Abstract
Adhesion and proliferation of vascular endothelial cells are important parameters in the endothelialization of biomedical devices for vascular applications. Endothelialization is a complex process affected by endothelial cells and their interaction with the extracellular microenvironment. Although numerous approaches are taken to study the influence of the external environment, a systematic investigation of the impact of an engineered microenvironment on endothelial cell processes is needed. This study aims to investigate the influence of topography, initial cell seeding density, and collagen coating on human umbilical vein endothelial cells (HUVECs). Utilizing the MultiARChitecture (MARC) chamber, the effects of various topographies on HUVECs are identified, and those with more prominent effects were further evaluated individually using the MARC plate. Endothelial cell marker expression and monocyte adhesion assay are examined on the HUVEC monolayer. HUVECs on 1.8 μm convex and concave microlens topographies demonstrate the lowest cell adhesion and proliferation, regardless of initial cell seeding density and collagen I coating, and the HUVEC monolayer on the microlens shows the lowest monocyte adhesion. This property of lens topographies would potentially be a useful parameter in designing vascular biomedical devices. The MARC chamber and MARC plate show a great potential for faster and easy pattern identification for various cellular processes.
Keywords: microtopography, high throughput screening, endothelial cells, monocytes adhesion assay, proliferation, extracellular matrix coating
1. Introduction
Endothelial cells make up a dynamic, heterogeneous organ that possesses vital secretory, synthetic, metabolic, and immunologic functions [1,2]. The properties and functions of vascular endothelial cells are influenced by the complex interactions that exist between the cells and the extracellular matrix (ECM). These interactions involve various physical and biochemical cues. One of the principal physical cues is the vascular ECM, also termed the basement membrane [3]; it provides critical support for the vascular endothelial cells and consists of laminin-rich fibril-like structures [4–6]. Any geometrical changes of the basement membrane may lead to vascular abnormalities, thereby exposing the influence that physical cues have on vascular endothelial cells [7].
Cell-matrix interactions that occur and their effect on vascular endothelial cell processes were evaluated by the assessment of various parameters including: the influence of substrate stiffness [8], stretching force [9], and shear stress [10–12]. Topographical cues such as pillars [13], gratings [14–16], and fibers [17,18] appeared to influence adhesion, proliferation, cell morphology, and alignment of vascular endothelial cells significantly. Despite the fact that there are a large number of studies focused on the interaction between vascular endothelial cells and their biochemical and biophysical cues, general trends on cell response towards topographies were difficult to determine. This is due to the wide range of measurement parameters, cell types, substrate materials, and topographical features examined [19]. Thus, a systematic analysis was needed for verification of topographical cues influence on cellular processes.
It is hypothesized that topographical cues influence the cell processes of single vascular endothelial cells, and function of endothelial monolayers. The following study represents a systematic screening of a range of topographical cues on the MultiARChitecture (MARC) chamber, which is a variation of the MARC chip [20,21] enabling easier handling and reduced volume of cell culture reagents. The effect of extracellular matrix collagen I coating, cell seeding density, and topographical cues demonstrated a range of positive and negative influences on adhesion and/or proliferation of endothelial cells. The most influential topographies were selected and the individual patterns were scaled-up for further investigation. A verification of adhesion and proliferation of single endothelial cells was performed in the MARC plate, a multi-well plate with a different topographical pattern in each well. The influence of topographical cues on endothelial cell monolayer and the interaction with the immune cells was examined by analyzing the expression of endothelial markers and in the monocyte adhesion assay.
2. Materials and Methods
2.1. Polydimethylsiloxane (PDMS) micro- and nano-pattern fabrication
2.1.1. Fabrication of MultiARChitecture (MARC) chip master, MARC chips and MARC chambers
The MARC chip was fabricated as previously described [20,21]. In short, 41 micro- and nano-sized isotropic and anisotropic topographies (Table 1, Figure 1A) were imprinted on polycarbonate (PC) via nanoimprint lithography (NIL). Selected patterns were cut into 2 mm × 2 mm pieces and assembled into a multi-array on a silicone substrate with five unpatterned fields. Polydimethylsiloxane (PDMS, Sylgard 184, Corning) was used as a binding material and the MARC chip master was surface-treated with perfluorodecyltrichlorosilane (FTDS) prior to usage. In order to fabricate a MARC chip with the same set of topographies, a double replication was performed. The MARC chip master was initially replicated with PDMS (3g) in a 5:1 ratio of elastomer to crosslinking agent and cured overnight (approximately 12 hrs) at 60°C, and demolded at room temperature. The obtained mirror MARC chip was surface-treated with FTDS and Triton X (0.01 % solution, Sigma-Aldrich) to be used as a master mold for the replication of the MARC chamber with PDMS (4g) in a 10:1 ratio of elastomer to crosslinking agent. Similar to the master, the sample was cross-linked overnight (approximately 12 hrs) at 60°C, demolded at room temperature, and stored at 60°C prior to utilization in experiments. The MARC chip was examined with scanning electron microscopy (SEM) and atomic force microscopy (AFM).
Table 1.
Table of patterns. List of 41 patterns replicated on 49 chip fields in the MultiARChitecture (MARC) chamber. Pattern number, isotropy and size are indicated.
| Position No | ID No. | Topography geometry | Dimension parameters [ Diameter or Width x Pitch (center to center) x Height or Depth] |
|---|---|---|---|
| 1 | U | Unpattern | - |
| 2 | 1 | Gratings | 2 μm × 4 μm × 2 μm |
| 3 | 2 | Gratings | 2 μm × 3 μm × 80 nm |
| 4 | 3 | Gratings | 1 μm × 3 μm × 120 nm |
| 5 | 4 | Gratings | 250 nm × 500 nm × 250 nm |
| 6 | 5 | Pillars (Square array) | 10 μm × 10 μm × 10 μm |
| 7 | 6 | Pillars (Square array) | 2 μm × 12 μm × 2 μm |
| 8 | 7 | Pillars (Square array) | 2 μm × 4 μm × 2 μm |
| 9 | 8 | Pillars (Square array) | 500 nm × 10 μm × 500 nm |
| 10 | 9 | Pillars (Square array) | 250 nm × 500 nm × 250 nm |
| 11 | 10 | Pillars (Square array) | 1 μm × 7.5 μm × 1 um |
| 12 | 11 | Convex lens (Hexagonal array) | 1.8 μm × 2 μm × 700 nm |
| 13 | U | Unpattern | - |
| 14 | 12 | Convex lens (Square array) | 800 nm × 1 μm × 300 nm |
| 15 | 13 | Pillars (Square array) | 2 μm × 12 μm × 700 nm |
| 16 | 14 | Pillars (Square array) | 2 μm × 12 μm × 500 nm |
| 17 | 15 | Pillars (Square array) | 2 μm × 12 μm × 450 nm |
| 18 | 16 | Pillars (Square array) | 2 μm × 12 μm × 350 nm |
| 19 | 17 | Pillars (Square array) | 2 μm × 12 μm × 200 nm |
| 20 | 18 | Pillars (Square array) | 500 nm × 10 μm × 300 nm |
| 21 | 19 | Pillars (Square array) | 500 nm × 10 μm × 200 nm |
| 22 | 20 | Pillars (Square array) | 500 nm × 10 μm × 100 nm |
| 23 | 21 | Pillars (Square array) | 250 nm × 500 nm × 120 nm |
| 24 | 22 | Concave lens (Hexagonal array) | 1.8 μm × 2 μm × 700 nm |
| 25 | U | Unpattern | - |
| 26 | 23 | Concave lens (Square array) | 800 nm × 1 μm × 300 nm |
| 27 | 24 | Hierarchical Mircon gratings ∟ nano gratings |
(Pri) 2 μm × 4 μm × 2 μm (Sec) 250 nm × 500 nm × 150 nm |
| 28 | 25 | Hierarchical Mircon gratings // nano gratings |
(Pri) 2 μm × 4 μm × 2 μm (Sec) 250 nm × 500 nm × 150 nm |
| 29 | 26 | Hierarchical Mircon gratings w/ nano pillars |
(Pri) 2 μm × 4 μm × 2 μm (Sec) 250 nm × 500 nm × 250 nm |
| 30 | 27 | Bumps | 300 nm × 300 nm × 200 nm |
| 31 | 28 | Cones | 270 nm × 300 nm × 350 nm |
| 32 | 29 | Inverse cones | 270 nm × 300 nm × 350 nm |
| 33 | 30 | V-gratings | 2 μm × 2μm × 1.5 μm |
| 34 | 31 | V-gratings | 500 nm × 500 nm × 350 nm |
| 35 | 32 | U-gratings | 2 μm × 2 μm × 700 nm |
| 36 | 33 | U-gratings | 500 nm × 500 nm × 200 nm |
| 37 | U | Unpattern | - |
| 38 | 34 | Gratings | 10 μm × 20 μm × 10 μm |
| 39 | 24 | Hierarchical Mircon gratings ∟ nano gratings |
(Pri) 2 μm × 4 μm × 2 μm (Sec) 250 nm × 500 nm × 150 nm |
| 40 | 25 | Hierarchical Mircon gratings // nano gratings |
(Pri) 2 μm × 4 μm × 2 μm (Sec) 250 nm × 500 nm × 150 nm |
| 41 | 26 | Hierarchical Mircon gratings w/ nano pillars |
(Pri) 2 μm × 4 μm × 2 μm (Sec) 250 nm × 500 nm × 250 nm |
| 42 | 35 | Convex lens (Square array) | 10 μm × 20 μm × 200 nm |
| 43 | 36 | Convex lens (Square array) | 10 μm × 20 μm × 2 μm |
| 44 | 37 | Convex lens (Square array) | 2 μm × 4 μm × 400 nm |
| 45 | 38 | Convex lens (Square array) | 2 μm × 4 μm × 800 nm |
| 46 | 39 | Convex hexagon lens (Square array) | 10 μm × 20 μm × 800 nm |
| 47 | 40 | Convex hexagon lens (Square array) | 10 μm × 20 μm × 2 μm |
| 48 | U | Unpattern | - |
| 49 | 41 | Hexgonal donut | 50 μm (outer), 10 μm (inner) × μm × 800 nm |
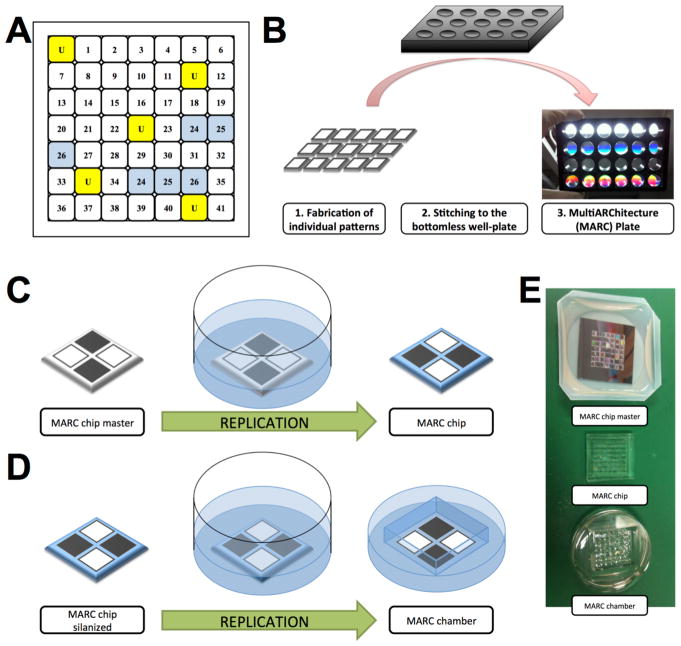
Figure 1. Schematic diagrams of MultiARChitecture (MARC) chamber and the fabrication of MARC chip, MARC chamber and MARC plate.
(A) MARC chamber topography map. Numbers indicate various topographies as shown in Table 1, and (U) is unpatterned control in randomized position. Unpatterned control and technical duplicates are highlighted in yellow and blue respectively. (B) Schematic diagram of MARC plate fabrication. 1) Soft lithographic replication of single patterned PDMS, 2) Attachment of single patterned PDMS pieces to a bottomless well-plate and 3) final product - MARC plate. (C–E) Schematic diagram of MARC chip master soft lithography replication into MARC chip and MARC chamber. (C) Replication process of MARC chip master into MARC chip. (D) Replication process of silanized MARC chip into MARC chamber. (D) Photographic images of MARC chip master, MARC chip and MARC chamber.
For a robust evaluation of topographies with uniform cell density, the MARC chamber, which is a variation of the MARC chip, was developed as a double negative replica of the MARC chip master. The fabrication process involves the replication of the MARC chip from the MARC chip master (Figure 1C–E). Subsequently, the MARC chip is surface-treated with FTDS and Triton X (0.01% solution) and utilized as a master-mold for a further replication. The replication of the MARC chip master was performed in a common 35 mm diameter culture dish (Figure 1D). The final product after demolding, the MARC chamber, mimicked the topographies on the MARC chip master, and it would be in a chamber form that fits robustly in 6 well-plates or 35mm dishes (Figure 1E). The MARC chamber possesses a number of advantages: (1) easy handling and fitting robustly into conventional cell culture plates or dishes, (2) enabling uniform cell seeding locally on the patterned area, (3) minimizing the volume of required reagent thus enabling economical use of media, growth factors, antibodies and other substances, and (4) enabling the utilization of up to 6 MARC chambers at the same time in a standard 6-well culture plate.
2.1.2. Fabrication of MARC plates
The MARC chip and the MARC chamber have been demonstrated to be efficient screening tools with microscopy-based analysis. The small patterned area, the challenge in harvesting cells from each individual cell area and the potential paracrine effects among patterns limit their use for further biological analyses. In order to verify the MARC chip/chamber screening assay, we designed and fabricated MARC plates. MARC plates were fabricated as bottomless well cell culture plates with attached 2 cm × 2 cm unpatterned and single-patterned PDMS replicas (Figure 1B). The single-patterned PDMS replicas were attached by utilizing PDMS in a 5:1 ratio of elastomer to crosslinking agent. The MARC plates were then cured overnight (approximately 12 hrs) at 60°C. The wells were washed with absolute ethanol to ensure that proper attachment of the single PDMS replicas occurred.
2.1.3. Preparation of MARC chambers, MARC plates and PDMS single replicas
The replicas of MARC chambers, MARC plates, and single-patterned PDMS (which will be denoted as “single PDMS”) were washed with absolute ethanol and treated with oxygen plasma for 1 min, 85W and 800 cc/min before UV-sterilized for 20 min. Selected MARC chambers and MARC plates were coated with bovine collagen type I (collagen I, 2.5 μg/cm2, Gibco) in acetic acid solution (0.02 M solution) for one hour at 37°C. Excess collagen was washed off with phosphate buffered saline (PBS).
2.1.4. Quantification of collagen binding in MARC chambers
To quantify the collagen binding on different patterned substrates, the absorbed collagen on MARC chambers was fixed with 4% paraformaldehyde (PFA), and then immunofluorescently stained with Alexa Fluor®488 conjugated anti-collagen I antibody (1:200, SouthernBiotech).
2.2. Cell assays
2.2.1. Vascular endothelial cell culture
As recommended by the supplier’s protocol at 5000 cells/cm2 seeding density, the human umbilical vein endothelial cells (HUVECs, Lonza, passage 4–6) were cultured for approximately 3–4 days in endothelial growth media (EGM-2, Lonza). The cells were maintained in an incubator at 37°C with stable 5% CO2. The medium was changed every other day. HUVECs were trypsinized and harvested according to the supplier’s instructions. For the adhesion and proliferation assays in the MARC chambers and adhesion assay in the MARC plates, the cells were cultured in EGM-2 at either 3000 or 10000 cells/cm2. Prior to staining, cells were washed with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffered saline solution and fixed with 4% PFA; this was performed at the 4 or 24-hour time point. For the single pattern study, HUVECs were cultured in EGM-2 on the single PDMS for 2–3 days until a monolayer formed. Human monocytes (U-937, ATCC) were cultured in a medium composed of Rosswell Park Memorial Institute Medium (RPMI, Gibco) supplemented with 10% fetal bovine serum (FBS, HyClone), 1% sodium pyruvate (Sigma-Aldrich), penicillin, and streptomycin (100 U/mL ml and 100 μg/mL ml respectively, GE Health Science). The U-937 cells were cultured in suspension for 4–5 days; medium was added every other day and harvested according to the supplier’s protocol.
2.2.2. Adhesion, proliferation and monocyte cell assays
2.2.2.1. Adhesion and proliferation assays in MARC chambers
For the adhesion assay in the MARC chambers, samples were fixed with 4% PFA, washed with HEPES buffered saline solution (HBSS), and stained with a nuclear stain 4′, 6-diamidino-2 phenylindole (DAPI, 1:3000, Life Technologies), and phalloidin 546 (1:300, Life Technologies). For the proliferation assay in the MARC chambers, the cells were incubated for 4 hrs (prior to the 24-hour endpoint) in the EdU-kit solution (Click-iT EdU Alexa Fluor 488 HCS assay, Invitrogen) according to the manufacturer’s instructions. Samples were fixed with 4% PFA and stained with DAPI (1:3000).
2.2.2.2. Adhesion assay in MARC plates
For the adhesion assay in the MARC plates, the cells were washed with HBSS, trypsinized, and neutralized to obtain a cell pellet. The cell pellet was then snap frozen in liquid nitrogen and stored at −80°C prior to further processing.
The CyQUANT® cell assay kit (CyQUANT® NF assay, Invitrogen) was used for the quantification of the cellular DNA content present in each well. The assay was performed according to the manufacturer’s protocol. The fluorescence intensity of each sample was measured with a microplate reader (xMark™ microplate absorbance spectrophotometer, Bio-Rad). Cell density was calculated from measured immunofluorescence values utilizing the cell ladder with known cell densities.
2.2.2.3. Cell marker expression assay on single PDMS replicas
For cell marker expression analysis, the HUVEC monolayers were washed with PBS with calcium and magnesium, fixed with 4% PFA before blocking with 10% goat serum (Thermo Fischer Scientific). The monolayers were stained with VE-cadherin (rabbit anti-VE-cadherin, 1:200, Cell Signaling Technology) and ICAM-1 (mouse anti-ICAM-1, 1:200, R&D Systems) and then secondary antibodies (Alexa Fluor™ 488 goat anti-rabbit and Alexa Fluor™ 546 goat anti-mouse), with DAPI as counter staining for the nuclei. Images were captured from each sample (CD31, VE-cadherin, and ICAM-1) with consistent image acquisition parameters, processed with Image J (Image J 1.47v) and evaluated for the endothelial cell markers.
2.2.2.4. Monocyte adhesion assay on single PDMS replicas
For the monocyte adhesion assay, the HUVEC monolayers were activated by incubation with tumor necrosis factor α (TNF- α, 2.5 ng/ml, Life Technologies) in EGM-2 for 5 hours at 37°C with stable 5% CO2. Monocytes were centrifuged and labeled with cell tracker dye (CellTracker™ Green CMFDA dye, Thermo Fisher Scientific) according to the manufacturer’s instructions. The single PDMS samples were washed with RPMI to remove EGM-2 with TNF- α. The monocytes were seeded at 150000 cells/cm2. Samples were incubated on a shaker for 45 min and washed with phosphate buffered saline (PBS) prior staining with DAPI (1:3000) and the fluorescently conjugated endothelial cell marker, CD31 (1:400, Miltenyi-Biotec).
2.3. MARC chamber and single PDMS pattern sample imaging
Samples were imaged with a Leica epifluorescence microscope (Leica DMi8) equipped with a Q-imaging camera and Q-Capture Pro software and Zeiss microscope (Axio Observer Z1) equipped with Zen software. During the imaging process, all acquisition parameters remained constant. Images were captured from each sample at 10x and 20x magnification and processed with Image J (Image J 1.47v) for particle analysis. On single cells that not engaged in any type of cell-cell interaction, cell circularity and cell area were analyzed on evaluated patterned surface and on images with 20x magnification. Cell circularity was calculated with a plugin built within Image J using the following equation: Circularity = 4 π (Area/Perimeter2). A circularity index value of 1.0 indicates a perfect circle and a value that approaches 0.0 indicates an elongated polygon. Cell circularity values were normalized to the number of analyzed cells per pattern (n = 200–400). The mean immunofluorescence intensity of CD31, VE-cadherin and ICAM-1 on each patterned surface was measured with Image J by taking the sum of the mean gray values of the monolayer, normalized to the total number of cells counted. The process was repeated for all images and patterned surfaces. Values were normalized to number of cells and presented in arbitrary units (A.U.) per cell.
2.4. Statistical analysis
In the MARC-chamber topography screening assay, including the values for adhesion and proliferation, the analysis of cell area and cell circularity, intensity analysis of immunofluorescence staining of collagen I and endothelial cell markers, and the monocyte cell density values were evaluated on outliers using Grubb’s test. The values of all data were presented as mean values, plus or minus the calculated standard deviation. Statistical analysis for collagen I adsorption in MARC chamber (n=30 for all surfaces), topography screening assay for adhesion and proliferation in MARC chamber (n=20 for unpatterned and n=4 for patterned surfaces), topography screening assay for adhesion and proliferation in MARC plate (n=10 for all surfaces), the analysis of cell area and cell circularity assay (n=240 for unpatterned and n=60 for patterned surfaces), the quantification of cell marker expression (n=15 for all surfaces) and monocyte adhesion assay (n=20 for all surfaces) was performed using an one-way ANOVA test with Tukey’s post-hoc test. Statistical tests were considered significant when P≤0.05 (noted as one asterisks), P≤0.01 (noted as two asterisks), P≤0.001 (noted as three asterisks), and P≤0.0001 (noted as four asterisks). All statistical analysis was performed using QuickCalcs and GraphPad Prism through GraphPad Software.
3. Results
3.1. MARC chamber and single pattern PDMS replica characterization
The fidelity of the MARC chip, MARC chamber, MARC plates, and MARC chamber pattern replication was verified by scanning electron microscopy (SEM) (Supplementary Figure 1) and atomic force microscopy (AFM) (Supplementary Figure 2). The fidelity of the single pattern PDMS replicas was verified by SEM (Supplementary Figure 3).
3.2. Collagen adsorption quantification on MARC chambers
The adsorbed collagen type I on MARC chambers was quantified to assess the influence of different topographies on collagen coating. The adsorbed collagen on different patterns was visualized by Alexa Fluor®488 conjugated anti-collagen I antibody, and was quantified by fluorescence intensity, as shown in Supplementary Figures 4 and Supplementary Figure 5 respectively. Though the collagen adsorption varied on different patterns, there were no observable variation trends, and the collagen adsorption on most patterned substrates was not significantly different from the unpatterned surfaces. Only the amount of adsorbed collagen on topography 19 (500 nm pillars, with 10 μm pitch, 500 nm height) appeared significantly lower than those on unpatterned surfaces. Grating patterns (Topography 1 to 5, Table 1) and convex lens (Topography 11) showed higher collagen binding compared with other patterns, while micro-sized hierarchical gratings (Topography 24 and 25), and microsized pillars (Topography 15) resulted in lower collagen adsorption.
3.3. Cell density and cell proliferation on unpatterned substrates
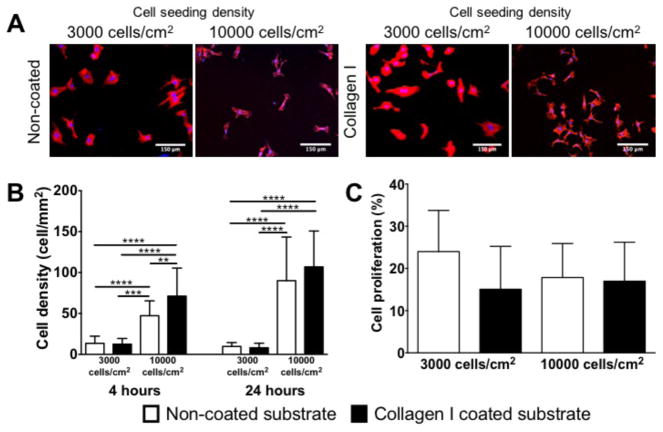
To evaluate the influence of ECM coating and cell seeding density on cell adhesion and proliferation on unpatterned PDMS substrates, we compared the unpatterned substrate samples in different conditions (Figure 2). The cell density was observed at 4 hours and 24 hours after cell seeding, while the cell proliferation was analyzed at 24 hours after cell seeding. At the 4-hour time point, significant difference in cell density on the non-coated and collagen I coated substrates was observed at the higher initial cell seeding density (10000 cell/cm2, Figure 2A and 2B). At the 24-hour time points, the cell density on the substrate with higher initial seeding density was significantly higher than the samples with lower initial seeding density (3000 cell/cm2), regardless if the substrate was pre-coated with collagen. Interestingly, the difference in cell density observed on the non-coated versus collagen I coated substrates at the 24-hour time points was not significantly different.
Figure 2.
Comparison of cell density and cell proliferation of HUVECs on unpatterned substrates at 4 and 24 hr time point. (A) Comparison of representative immunofluorescence images of HUVECs on non-coated or collagen I coated substrate at 3000 cells/cm2 or 10000 cells/cm2 seeding density at 4 hr time point. (B) Quantification of cell density of HUVECs at 4 hr and 24 hr time point. (C) Comparison of cell proliferation of HUVECs on non-coated and collagen I coated unpatterned substrates at 3000 and 10000/cm2 cells seeding density at 24hrs time point. No significant differences were observed in cell proliferation. White bar represents 150 μm. HUVECs were stained for Phalloidin (in blue) and the nuclear marker – DAPI (in blue). Data (n=20) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
Additionally, we evaluated the cell proliferation of HUVECs on unpatterned PDMS substrates at the 24-hour time point with different ECM coatings and cell seeding densities. No significant difference was observed on the cell proliferation on non-coated versus collagen I coated unpatterned substrates at both cell seeding densities (Figure 2C).
3.4. Cell density quantification at 4-hour time point
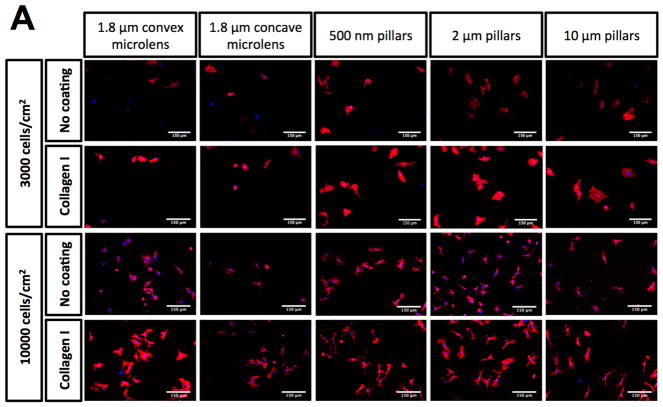
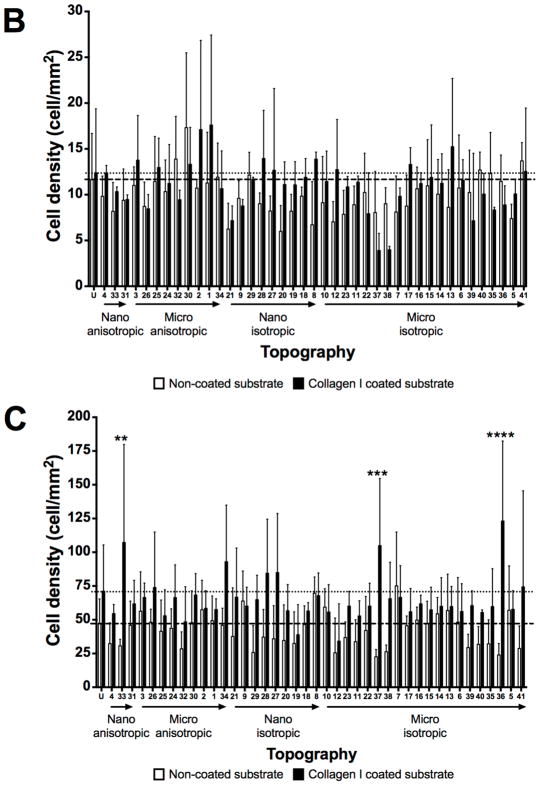
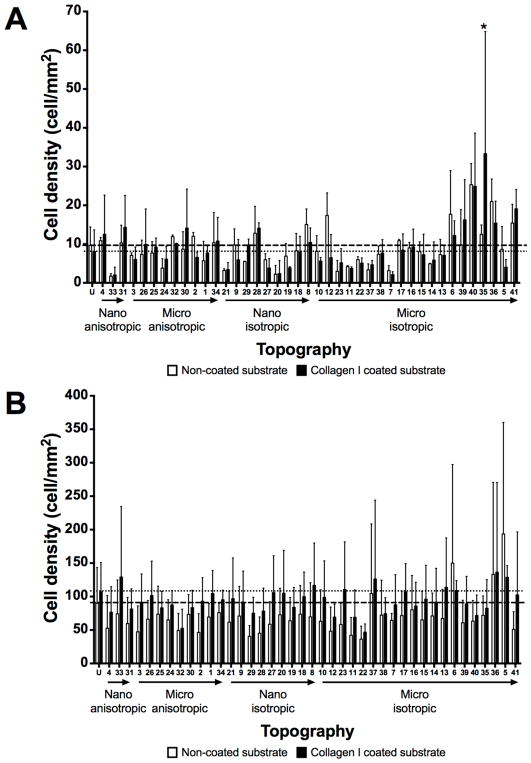
The HUVECs were seeded on a variety of topographies presented in the MARC chamber in order to evaluate the interaction between the topographies and endothelial cells. To examine the influence of ECM coating on endothelial cell adhesion, cells were seeded on either bovine collagen I coated or non-coated topographies at two different cell seeding densities. Representative images of the HUVECs, which were visualized by phalloidin and DAPI staining for F-actin and nuclei, on the various samples and topographies are shown in Supplementary Figures 6 – 9. Overall, the cell density at 4-hour post cell seeding varied between (1) ECM coating – collagen I pre-coated versus non-coated surfaces, (2) different initial cell seeding densities, and (3) unpatterned versus patterned surfaces. Collagen I coating on patterns led to two observable trends. First, the collagen coating demonstrated a slight overall increase in cell adhesion on most of the patterns (Figure 3), regardless of seeding density. Second, the cell adhesion on collagen I coated surfaces showed larger variation and noise among biological replicas, in comparison to the cell adhesion values obtained from non-collagen I coated surfaces. Statistical significantly higher cell density (P≤0.01 and P≤0.001) was observed only on three patterns at 10000 cells/cm2 seeding density (Topography 33 – 500 nm U-groove, with 500 nm pitch, 200 nm height; Topography 36 - 10 μm lens (square array) with 10 μm pitch, 2 μm height; Topography 37 - 2 μm lens (square array) with 2 μm space, 400 nm sag) when compared with the values of the corresponding non-coated substrates (Figure 3B–C). Although higher cell density at 4-hour was expected on samples with higher initial cell seeding compared to samples with lower cell seeding, significant differences in the 4-hour cell density were only observed on some of the patterns (Supplementary Figure 10). Significant differences in cell density at 4-hour between lower and higher initial cell seeding on patterns was observed on more number of patterns without collagen I coating (21 patterns) than on patterns with collagen I coating (7 patterns). Moreover, micro-sized hierarchical 2 μm gratings (Topography 25 and 26) and 10 μm gratings (Topography 34) with higher seeding density showed significantly higher 4-hour cell density compared to the corresponding samples with lower seeding density, regardless of collagen I coating. Overall, regardless of collagen I coating, the anisotropic micro-sized patterned surfaces, such as gratings, showed higher cell adhesion in comparison to other isotropic nano- and micro-sized patterned surfaces (such as well and pillars). A few anisotropic micro-gratings (such as Topography 1 - 2 μm lines, 2 μm space, 2 μm height; Topography 2 - 2 μm lines, 1 μm space, 80 nm height; Topography 30 - 2 μm V-groove, 2 μm pitch, 1.5 μm height) showed higher cell density at 4 hour compared to the unpatterned surfaces. Lens topographies, characterized as curved geometry, showed a lower cell adhesion on coated and non-coated surfaces at both cell seeding densities. However, the higher cell density on micro-gratings and lower cell density on lens topographies were not significantly different from unpatterned surfaces.
Figure 3.
Cell adhesion evaluation at 4 hour time point on unpatterned and 41 patterned surfaces (MARC chamber) with different ECM coatings and cell seeding densities. (A) Representative immunofluorescence images of selected patterns on non-collagen I and Collagen I coated surfaces at 3000 and 10000 cells/cm2 initial cell seeding density. White bar represents 150 μm. HUVECs were stained for phalloidin and the nuclear marker – DAPI. Comparison of cell density quantification on non-coated surfaces at initial cell seeding density 3000 cells/cm2 (B) and 10000 cells/cm2 (C). Geometry size, isotropy and anisotropy are indicated. Arrows represent increase in geometry size. Mean values of non-collagen I and collagen I coated unpatterned surfaces are represented by dashed and dotted lines respectively. Data (n=20 for unpatterned surfaces and n=4 for patterned surfaces) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
3.5. Cell proliferation and adhesion quantification at 24 hours after cell seeding
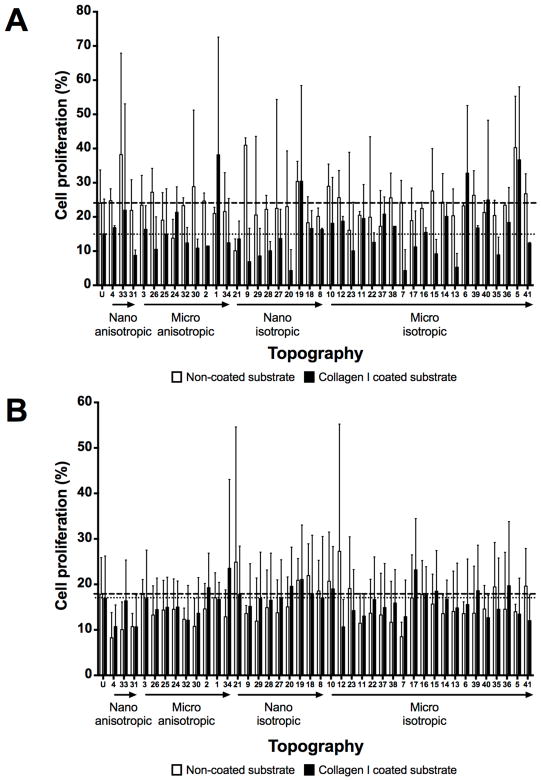
As the formation of the endothelial cell monolayer depends on cell adhesion and cell proliferation, the cell density and proliferation of HUVECs at the 24-hour time point (post cell seeding) were quantified. Similar to the cell adhesion results at the 4-hour time point, the effects of cell density at the 24-hour time point varied between (1) ECM coating – collagen I coated versus non-coated surfaces, (2) different initial cell seeding densities, and (3) unpatterned versus patterned surfaces (Figure 4 and 5). Higher cell densities at 24-hour were observed on patterns with higher initial cell seeding density when compared to corresponding patterns with lower initial cell seeding density (Figure 4). A small difference between patterned substrates with higher and lower cell seeding densities was observed on collagen I coated patterns; in contrast, a close to 10-fold higher cell density was observed on non-coated unpatterned substrates with higher initial seeding density compared to the corresponding pattern with lower seeding density (Supplementary Figure 11). A 10-fold increase in cell densities was observed on the non-coated patterns, whereas only two micro-sized isotropic patterns (Topography 5 - 10 μm pillars, 10 μm pitch, 10 μm height; Topography 6 - 2 μm pillars, 12 μm pitch, 2 μm height) demonstrated significantly higher 24-hour cell density on the samples with higher seeding density, compared to the corresponding patterns with lower seeding density (Supplementary Figure 11A). Topography 5 and 6 were also among the 21 non-coated patterns with higher 4-hour density on the samples with higher initial seeding density (Supplementary Figure 10A).
Figure 4.
Cell adhesion evaluation at 24 hrs time point on unpatterned and 41 patterned surfaces (MARC chamber) with different ECM coatings and cell seeding densities. (A) Comparison of cell density quantification on non-coated surfaces at initial cell seeding density 3000 cells/cm2 (A) and 10000 cells/cm2 (B). Geometry size, isotropy and anisotropy are indicated. Arrows represent increase in geometry size. Mean values of non-collagen I and collagen I coated unpatterned surfaces are represented by dashed and dotted lines respectively. Data (n=20 for unpatterned surfaces and n=4 for patterned surfaces) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
Figure 5.
Cell proliferation evaluation at 24 hour time point on unpatterned and 41 patterned surfaces (MARC chamber) with different ECM coatings and cell seeding densities. (A) Comparison of cell proliferation on non-coated surfaces at initial cell seeding density 3000 cells/cm2 (A) and 10000 cells/cm2 (B). Geometry size, isotropy and anisotropy are indicated. Arrows represent increase in geometry size. Mean values of non-collagen I and collagen I coated unpatterned surfaces are represented by dashed and dotted lines respectively. Data (n=20 for unpatterned surfaces and n=4 for patterned surfaces) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD. No significant differences were observed.
In terms of cell proliferation measured by EdU assay, the cell proliferation was higher on non-coated patterns with lower initial cell seeding density, when comparing with on patterns with collagen I coating (Figure 5A) and patterns with higher initial cell seeding either with or without collagen I coating (Figure 5B, Supplementary Figure 12). Moreover, regardless of the collagen I coating, the lens topographies (curved geometry patterns: Topography 11 - 1.8 μm diameter, 2 μm pitch, 0.7 μm sag convex microlens; Topography 12 - 1 μm diameter, 1 μm pitch, 0.3 μm sag convex microlens; Topography 22 - 1.8 μm diameter, 2 μm pitch, 0.7 μm sag concave microlens; Topography 23 - 1 μm diameter, 1 μm pitch, 0.3 μm sag concave microlens) exhibited lower cell density and cell proliferation in comparison to other patterned and unpatterned substrates; however, most of the values did not exhibit any significant difference.
3.6. Cell circularity and cell area quantification
To evaluate the interaction of vascular endothelial cells with the patterned substrates, a morphometric analysis to quantify the cell area and cell circularity of individual HUVECs at the 4-hour time point was performed. Based on the results observed in the cell adhesion and cell proliferation in the above sections, the morphometric evaluation of the HUVECs was performed on non-coated samples at the 4-hour time point. The selection rationales to simplify the analysis were based on the following parameters:
Non-coated patterned substrates cell density data were selected. The HUVEC-substrate interaction was evaluated on substrate with serum-protein, without the influence of the pre-coated collagen I.
The 4 hour time point with an initial cell seeding density of 10000 cells/cm2 were selected because the cell culture would remain sub-confluent, which allow the morphometric analysis of individual cells, while a sufficient cell density would be present on each of the pattern on the MARC chip for data and statistical analysis.
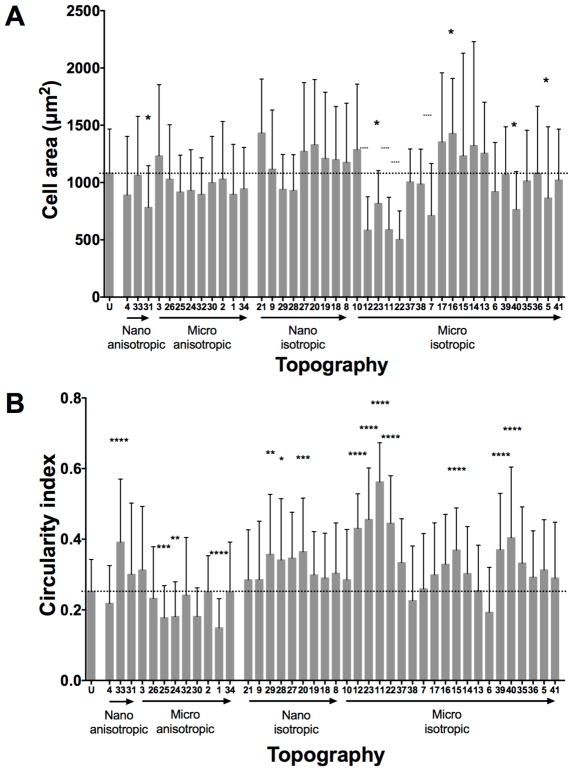
The HUVEC area varied greatly amongst the different patterned and unpatterned surfaces, either with a significantly higher cell area or a significantly lower cell area (Figure 6A, Supplementary Figure 7). In general, the HUVECs showed either more rounded cell morphology with a smaller cell area, or larger cell morphology with numerous filopodia, while some of the HUVECs were elongated in the direction of the underlying topography (Topography 31 - 500 nm V-groove gratings).
Figure 6.
Cell area and cell circularity quantification of HUVECs at 4 hour time point with initial 10000 cell/cm2 seeding density on non-coated patterned substrates. (A) Quantification of cell area and (B) cell circularity index of HUVECs on unpatterned and patterned surfaces. Geometry size, isotropy and anisotropy are indicated. Arrows represent increase in geometry size. Dotted line represents unpatterned surface mean value. Data (n=240 for unpatterned surfaces and n=60 for patterned surfaces) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
Topography 16 (2 μm pillars with 12 μm pitch and 350 nm height) led to a significantly larger cell area of HUVECs. HUVECs on the gratings, pillars, and lens structures of different sizes also led to significantly smaller cell areas, compared to HUVECs on unpatterned substrates. However, most of the patterned surfaces did not significantly influence the HUVEC cell area when compared to the cell area of HUVECs on the unpatterned surface (Figure 6A, Supplementary Figure 8).
Cell circularity is another morphometric parameter that was analyzed (Figure 6B, Supplementary Figure 8). Notably, HUVECs on anisotropic micro-sized patterned surfaces (Topography 1 - 2 μm lines, 2 μm space, 2 μm height; Topography 24 - 2 μm lines with perpendicular 250 nm lines on the ridge; Topography 25 - 2 μm line with parallel 250 nm lines on the ridge) showed more elongated cell bodies, with significantly lower cell circularity. HUVECs adherent on the nano-sized anisotropic patterned substrate, Topography 33 (500 nm U-groove with 500 nm pitch and 200 nm height), demonstrated round shaped cell bodies. While on round and curved isotropic nano- and micro-sized geometry such as pillars, wells and lenses, the HUVECs tended to be less elongated and appeared rounder; and therefore exhibited a significantly higher cell circularity index (Figure 6B).
Overall, micro-sized lens topographies influenced the endothelial cells to have rounder and smaller cell bodies in comparison to other isotopic and anisotropic patterned substrates. The comparison of cell adhesion, proliferation, cell area, and circularity lead to the selection of a set of patterns for further verification on individual patterns by using the MARC plates, and the analysis of their influence on endothelial cell monolayer. The pattern selection criteria were as followed:
Selected patterned substrates should demonstrate consistent influences, either a consistent increase or decrease compared to the unpatterned control, on the adhesion and cell density at the 4-hour and 24-hour time points.
Selected patterned substrates should affect cell proliferation, either higher or lower values of cell proliferation, when compared to the unpatterned control.
Selected patterned substrates should induce changes in cell morphology, either more elongated or round cell morphology, when compared to the unpatterned control.
Selected patterned substrates should affect the cell area, either larger or small cell area, when compared to the unpatterned control.
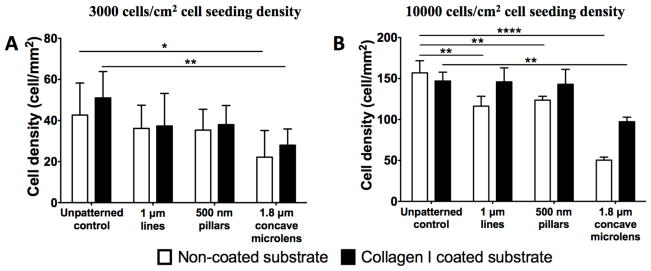
3.7. Cell density quantification in MARC plates at 24 hours after cell seeding
To verify the data obtained in the MARC chamber-screening assay, HUVECs were seeded on selected patterns on a MARC plate and cultured for 24 hours. Overall, cell density was lower on the substrates with initial 3000 cells/cm2 cell seeding density, in comparison to the substrates seeded with 10000 cells/cm2 (Figure 7). The 1.8 μm diameter concave microlens demonstrated a significantly lower cell density compared to the unpattern controls, regardless of the collagen coating or initial cell seeding density. At 3000 cells/cm2 seeding density, no comparable difference between samples with and without collagen I coatings was revealed at the 24 hour (Figure 7A). The influence of the topographies on HUVECs were consistent. The topographies yielded a lower cell density at 24 hours after cell seeding, regardless of the collagen coating. At a higher initial cell density, collagen I coating enhanced the cell density on the various topographies. The cell density on the collagen-coated 1 μm lines and 500 nm pillars were comparable to the collagen coated unpatterned control. On the patterns without collagen I coating, cell density on 1 μm lines, 500 nm pillars and 1.8 μm concave lenses were significantly lower than the unpatterned control. Overall, collagen I coating yielded an additive effect in enhancing endothelial cell density (Figure 7B).
Figure 7.
Cell density quantification post 24 hour cell seeding on MARC plates. Comparison of cell density of HUVECs seeded on non-coated surfaces (A), collagen I coated surfaces (B), in cell seeding density 3000 cells/cm2 (A) and 10000 cells/cm2 (B). Data (n=10 for all surfaces) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
3.8. Cell markers expression of endothelial cell monolayer
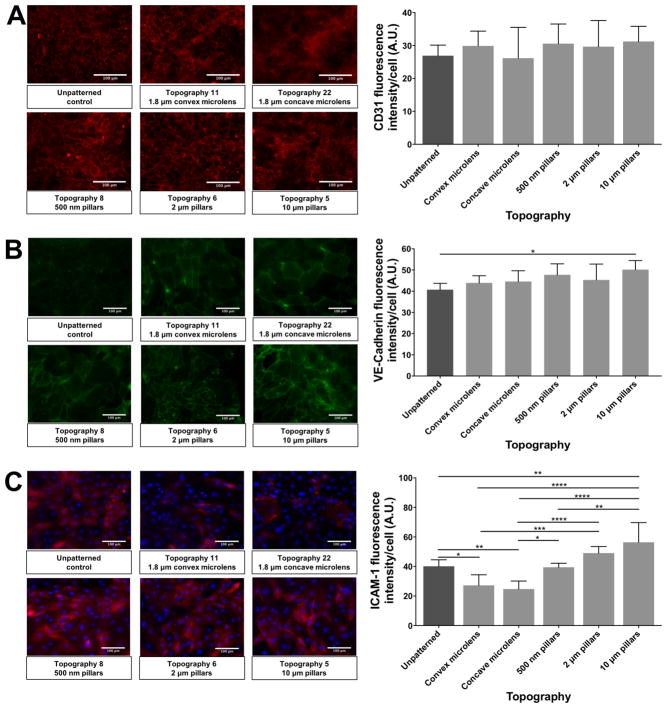
The expression of CD31, VE-cadherin and ICAM-1 of HUVEC monolayers formed on selected patterned substrates were assessed to evaluate the influence of topographic factors on endothelial cell markers expression and cell-cell junction.
The CD31 expression of the HUVEC monolayers on selected patterned surfaces was slightly different among the different patterns (Figure 8A). HUVEC monolayer on convex microlens (1.8 μm diameter with 2 μm pitch, and 0.7 μm sag convex microlens) and the pillar structures (500 nm pillars with 10 μm pitch and 500 nm height, 2 μm pillars with 12 μm pitch and 2 μm height and 10 μm pillars with 10 μm pitch and 10 μm height) showed a higher CD31 expression compared to the unpatterned control. The CD31 expression of HUVEC monolayer on concave microlens (1.8 μm diameter with 2 μm pitch, and 0.7 μm sag concave microlens) samples was similar to that of unpatterned control. However, all the differences were not statistically significant.
Figure 8.
Comparison of representative immunofluorescence images and quantified values of cell markers expression of HUVEC monolayer on selected patterned substrates. (A) CD31 expression (Red) of HUVEC monolayer on selected patterned substrates. (B) VE-cadherin expression (Green) of HUVEC monolayer on selected patterned substrates. (C) ICAM-1 expression (Red) of HUVEC monolayer on selected patterned substrates. Due to the uneven expression of ICAM-1 on the HUVEC monolayers, DAPI stained nuclei (Blue) was showed with ICAM-1 to help confirm the monolayer. In all panels, white bar represents 100 μm. Data (n=15 for all surfaces) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
VE-cadherin is an important cell marker for endothelial cell-cell junction and vascular monolayer permeability. As shown in Figure 8B, the HUVEC monolayer on all topographies (1.8 μm diameter with 2 μm pitch, and 0.7 μm sag convex and concave microlens, 500 nm pillars with 10 μm pitch and 500 nm height, 2 μm pillars with 12 μm pitch and 2 μm height and 10 μm pillars with 10 μm pitch and 10 μm height) exhibited higher VE-cadherin expression, and the 10 μm pillar topography revealed a statistically significant up-regulation compared to unpatterned control.
The expression level of ICAM-1 varied a lot among different patterned substrates (Figure 8C). Micro-sized lens topographies (1.8 μm diameter with 2 μm pitch, and 0.7 μm sag convex and concave microlens) showed lower ICAM-1 expression compared to unpatterned substrates, and pillar structures (500 nm pillars with 10 μm pitch and 500 nm height, 2 μm pillars with 12 μm pitch and 2 μm height and 10 μm pillars with 10 μm pitch and 10 μm height). An upward trend of ICAM-1 expression was found on pillar substrates, among which the 10 μm pillars topography exhibited the highest expression.
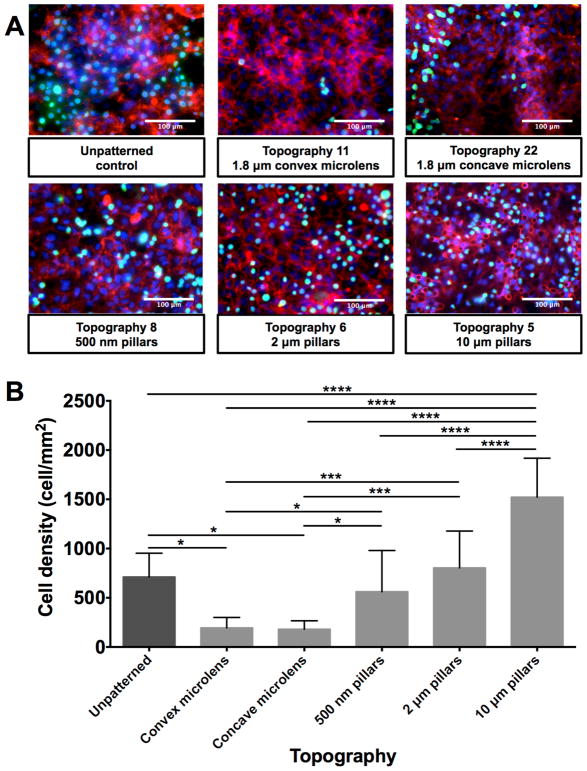
3.9. Evaluation of monocyte adhesion on endothelial cell monolayer
To evaluate the potential influence of underlying patterned substrates on the functions of the vascular endothelial cell monolayer, the interaction of the vascular endothelial monolayer with the immune system was assessed. The adhesion of activated monocyte cells by tumor necrosis factor α (TNF- α) on the HUVEC monolayer was tested as previously described [22–24]. The monocytes revealed different adherence to the HUVEC monolayers on the selected patterned substrates (Figure 9), and the variation trend is similar to the ICAM-1 expression results.
Figure 9.
Comparison of adhesion of monocytes on HUVEC monolayer on selected patterned substrates. (A) Comparison of representative immunofluorescence images of monocytes adhesion on HUVEC monolayer on selected patterned substrates. (B) Comparison of quantified values of monocyte cell adhesion on HUVEC monolayer on selected patterned substrates. White bar represents 100 μm. HUVECs were stained for CD 31 (in red), nuclear marker - DAPI (in blue) and monocytes were labeled with CellTracker dye (in green). Data (n=20 for all surfaces) were evaluated on outliers using Grubb’s test. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-hoc test. Data represent mean±SD, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
Micro-sized lens topographies (1.8 μm diameter with 2 μm pitch, and 0.7 μm sag convex and concave microlens) exhibited lower monocyte adhesion than other pillars structures (500 nm pillars with 10 μm pitch and 500 nm height, 2 μm pillars with 12 μm pitch and 2 μm height and 10 μm pillars with 10 μm pitch and 10 μm height) and the unpatterned surface control. The pillar topographies showed an increasing trend of monocyte amounts adherent to the endothelial monolayer (Figure 9B). 500 nm pillar topography was found to have comparable monocyte adhesion to the unpatterned substrate, the 2 μm pillars and the 10 μm pillar topography demonstrated a higher monocyte adhesion in comparison to all other topographies and the unpatterned substrate.
4. Discussion
Numerous studies demonstrated the significance of various aspects of physical cues and their influence on vascular endothelial cells. These physical cues include ECM [25,26], shear stress [10,11], substrate stiffness [8], stretching force [9] and topography [13–18]. However, the evaluations of physical cues on endothelial cells were performed in assays evaluating one parameter at a time, unlike high throughput studies evaluating the influence of physical cues on other cell types [21,27–34]. In this study, we focused on exploring the high throughput screening of the influence of topographies on endothelial cell behavior. The study was designed to be two parts: first, single cell studies on the MARC chip, in which cell adhesion, proliferation and morphology were assessed to investigate the influence of external environment on the initial endothelial cell response; subsequently, the endothelial monolayer formation and function on the selected patterns, in which cell markers expression and monocyte adhesion were analyzed to assess the endothelialization under different external environment.
The evaluation of topographic influence on different cells types with the MARC chip assay was studied previously [21,34]. To improve the utilization of the MARC chip, it was redesigned to a chamber which requires a smaller amount of culture medium, growth factors, and other substances. Such a MARC chip modification increased the application potential of the MARC assay. The MARC plate, with one pattern in each well of a multi-well plate, was designed and fabricated for a downstream investigation of selected patterns for various cell response assays such as adhesion and proliferation. A range of micro- and nano-patterned PDMS substrates was utilized to form a unified multi-topographical assay, the MARC plate. It provided a larger cell culture area per topography than the screening device MARC chip and MARC chamber. In addition, the MARC plate could also enable molecular biology assay in each of the individual well, which is conceptually similar to the Integrated mechanobiology platform (IMP) [32]. In this study, the MARC chamber and MARC plate were used for screening and downstream verification of endothelial cell adhesion and proliferation, respectively.
The vascular endothelial cell interaction with the native extracellular matrix, or with biomaterials of implantable device, is an increasingly important subject in studies endothelial dysfunction and vascular tissue engineering. The cell-interaction with biomaterials is essential in designing vascular devices. Much previous research focused on the biochemical cue interaction with vascular endothelial cells. However, it was shown that the physical parameters of the extracellular matrix, such as rigidity and topography, have an impact on the vascular cell behavior as well. For instance, a previous study was conducted identifying the influence of ECM coating on vascular endothelial cells, focusing on collagen I coated substrates [25,26]. Vascular endothelial cells are capable of inducing the production of collagen I through the reduction of autocrine and paracrine nitric oxide (NO) signaling [35]. The NO paracrine effect influences vascular endothelial cells and smooth muscle cells on vasculogenesis [35] which is a process that leads to de novo formation of vessels [36,37]. Although collagen I plays an important role in supporting endothelialization [38,39] and vasculogenesis [40], other ECM proteins and the combination of other ECM proteins with collagen I are all crucial for vascular endothelial cells. Other physical cues such as topography were investigated extensively, especially the influence of topographical cues on vascular endothelial adhesion[41]. Several studies focused on either anisotropic [16,18,42–45] or isotropic topographies [13,46–48]. This study demonstrated a systematic screening of various nano- and micro-sized patterned substrates with different isotropies, with or without collagen I coating.
Extracellular matrix serves as critical support for vascular endothelial cells, and ECM will influence cell behaviors during the long-term cell-interaction with materials. However, the turnover of ECM for endothelial cells usually takes hours to a day [49,50]. In our study of single cell behaviors, we aimed to evaluate the initial interaction between cells and substrates. Thus, the first 4 hours and 24 hours were investigated, during these periods, the adsorbed ECM proteins from serum, or the collagen I coating would be more influential for cell behaviors. The stability and the adsorption of collagen I on substrates were influenced by micro- and nano-sized topographies due to the fibrous nature of collagen I. Therefore, not surprisingly, the collagen binding on the MARC chip showed variations among differently patterned substrates although the difference was not statistically significant. This observation was similar and consistent with our previous studies demonstrating non-significant differences of fibronectin coating adsorption quantification on nano-patterned and unpatterned PDMS substrates [51].
In the single cell study, collagen I coating was observed to have an additive influence on enhancing cell adhesion on patterned substrates. This observation was similar to a recent study, in which the influence of collagen I, fibronectin, or their combination on nano-fibrous membranes showed an additive effect on cell-cell interactions [52]. However, the augmenting influence of patterned substrates with collagen I coating was not consistent among experimental replicas or patterns in the current study. Meanwhile, it should be noted that the non-coated substrates would also be coated with a layer of ECM proteins such as fibronectin adsorbed from the serum in the cell culture medium. Interestingly, a more consistent topographical effect on cell adhesion and proliferation was observed on the substrates without collagen I, but only with adsorbed serum protein. These results imply that the combination of collagen I or fibronectin with topographical cues could exert different influences on vascular endothelial cell processes.
Nano- and micro-sized patterns such as pillars, wells, and the curved structures (lens topography), yielded a lower cell density and cell proliferation when compared to other patterned and unpatterned substrate. Similar results of a selective influence on the cell adhesion of vascular cells was previously observed on pillars and grooves [53]. Although the reasons for lower cell adhesion are unclear, it is speculated that the characteristics of the pillars, wells and lens topographies could influence the cell sensing machinery. Micro- and nano-topography influence on cell-matrix interaction such as focal adhesion (FA) formation was demonstrated by various studies including the work of our group [48,54,55]. The cell sensing machinery is potentially tuned to the characteristic lengths of FA of vascular endothelial cells as previously described [56]. Interestingly, in this study, even the quantity of collagen I adsorbed on the various patterns was not significantly different, the topographies and the potential differences in the conformation of ECM protein may affect the FA formation and modulate cell behaviors. The FA interaction with specific patterns could in turn result in different cell morphology and cell area. It was observed that the curved and round structures influenced the HUVECs to exhibit smaller cell area and round morphology. Therefore, the reduction in cell density and proliferation could be affected by the reduced FA interaction with round or curved topography.
In terms of proliferation, regardless of the ECM coating, no consistent trends of augmenting or inhibitory influence were observed across the patterned and unpatterned substrates. This observation could be attributed to vascular endothelial cell-cell contact that has an inhibitory influence on cell proliferation [57–59]. To minimize the effect of cell-cell contact inhibition on the proliferation studies, two different initial seeding densities were chosen. With a lower cell seeding density, a higher cell proliferation was observed on the patterned surfaces without collagen I coating (Supplementary Figure 12A). However, on patterns that yielded higher cell density at 24 hour time point, the cell proliferation rate, in terms of low EdU percentage, remained low. For example, in Figure 3 and Figure 4, pattern 41 showed a high cell density, but not a high proliferation percentage (Figure 5, Supplementary Figure 12). Thus, it was speculated that the cell-cell contact inhibition of the vascular endothelial cells could affect the proliferation assessment. The assessment could be improved by analyzing the proliferation at an earlier time-point. Using the individual pattern study setup such as in the MARC plate could also provide a more precise control of the starting density in each of the patterns.
Besides the substrate geometry and the ECM coating, the initial cell seeding density plays an important role in the adhesion of vascular endothelial cells. Cell seeding density, especially in cell therapy and tissue regeneration, is an important factor regardless of the cell type and applications [60–62]. On all patterned substrates, including the micro-sized lens topographies that exhibited low cell adhesion and cell proliferation, a higher initial cell seeding density led to higher cell survival and cell adhesion of HUVECs. Thus it was confirmed that the initial cell seeding density is an important parameter in vascular applications [63].
The formation and function of endothelial monolayer will be important to understand vascular endothelial cell-interaction with the underlying substrate. The MARC chamber screening assay investigated the endothelial cell behavior of sub-confluence cell culture. As there is no physical barrier between topographical patterns on a MARC chip, confluence monolayer of cells on MARC chip could potential receive influences and signaling from the nearby patterns through cell-cell signaling and paracrine signals. Thus, the topographical influence on the vascular endothelial cell monolayer was additionally investigated using the MARC plate.
CD31 is a cell junctional molecule that is known to have diverse vascular functions, including inflammatory response and platelet function [64]. Various studies [65,66] have investigated the regulation of expression of CD31 on endothelial cells. In this study, the topographic cues didn’t significantly affect the expression of CD31. Cell-cell junction is another important parameter and function of endothelial monolayer. VE-cadherin is an endothelial specific adhesion molecule located between endothelial cells that regulates the permeability of vascular cell monolayer [67]. All the patterns showed a promotion of VE-cadherin expression, whereby the most significant upregulation of VE-cadherin expression was found on 10 μm pillar substrates. In addition to CD31 and VE-cadherin expression, another important cellular junction molecule is ICAM-1. ICAM-1 is an intercellular adhesion molecule that is largely related to the cell immune response. Substrates with pillar structures showed higher expression of ICAM-1 compared to lens topographies. The most significantly higher expression was found on 10 μm pillar substrates. Such results indicated that the 10 μm pillar topography has the most significant modulation of HUVEC monolayer.
Another major function of an intact vascular endothelial cell monolayer is the interaction with the immune system [1,2]. Various physical cues were previously studied as effectors in the interaction between the vascular endothelial cells and the immune system [68,69]. To investigate the influence of the underlying patterned substrate on the function of the vascular endothelial cell monolayer, the adhesion of monocyte cells to the vascular endothelial cell monolayer was evaluated. Lower monocyte cell adhesion was observed on vascular endothelial cell monolayer cultured on lens topographies compared to the vascular endothelial cells cultured on unpatterned surfaces. On the other hand, the pillar topographies (500 nm, 2 μm pillars and 10 μm pillars with diameter to height 1:1 ratio) demonstrated higher adhesion of monocytes. The results were in line with the ICAM-1 expression data, and verified the data obtained from cell marker expression. Such results led to a speculation that the height, diameter and spacing could be the main parameters in modulating the function of vascular endothelial cells in terms of the interaction with monocytes. Additionally, the curved character of the lens topography could play a crucial role in the monocyte-vascular endothelial cells interaction too. The influence of lens topographies was investigated in few studies. It was demonstrated that 1.8 μm convex microlens can induce differentiation of mesenchymal stem cells [34] and 10 μm convex and concave micro-structures influence the shape of human macrophages [70]. Large scale (250 – 750 μm) convex and concave lens substrates were shown to alter the cell migration [71], adhesion and proliferation [72] of mesenchymal stem cells. It was speculated that such curved geometry leads to nucleus and cell membrane deformations, which influences the cellular behavior [71,72]. However, none of the lens topography studies examined the influence on cellular behavior in terms of interaction with other cells such as monocytes. The answer for the question of what parameters of convex and concave topographies influences the cellular behavior could be found in paracrine substances such as cytokines and their release and ECM modulation. It was previously shown that the spatial ratio of nanogratings and grooves decreases the secretion of inflammatory cytokines, such as IL-1β, IL-3 and MCP-1, from NIH3T3 fibroblasts when compared to the inflammatory cytokines release from NIH3T3 fibroblasts on unpatterned substrates [73]. The mechanosensory complex of endothelial cells and integrin activation were also shown to be influenced by physical cues [74]. The decreased α5β1 integrin binding on microlens topography without collagen I coating could lead to a decreased interaction with the fibronectin contained in the culture medium. Endothelial cells must be anchored to their extracellular substrate in order to transduce mechanical signals and respond to them [75]. Therefore, a lower interaction with fibronectin will lead to structural changes and cytoskeleton remodeling, which subsequently influences the capability of the apical site of endothelial cells to interact with other cells [45]. Such a reaction would result in a lower adhesion of monocytes to the endothelial cell monolayer as observed on lens topographies. The opposite process of an increased monocyte cell adhesion was observed on the pillar topographies. The increase of diameter and height of pillars with diameter to height 1:1 ratio also demonstrated an increasing trend of monocyte adhesion. Such influence of pillar topographies with various diameters on endothelial cell adhesion was shown previously [54]. However, to investigate these processes further, pillar topographies with various spatial ratios and lens topographies with various diameter and heights must be examined.
5. Conclusions
This study investigated the effects of physical cues on vascular endothelial cells including cell adhesion, cell proliferation, cell morphology, and cell function, such as immune-modulation.
A systematic screening of various nano- and micro-sized patterned substrates with or without collagen I coating at two different initial cell-seeding densities was implemented. The study was preformed using two high throughput platforms: the MARC chamber and the MARC plate. Regardless of the ECM coating and cell seeding density, patterned substrates demonstrated either promoting or inhibitory influence on cell adhesion and cell proliferation. The cell morphology in the process was influenced as well. Taken together, the data suggests the importance of topographical cue interaction with endothelial cells.
This study emphasized an effective, systematic screening process of topographies. It also demonstrated the promoting or inhibitory effects of topographical cues on the cell marker expression of vascular endothelial cell monolayers, and the endothelial monolayer interaction with monocytes. Findings presented in this study emphasize the importance of selecting specific topographical patterns for applications in vascular biomedical devices, such as grafts and stents, where low immunomodulation is required.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation, Prime Minister’s Office, Singapore under its Research Centre of Excellence programme administered by the Mechanobiology Institute, Singapore and partially supported by National Institute of Health (NIH R01 HL130274-01A1). The authors would like to thank M. Sheetz, KK Teo, D Wong and M Cutiongco for their helpful discussion and technical support. M.K.’s scholarship was supported by the Mechanobiology Institute scholarship, which was funded by the National Research Foundation, Singapore and Ministry of Education, Singapore. Y.Y. is supported by National Institute of Health (NIH R01 HL130274-01A1). S.H.G. was supported by A*STAR Scientific Staff Development Award Program.
Contributor Information
Marek Kukumberg, Mechanobiology Institute, National University of Singapore, #05-01 T-lab, 5A Engineering Drive 1, Singapore 117411.
Yuan Yao, Department of Chemical Engineering, University of Waterloo, 200 University Avenue West, Waterloo, Ontario, N2L 3G1, Canada.
Seok Hong Goh, Institute of Materials Research and Engineering, Agency for Science, Technology and Research (A*STAR), 2 Fusionopolis Way, Innovis, 138634, Singapore, Department of Biomedical Engineering, National University of Singapore, E4, #04-10,4 Engineering Drive 3, Singapore 117583.
Dawn JH Neo, Mechanobiology Institute, National University of Singapore, #05-01 T-lab, 5A Engineering Drive 1, Singapore 117411.
Jia Yi Yao, Department of Biomedical Engineering, National University of Singapore, E4, #04-10,4 Engineering Drive 3, Singapore 117583.
Evelyn KF Yim, Mechanobiology Institute, National University of Singapore, #05-01 T-lab, 5A Engineering Drive 1, Singapore 117411, Department of Biomedical Engineering, National University of Singapore, E4, #04-10,4 Engineering Drive 3, Singapore 117583, Department of Surgery, National University of Singapore, NUHS Tower Block, Level 8,1E Kent Ridge Road, Singapore 119228, Department of Chemical Engineering, University of Waterloo, 200 University Avenue West, Waterloo, Ontario, N2L 3G1, Canada.
References
- 1.Michiels C. J Cell Physiol. 2003;196:430. doi: 10.1002/jcp.10333. [DOI] [PubMed] [Google Scholar]
- 2.Dudley AC. Cold Spring Harbor Perspectives in Medicine. 2012;2:a006536. doi: 10.1101/cshperspect.a006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis GE. Circulation Research. 2005;97:1093. doi: 10.1161/01.RES.0000191547.64391.e3. [DOI] [PubMed] [Google Scholar]
- 4.Inoue S, Leblond CP, Laurie GW. The Journal of Cell Biology. 1983;97:1524. doi: 10.1083/jcb.97.5.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leblond CP, Inoue S. American Journal of Anatomy. 1989 doi: 10.1002/aja.1001850403/full. [DOI] [Google Scholar]
- 6.LeBleu VS, Macdonald B, Kalluri R. Exp Biol Med (Maywood) 2007;232:1121. doi: 10.3181/0703-MR-72. [DOI] [PubMed] [Google Scholar]
- 7.Baluk P, Morikawa S, Haskell A, Mancuso M, McDonald DM. The American Journal of Pathology. 2003;163:1801. doi: 10.1016/S0002-9440(10)63540-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood JA, Shah NM, McKee CT, Hughbanks ML, Liliensiek SJ, Russell P, Murphy CJ. Biomaterials. 2011;32:5056. doi: 10.1016/j.biomaterials.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W, Sakamoto N, Miyazawa R, Sato M. Biochemical and Biophysical Research Communications. 2012;418:708. doi: 10.1016/j.bbrc.2012.01.083. [DOI] [PubMed] [Google Scholar]
- 10.Ahmann KA, Johnson SL, Hebbel RP, Tranquillo RT. Tissue Engineering Part A. 2011;17:2511. doi: 10.1089/ten.tea.2011.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stolberg S, McCloskey KE. Biotechnol Prog. 2008;25:10. doi: 10.1002/btpr.124. [DOI] [PubMed] [Google Scholar]
- 12.Egginton S. Biochem Soc Trans. 2011;39:1633. doi: 10.1042/BST20110715. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson LE, Rand DR, Tsao J, Eberle W, Gerecht S. J Biomed Mater Res. 2012;100A:1457. doi: 10.1002/jbm.a.34059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan JT, Wood JA, Shah NM, Hughbanks ML, Russell P, Barakat AI, Murphy CJ. Biomaterials. 2012;33:4126. doi: 10.1016/j.biomaterials.2012.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biela SA, Su Y, Spatz JP, Kemkemer R. Acta Biomaterialia. 2009;5:2460. doi: 10.1016/j.actbio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Dreier B, Gasiorowski JZ, Morgan JT, Nealey PF, Russell P, Murphy CJ. AJP: Cell Physiology. 2013;305:C290. doi: 10.1152/ajpcell.00264.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L, Aid R, Le Visage C, Chew SY. Macromol Biosci. 2011;12:395. doi: 10.1002/mabi.201100336. [DOI] [PubMed] [Google Scholar]
- 18.Whited BM, Rylander MN. Biotechnol Bioeng. 2013;111:184. doi: 10.1002/bit.24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez E, Engel E, Planell JA, Samitier J. Annals of Anatomy - Anatomischer Anzeiger. 2009;191:126. doi: 10.1016/j.aanat.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Ankam S, Suryana M, Chan LY, Moe AAK, Teo BKK, Law JBK, Sheetz MP, Low HY, Yim EKF. Acta Biomaterialia. 2013;9:4535. doi: 10.1016/j.actbio.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Moe AAK, Suryana M, Marcy G, Lim SK, Ankam S, Goh JZW, Jin J, Teo BKK, Law JBK, Low HY, Goh ELK, Sheetz MP, Yim EKF. Small. 2012 doi: 10.1002/smll.201200490. [DOI] [PubMed] [Google Scholar]
- 22.Giuffrè L, Cordey AS, Monai N, Tardy Y, Schapira M, Spertini O. The Journal of Cell Biology. 1997;136:945. doi: 10.1083/jcb.136.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber J, Fürnkranz A, Bochkov VN, Patricia MK. The Journal of Lipid Research. 2006;47:1054. doi: 10.1194/jlr.M500555-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz B, Vischer P, Brand E, Schmidt-Petersen K, Korb-Pap A, Guske K, Nedele J, Schelleckes M, Hillen J, Rötrige A, Simmet T, Paul M, Cambien F, Brand SM. Atherosclerosis. 2013;230:185. doi: 10.1016/j.atherosclerosis.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 25.Wissink MJ, van Luyn MJ, Beernink R, Dijk F, Poot AA, Engbers GH, Beugeling T, van Aken WG, Feijen J. Thromb Haemost. 2000;84:325. [PubMed] [Google Scholar]
- 26.Shamloo A, Mohammadaliha N, Heilshorn SC, Bauer AL. Annals of Biomedical Engineering. 2015:1. doi: 10.1007/s10439-015-1416-2. [DOI] [PubMed] [Google Scholar]
- 27.Markert LD, Lovmand J, Foss M, Lauridsen RH, Lovmand M, Füchtbauer EM, Füchtbauer A, Wertz K, Besenbacher F, Pedersen FS, Duch M. Stem Cells and Development. 2009;18:1331. doi: 10.1089/scd.2009.0114. [DOI] [PubMed] [Google Scholar]
- 28.Unadkat HV, Hulsman M. Proceedings of the National Academy of Sciences. 2011 [Google Scholar]
- 29.Hulsman M, Hulshof F, Unadkat H, Papenburg BJ, Stamatialis DF, Truckenmüller R, van Blitterswijk C, de Boer J, Reinders MJT. Acta Biomaterialia. 2015;15:29. doi: 10.1016/j.actbio.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Tang QY, Jadhav AD, Narang A, Qian WX, Shi P, Pang SW. Sci Rep. 2015;5:941. doi: 10.1038/srep08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neto AI, Vasconcelos NL, Oliveira SM, Ruiz-Molina D, Mano JF. Adv Funct Mater. 2016;26:2745. [Google Scholar]
- 32.Hu J, Gondarenko AA, Dang AP, Bashour KT, O’Connor RS, Lee S, Liapis A, Ghassemi S, Milone MC, Sheetz MP, Dustin ML, Kam LC, Hone JC. Nano Lett. 2016 doi: 10.1021/acs.nanolett.5b04364. acs.nanolett.5b04364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ankam S, Teo BK, Kukumberg M, Yim EK. organogenesis. 2013;9:128. doi: 10.4161/org.25425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kukumberg M, Yao JY, Neo DJH, Yim EKF. Biomaterials. 2017;131:68. doi: 10.1016/j.biomaterials.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 35.Myers PR, Tanner MA. Arteriosclerosis, Thrombosis, and Vascular Biology. 1998;18:717. doi: 10.1161/01.atv.18.5.717. [DOI] [PubMed] [Google Scholar]
- 36.Kubis N, Levy BI. Interv Neuroradiol. 2003;9:239. doi: 10.1177/159101990300900302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patan S. Journal of Neuro-Oncology. 2000;50:1. doi: 10.1023/a:1006493130855. [DOI] [PubMed] [Google Scholar]
- 38.Sgarioto M, Vigneron P, Patterson J, Malherbe F, Nagel MD, Egles C. Comptes rendus - Biologies. 2012;335:520. doi: 10.1016/j.crvi.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Peterson AW, Caldwell DJ, Rioja AY, Rao RR, Putnam AJ, Stegemann JP. Biomater Sci. 2014;2:1497. doi: 10.1039/C4BM00141A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vernon RB, Lara SL, Drake CJ, Iruela-Arispe ML, Angello JC, Little CD, Wight TN, Sage EH. In Vitro Cell Dev Biol Anim. 1995;31:120. doi: 10.1007/BF02633972. [DOI] [PubMed] [Google Scholar]
- 41.Chong DST, Lindsey B, Dalby MJ, Gadegaard N, Seifalian AM, Hamilton G. European Journal of Vascular and Endovascular Surgery. 2014;47:566. doi: 10.1016/j.ejvs.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Uttayarat P, Toworfe GK, Dietrich F, Lelkes PI, Composto RJ. J Biomed Mater Res. 2005;75A:668. doi: 10.1002/jbm.a.30478. [DOI] [PubMed] [Google Scholar]
- 43.Uttayarat PILARJCP. 2016:1. [Google Scholar]
- 44.Hatano R, Mercurio K, Luna JI, Glaser DE, Leppert VJ, McCloskey KE. J Biol Eng. 2013;7:18. doi: 10.1186/1754-1611-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeon H, Tsui JH, Jang SI, Lee JH, Park S, Mun K, Boo YC, Kim DH. ACS Appl Mater Interfaces. 2015;7:4525. doi: 10.1021/acsami.5b00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zawislak JD, Kolasinski KW, Helmke BP. Phys Status Solidi (a) 2009;206:1356. [Google Scholar]
- 47.Chua J, Liew L, Yim E. JFB. 2013;4:38. doi: 10.3390/jfb4010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muhammad R, Peh GSL, Adnan K, Law JBK, Mehta JS, Yim EKF. Acta Biomaterialia. 2015;19:138. doi: 10.1016/j.actbio.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 49.Mann BK, Tsai AT, Scott-Burden T, West JL. Biomaterials. 1999;20:2281. doi: 10.1016/s0142-9612(99)00158-1. [DOI] [PubMed] [Google Scholar]
- 50.Kusuma S, Zhao S, Gerecht S. The FASEB Journal. 2012;26:4925. doi: 10.1096/fj.12-209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teo BKK, Wong ST, Lim CK, Kung TYS, Yap CH, Ramagopal Y, Romer LH, Yim EKF. ACS Nano. 2013;7:4785. doi: 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]
- 52.Kang D, Kim JH, Jeong YH, Kwak JY, Yoon S, Jin S. Biofabrication. 2016;8:1. doi: 10.1088/1758-5090/8/2/025008. [DOI] [PubMed] [Google Scholar]
- 53.Ding Y, Yang Z, Bi CWC, Yang M, Xu SL, Lu X, Huang N, Huang P, Leng Y. ACS Appl Mater Interfaces. 2014;6:12062. doi: 10.1021/am502692k. [DOI] [PubMed] [Google Scholar]
- 54.Cutiongco MFA, Goh SH, Aid-Launais R, Le Visage C, Low HY, Yim EKF. Biomaterials. 2016;84:184. doi: 10.1016/j.biomaterials.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 55.Rizwan M, Peh GS, Adnan K, Naso SL, Mendez AR, Mehta JS, Yim EKF. Advanced Healthcare Materials. 2016:1. doi: 10.1002/adhm.201600848. [DOI] [PubMed] [Google Scholar]
- 56.Ventre M, Natale CF, Rianna C, Netti PA. Journal of The Royal Society Interface. 2014;11:20140687. doi: 10.1098/rsif.2014.0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vlodavsky I, Fielding PE, Fielding CJ, Gospodarowicz D. Proceedings of the National Academy of Sciences. 1977;75:356. doi: 10.1073/pnas.75.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz SM, Gajdusek CM. Biology of Endothelial Cells. Springer US; Boston, MA: 1984. pp. 66–73. [Google Scholar]
- 59.Suzuki E, Nagata D, Yoshizumi M, Kakoki M, Goto A, Omata M, Hirata Y. J Biol Chem. 2000;275:3637. doi: 10.1074/jbc.275.5.3637. [DOI] [PubMed] [Google Scholar]
- 60.Nirmalanandhan VS, Levy MS, Huth AJ, Butler DL. Tissue Eng. 2006;12:1865. doi: 10.1089/ten.2006.12.1865. [DOI] [PubMed] [Google Scholar]
- 61.Yassin MA, Leknes KN, Pedersen TO, Xing Z, Sun Y, Lie SA, Finne-Wistrand A, Mustafa K. J Biomed Mater Res. 2015;103:3649. doi: 10.1002/jbm.a.35505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gage BK, Webber TD, Kieffer TJ. PLoS ONE. 2013;8:e82076. doi: 10.1371/journal.pone.0082076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahota PS, Burn JL, Brown NJ, MacNeil S. Wound Repair Regen. 2004;12:635. doi: 10.1111/j.1067-1927.2004.12608.x. [DOI] [PubMed] [Google Scholar]
- 64.Woodfin A, Voisin MB, Nourshargh S. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;27:2514. doi: 10.1161/ATVBAHA.107.151456. [DOI] [PubMed] [Google Scholar]
- 65.Shaw SK, Perkins BN, Lim YC, Liu Y, Nusrat A, Schnell FJ, Parkos CA, Luscinskas FW. The American Journal of Pathology. 2010;159:2281. doi: 10.1016/s0002-9440(10)63078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mamdouh Z, Chen X, Pierini LM, Maxfield FR, Muller WA. Nature. 2003;421:748. doi: 10.1038/nature01300. [DOI] [PubMed] [Google Scholar]
- 67.Vestweber D. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;28:223. doi: 10.1161/ATVBAHA.107.158014. [DOI] [PubMed] [Google Scholar]
- 68.Rinker KD, Prabhakar V, Truskey GA. Biophysj. 2001;80:1722. doi: 10.1016/S0006-3495(01)76143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mestas J, Ley K. Trends in Cardiovascular Medicine. 2008;18:228. doi: 10.1016/j.tcm.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malheiro V, Lehner F, Dinca V, Hoffmann P, Maniura-Weber K. Biomater Sci. 2016;4:1562. doi: 10.1039/c6bm00425c. [DOI] [PubMed] [Google Scholar]
- 71.Werner M, Blanquer SBG, Haimi SP, Korus G, Dunlop JWC, Duda GN, Grijpma DW, Petersen A. Adv Sci. 2016;4:1600347. doi: 10.1002/advs.201600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park JY, Lee DH, Lee EJ, Lee SH. Lab Chip. 2009;9:2043. doi: 10.1039/b820955c. [DOI] [PubMed] [Google Scholar]
- 73.Jeon H, Koo S, Reese WM, Loskill P, Grigoropoulos CP, Healy KE. Nature Materials. 2015:1. doi: 10.1038/nmat4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tzima E, del Pozo MA, Shattil SJ, Chien S, Schwartz MA. The EMBO Journal. 2001;20:4639. doi: 10.1093/emboj/20.17.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takahashi M, Berk BC. J Clin Invest. 1996;98:2623. doi: 10.1172/JCI119083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.