohort. (B) Positive association between SPOPmut status and higher PSA via univariable analysis. The number of cases is shown in each cohort. (C) Positive association between ERG fusion status and lower PSA via univariable analysis. The number of cases is shown in each cohort.
SPOP Mutation Is Associated With Favorable Clinical Outcomes After RP
On Kaplan-Meier analysis, the SPOP mutant subclass had the highest biochemical-free, metastasis-free, and lowest prostate cancer–specific mortality compared with ERG-positive, ETS-positive, and other subtypes in the retrospective GRID cohort (Fig 5B and Data Supplement). Whereas long-term outcomes were not available for the prospective GRID cohort, we evaluated the association with metastasis risk using the Decipher score, a validated metric for prostate cancer metastatic potential.22,24,37,38 We found fewer SPOP mutant tumors in the Decipher high-risk score (> 0.6) group compared with the low- and average-risk groups (Data Supplement), which again is consistent with a favorable prognosis subtype. Together, these data support that SPOP mutant prostate cancer had favorable prognosis after RP, despite its association with higher preoperative PSA.
Fig 5.
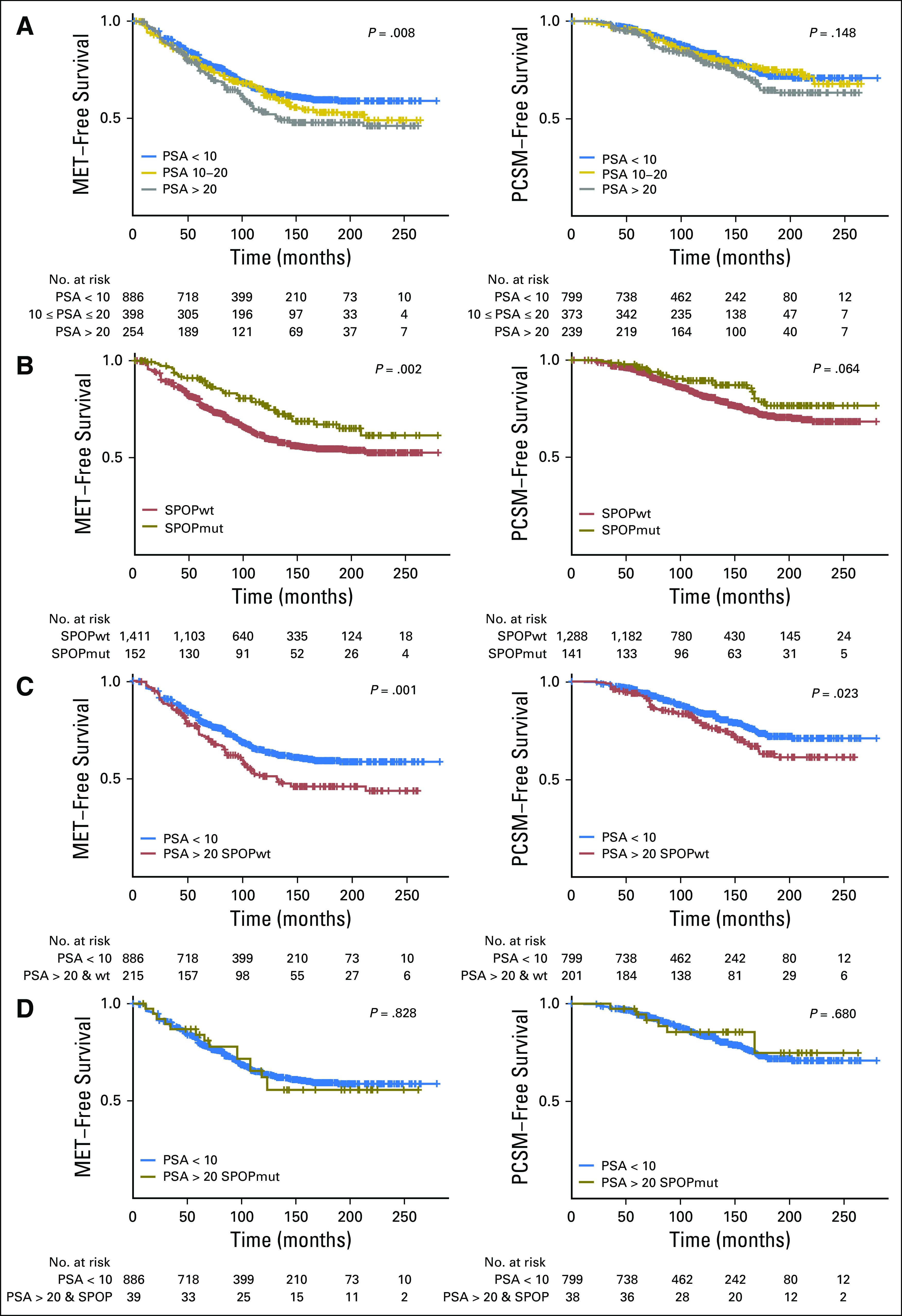
Favorable prognosis in high-risk prostate-specific antigen (PSA) subgroup in the SPOP mutant (SPOPmut) subclass. (A) Clinical outcome difference between lower, average, and higher PSA groups via Kaplan-Meier analyses for metastasis (MET) and prostate cancer–specific mortality (PCSM) –free survival rates. (B) Significant clinical outcome difference between SPOPmut and wild-type (wt) subclasses via Kaplan-Meier analysis of MET- and PCSM-free survival rates. (C) Significant clinical outcome difference between lower PSA (PSA < 10 ng/mL) and SPOP wild type (SPOPwt) subclass within higher PSA (PSA > 20 ng/mL) groups via Kaplan-Meier analysis of MET- and PCSM-free survival rates. (D) No clinical outcome difference between lower PSA (PSA < 10 ng/mL) and SPOPmut subclass within higher PSA (PSA > 20 ng/mL) groups via Kaplan-Meier analysis of MET- and PCSM-free survival rates.
Favorable Prognosis in High-Risk PSA Subgroup in the SPOP Mutant Subclass
Pretreatment PSA level is a standard component of risk stratification for prostate cancer, with a PSA level > 20 ng/mL considered to be high risk by a number of risk assessment methods.39-41 Consistent with this, higher PSA was associated with worse metastasis-free survival and prostate cancer–specific mortality in the retrospective cohorts (Fig 5A); however, SPOP mutant status had a dramatic effect on prognosis within the subgroup of patients with high PSA. Among patients with a PSA level > 20 ng/mL, SPOP mutant tumors were associated with better clinical outcomes, which were comparable to the lowest PSA subgroup (PSA < 10 ng/mL; Fig 5D). These data suggest that, among patients with high-risk PSA levels, the SPOP mutant subtype was associated with favorable prognosis in patients who underwent RP. More broadly, these data establish the principle that the identification of molecular subtype may impact the interpretation of PSA-based risk stratification in a variety of clinical scenarios.
DISCUSSION
PSA is a critical component of risk assessment systems in multiple clinical scenarios: risk of prostate cancer before diagnosis, stratification of newly diagnosed disease, monitoring for recurrence after initial therapy, and as a marker for response to therapy and prognosis in metastatic disease. Despite some controversy surrounding its clinical utility,42-44 PSA remains a key prostate cancer biomarker for the foreseeable future. The current work provides a framework for precision PSA interpretation by prostate cancer molecular subtype.
National Comprehensive Cancer Network, American Urological Association/American Society for Therapeutic Radiology and Oncology, and European Association of Urology–European Society for Radiotherapy and Oncology–International Society of Geriatric Oncology guidelines for clinically localized prostate cancer5-7 classify patients with a pretreatment PSA level of > 20 as high risk, even in the absence of other adverse prognostic features. This results in increased treatment burden, and it is recommended that patients classified as high risk who opt for radiotherapy undergo 2 to 3 years of concurrent androgen-deprivation therapy compared with a duration of 4 to 6 months for patients with intermediate risk disease. Knowledge of the molecular subtype of prostate cancer and its impact on PSA level could therefore improve risk stratification, sparing unnecessary treatment burden and directing higher-intensity therapy to patients who are truly at higher risk; however, whether molecular subtype will actually add clinical value to the current risk stratification tools—or if it adds additional information compared with such tests as Decipher—remains unclear. PSA is heavily used in many other settings as well. The data presented here have implications for defining subtype-specific thresholds for PSA recurrence after local therapy and for monitoring PSA responses in metastatic disease. Additional studies will be necessary to optimally deploy these strategies clinically, but it is clear that molecular subtyping should be considered in future clinical trial designs.
Recent advances in technology have increased our understanding of molecular subclasses of prostate cancer, but clinical and biologic differences among the key ERG-positive, ETS-positive, and SPOP mutant subclasses remain poorly understood. Our finding, that there are distinct associations with PSA and pathologic stage among subclasses, has both biologic and clinical implications. Biologically, prostate cancer cells that harbor mutant SPOP may produce more PSA on a per-cell basis as a result of enhanced androgen transcription, essentially leading to higher PSA from fewer cancer cells, whereas the opposite may be true from ERG-positive tumors. Clinically, this may lead to earlier detection of SPOP mutant cancers with lower pathologic stage because of lead-time bias. Alternatively, the underlying biology of these tumors may lead to different rates of progression or other impacts on patient outcomes. These hypotheses need to be rigorously tested in future functional and clinical studies.
As a result of the retrospective nature of the cohorts and small sample size of case cohort studies, survival analysis was inevitably affected by baseline risk. We grouped all individual studies from retrospective cohorts and performed survival analyses to study the clinical outcomes of the SPOP mutant subclass. Although the favorable prognosis was consistent with improved clinical outcomes in the SPOP mutant subclass, these survival results need to be independently validated in additional clinical trials to be generalizable.
In conclusion, we have developed the SCaPT model to predict SPOP mutant subclass purely on the basis of transcriptional data with high confidence and accuracy. The SPOP mutant subclass was associated with higher PSA but fewer adverse pathologic features and favorable prognosis. We believe this work not only builds a prediction model for SPOP mutant prostate cancers and expands the data types usable for the interrogation of clinical outcomes, but it also reinforces the concept that molecular subtyping of prostate cancer can alter the interpretation of the current standard of care risk stratification methods. More broadly, these data suggest a paradigm in which the interpretation of cancer biomarkers may be influenced by underlying molecular subtype.
ACKNOWLEDGMENT
We thank the patients with prostate cancer and families who contributed to this research. We thank The Cancer Genome Atlas Research Network for providing prostate cancer genomic and transcriptomic data. We thank the Weill Cornell Medicine Genomics and Epigenomics Core Facility and Memorial Sloan Kettering Cancer Center cBioPortal.
Footnotes
Supported by National Cancer Institute Grants No. K08-CA187417-01, R01-CA215040-01 and P50-CA211024-01 (C.E.B.), a Urology Care Foundation Rising Star in Urology Research Award (C.E.B.), Damon Runyon Cancer Research Foundation MetLife Foundation Family Clinical Investigator Award (C.E.B.), and the Prostate Cancer Foundation (C.E.B).
AUTHOR CONTRIBUTIONS
Conception and design: Deli Liu, Jonathan Shoag, Robert B. Jenkins, Mark A. Rubin, Scott A. Tomlins, Andrea Sboner, Christopher E. Barbieri
Financial support: Mark A. Rubin, Christopher E. Barbieri
Administrative support: Mark A. Rubin, Eric A. Klein, Daniel E. Spratt, Christopher E. Barbieri
Provision of study materials or patients: R. Jeffrey Karnes, Ashley E. Ross, Bruce Trock, Elai Davicioni, Christopher E. Barbieri
Collection and assembly of data: Deli Liu, Mandeep Takhar, Jonathan Shoag, Robert B. Jenkins, R. Jeffrey Karnes, Ashley E. Ross, Robert B. Den, Daniel E. Spratt, Elai Davicioni, Christopher E. Barbieri
Data analysis and interpretation: Deli Liu, Mandeep Takhar, Mohammed Alshalalfa, Nicholas Erho, Robert B. Jenkins, R. Jeffrey Karnes, Edward M. Schaeffer, Bruce Trock, Eric A. Klein, Robert B. Den, Daniel E. Spratt, Elai Davicioni, Andrea Sboner, Christopher E. Barbieri
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Deli Liu
No relationship to disclose
Mandeep Takhar
Employment: GenomeDx
Mohammed Alshalalfa
Employment: GenomeDx
Travel, Accommodations, Expenses: GenomeDx
Nicholas Erho
Employment: GenomeDx
Jonathan Shoag
No relationship to disclose
Robert B. Jenkins
Patents, Royalties, Other Intellectual Property: Abbott Molecular, GenomeDx
R. Jeffrey Karnes
Research Funding: GenomeDx
Patents, Royalties, Other Intellectual Property: GenomeDx
Travel, Accommodations, Expenses: GenomeDx
Ashley E. Ross
Stock and Other Ownership Interests: GenomeDx
Honoraria: Healthtronics
Consulting or Advisory Role: Healthtronics
Research Funding: Metamark Genetics
Edward M. Schaeffer
Consulting or Advisory Role: OPKO Diagnostics, AbbVie
Mark A. Rubin
Research Funding: Eli Lilly, Janssen Pharmaceuticals
Bruce Trock
Consulting or Advisory Role: GenomeDx
Consulting or Advisory Role: Myriad Genetics
Research Funding: Myriad Genetics, MDxHealth
Eric A. Klein
Consulting or Advisory Role: GenomeDx, Genomic Health
Speakers' Bureau: Genomic Health
Robert B. Den
Consulting or Advisory Role: GenomeDx
Speakers' Bureau: Bayer
Research Funding: Medivation, Astellas Pharma, GenomeDx
Travel, Accommodations, Expenses: GenomeDx
Scott A. Tomlins
Leadership: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
Consulting or Advisory Role: AbbVie, Janssen Pharmaceuticals, Astellas Pharma, Medivation, Strata Oncology, Sanofi, Almac Diagnostics
Research Funding: Astellas Pharma (Inst), Medivation (Inst), GenomeDx (Inst)
Patents, Royalties, Other Intellectual Property: Coauthor on a patent issued to the University of Michigan on ETS gene fusions in prostate cancer
Travel, Accommodations, Expenses: Strata Oncology
Daniel E. Spratt
No relationship to disclose
Elai Davicioni
Employment: GenomeDx
Leadership: GenomeDx
Stock and Other Ownership Interests: GenomeDx
Patents, Royalties, Other Intellectual Property: Cancer diagnostics using biomarkers 20140066323
Travel, Accommodations, Expenses: GenomeDx
Andrea Sboner
No relationship to disclose
Christopher E. Barbieri
Patents, Royalties, Other Intellectual Property: Coinventor on a patent application filed regarding SPOP mutations in prostate cancer by Weill Cornell Medicine
REFERENCES
- 1.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network NCCN guidelines: Prostate cancer (version 2.2017) https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- 6.Sanda MG, Cadeddu JA, Kirkby E, et al. Clinically localized prostate cancer: AUA/ASTRO/SUO guideline. Part I: Risk stratification, shared decision making, and care options. J Urol. 2017;6:27. doi: 10.1016/j.juro.2017.11.095. [DOI] [PubMed] [Google Scholar]
- 7.Mottet N, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Lapointe J, Li C, Giacomini CP, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 9.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 10.Blattner M, Lee DJ, O’Reilly C, et al. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia. 2014;16:14–20. doi: 10.1593/neo.131704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blattner M, Liu D, Robinson BD, et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell. 2017;31:436–451. doi: 10.1016/j.ccell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boysen G, Barbieri CE, Prandi D, et al. SPOP mutation leads to genomic instability in prostate cancer. eLife. 2015;4:e09207. doi: 10.7554/eLife.09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoag J, Liu D, Blattner M, et al. SPOP mutation drives prostate neoplasia without stabilizing oncogenic transcription factor ERG. J Clin Invest. 2018;128:381–386. doi: 10.1172/JCI96551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson MH, Ross AE, Alshalalfa M, et al. SPINK1 defines a molecular subtype of prostate cancer in men with more rapid progression in an at risk, natural history radical prostatectomy cohort. J Urol. 2016;196:1436–1444. doi: 10.1016/j.juro.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross AE, Johnson MH, Yousefi K, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69:157–165. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 16.Zhao SG, Evans JR, Kothari V, et al. The landscape of prognostic outlier genes in high-risk prostate cancer. Clin Cancer Res. 2016;22:1777–1786. doi: 10.1158/1078-0432.CCR-15-1250. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen PL, Haddad Z, Ross AE, et al. Ability of a genomic classifier to predict metastasis and prostate cancer-specific mortality after radiation or surgery based on needle biopsy specimens. Eur Urol. 2017;72:845–852. doi: 10.1016/j.eururo.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Spratt DE, Yousefi K, Deheshi S, et al. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol. 2017;35:1991–1998. doi: 10.1200/JCO.2016.70.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao SG, Chang SL, Erho N, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 2017;3:1663–1672. doi: 10.1001/jamaoncol.2017.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at-risk patient population. J Urol. 2013;190:2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Den RB, Feng FY, Showalter TN, et al. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys. 2014;89:1038–1046. doi: 10.1016/j.ijrobp.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. 2015;67:778–786. doi: 10.1016/j.eururo.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 23.Boormans JL, Korsten H, Ziel-van der Made AJ, et al. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer. 2013;133:335–345. doi: 10.1002/ijc.28025. [DOI] [PubMed] [Google Scholar]
- 24.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomlins SA, Alshalalfa M, Davicioni E, et al. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur Urol. 2015;68:555–567. doi: 10.1016/j.eururo.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer DF. Constructing confidence sets using rank statistics. J Am Stat Assoc. 1972;67:687–690. [Google Scholar]
- 27.Romano JP, Shaikh AM, Wolf M. Control of the false discovery rate under dependence using the bootstrap and subsampling. Test. 2008;17:417–442. [Google Scholar]
- 28.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 29.Bennett KP, Campbell C. Support vector machines: Hype or hallelujah? SIGKDD Explor. 2000;2:1–13. [Google Scholar]
- 30.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1–27. [Google Scholar]
- 31.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fine SW, Gopalan A, Leversha MA, et al. TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol. 2010;23:1325–1333. doi: 10.1038/modpathol.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou CK, Young D, Yeboah ED, et al. TMPRSS2:ERG gene fusions in prostate cancer of West African men and a meta-analysis of racial differences. Am J Epidemiol. 2017;186:1352–1361. doi: 10.1093/aje/kwx235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer G, Mosquera JM, Ramoner R, et al. Distinct ERG rearrangement prevalence in prostate cancer: Higher frequency in young age and in low PSA prostate cancer. Prostate Cancer Prostatic Dis. 2013;16:132–138. doi: 10.1038/pcan.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Font-Tello A, Juanpere N, de Muga S, et al. Association of ERG and TMPRSS2-ERG with grade, stage, and prognosis of prostate cancer is dependent on their expression levels. Prostate. 2015;75:1216–1226. doi: 10.1002/pros.23004. [DOI] [PubMed] [Google Scholar]
- 37.Marrone M, Potosky AL, Penson D, et al. A 22 gene-expression assay, Decipher (GenomeDx Biosciences) to predict five-year risk of metastatic prostate cancer in men treated with radical prostatectomy. PLoS Curr. doi: 10.1371/currents.eogt.761b81608129ed61b0b48d42c04f92a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein EA, Haddad Z, Yousefi K, et al. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology. 2016;90:148–152. doi: 10.1016/j.urology.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Cooperberg MR, Pasta DJ, Elkin EP, et al. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: A straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173:1938–1942. doi: 10.1097/01.ju.0000158155.33890.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooperberg MR, Broering JM, Carroll PR. Risk assessment for prostate cancer metastasis and mortality at the time of diagnosis. J Natl Cancer Inst. 2009;101:878–887. doi: 10.1093/jnci/djp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 42.Adhyam M, Gupta AK. A review on the clinical utility of PSA in cancer prostate. Indian J Surg Oncol. 2012;3:120–129. doi: 10.1007/s13193-012-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prensner JR, Rubin MA, Wei JT, et al. Beyond PSA: The next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dellavedova T. Prostatic specific antigen. From its early days until becoming a prostate cancer biomarker. Arch Esp Urol. 2016;69:19–23. [PubMed] [Google Scholar]