Abstract
Spermatogonia represent a diploid germ cell population that includes spermatogonial stem cells. In this report, we describe new methods for isolation of highly enriched porcine spermatogonia based on light scatter properties, and for targeted mutagenesis in porcine spermatogonia using nucleofection and TALENs. We optimized a nucleofection protocol to deliver TALENs specifically targeting the DMD locus in porcine spermatogonia. We also validated specific sorting of porcine spermatogonia based on light scatter properties. We were able to obtain a highly enriched germ cell population with over 90% of cells being UCH-L1 positive undifferentiated spermatogonia. After gene targeting in porcine spermatogonia, indel (insertion or deletion) mutations as a result of non-homologous end joining (NHEJ) were detected in up to 18% of transfected cells. Our report demonstrates for the first time an approach to obtain a live cell population highly enriched in undifferentiated spermatogonia from immature porcine testes, and that gene targeting can be achieved in porcine spermatogonia which will enable germ line modification.
Keywords: gene editing, germ cells, nuclease, nucleofection, pig, sorting
1 | INTRODUCTION
Spermatogonia represent a diploid germ cell population in the seminiferous tubules which undergo mitotic division during spermatogenesis to give rise to primary spermatocytes. Spermatogonial stem cells (SSCs) are a subset of undifferentiated type A spermatogonia which have the capacity to self-renew to maintain the SSC pool as well as to differentiate to ultimately form sperm. SSCs represent a rare cell population, constituting ~0.03% of all germ cells in the adult mouse testis (Tegelenbosch & de Rooij, 1993). The lack of definite phenotypical, morphological, or biochemical markers that can unequivocally identify SSCs makes it impossible to isolate SSCs for in vitro studies. However, SSCs can be relatively enriched from a testicular cell suspension by various in vivo and/or in vitro approaches.
For in vivo enrichment, neonatal, and prepubertal testes serve as the preferred source for harvesting germ cells as gonocytes/spermatogonia are the only type of germ cells present in the seminiferous tubules during those developmental stages (Bellve et al., 1977). Surgical induction of cryptorchidism, Vitamin A deficiency and hyperthermic treatment in adult mouse testis resulted in enrichment of SSCs in a testicular cell preparation by eliminating differentiating germ cells (McLean, Russell, & Griswold, 2002; Shinohara, Avarbock, & Brinster, 2000). For in vitro enrichment of spermatogonia, several approaches that take advantage of differences in density and size of various cell populations (such as Percoll gradient centrifugation) or that rely on different adhesion of somatic cells and germ cells to a substratum in culture (such as differential plating) have been implemented (Bellve et al., 1977; Luo, Megee, Rathi, & Dobrinski, 2006; Morena, Boitani, Pesce, De Felici, & Stefanini, 1996; van Dissel-Emiliani, de Rooij,&Meistrich, 1989). With the identification of surface markers present on a subset(s) of spermatogonia such as Thy1, GFR-α1, c-kit, CD9, Integrin-α6, and Integrin-β1, magnetic-activated cell sorting (MACS), and fluorescence-activated cell sorting (FACS) have been used to isolate a cell population enriched with SSCs in rodents (Ebata, Zhang, & Nagano, 2005; Kanatsu-Shinohara, Toyokuni, & Shinohara, 2004; Kubota, Avarbock, & Brinster, 2004; Shinohara, Orwig, Avarbock, & Brinster, 2000).
For domestic animals, it is challenging to identify a surface marker that can be reliably and consistently used for enriching spermatogonia containing SSCs. So far, among all the surface markers reported for rodents, only Thy1 has been described for enriching undifferentiated spermatogonia in prepubertal goats, pigs and cattle (Abbasi et al., 2013; Reding, Stepnoski, Cloninger, & Oatley, 2010; Zheng et al., 2014). Currently, the prevailing methods for enriching SSCs from large animals are differential plating and Percoll density gradient centrifugation, which do not rely on the presence of specific surface protein(s) on spermatogonia. The combination of those methods has enabled a high enrichment of gonocytes/spermatogonia (up to >70%) from neonatal/prepubertal donors in pigs and cattle (Aponte et al., 2008; Fujihara, Kim, Minami, Yamada, & Imai, 2011; Goel, Sugimoto, Minami, Yamada, Kume, & Imai, 2007). Here, we show that a highly pure population of live spermatogonia can be reliably obtained by sorting based only on light scatter properties of spermatogonia.
One unique characteristic of SSCs is that when transplanted into the seminiferous tubules of a recipient animal, SSCs are able to colonize the stem cell niche at the basement membrane and establish long-term donor-derived spermatogenesis in recipients (Brinster & Zimmermann, 1994). This feature allows transmission of the donor haplotype to the offspring by recipient animals, which makes SSCs an attractive vehicle for germline genetic modifications (Brinster & Avarbock, 1994). Rodent SSCs that carried randomly integrated transgenes in their genome have resulted in transgenic mouse and rat offspring upon germ cell transplantation (Hamra et al., 2002; Nagano et al., 2001; Nagano, Shinohara, Avarbock, & Brinster, 2000; Ryu et al., 2007; Takehashi et al., 2007). With the establishment of a long-term culture system for rodent SSCs which allows in vitro selection, expansion, and screening of SSCs, gene targeting in SSCs became possible (Iwamori, Iwamori, & Matzuk, 2012; Izsvak et al., 2010; Kanatsu-Shinohara et al., 2006; Kanatsu-Shinohara, Toyokuni, & Shinohara, 2005). Mouse SSCs that carried targeted mutations and withstood extensive in vitro manipulation were able to colonize the recipient testis, produce mutant sperm, and result in mutant offspring (Kanatsu-Shinohara et al., 2006). In addition, homologous recombination-mediated gene correction was feasible in mouse SSCs, which revealed their potential in gene therapy (Iwamori, Iwamori, & Matzuk, 2012). Genome-wide mutagenesis has also been achieved in rat SSCs by using Sleeping Beauty transposon-based gene trap vectors (Izsvak et al., 2010). Transplantation of a polyclonal library of targeted SSCs or individually picked monoclonal targeted SSC lines into the recipient rat testis resulted in germline transmission of the mutations and generation of KO rat offspring (Izsvak et al., 2010).
For domestic animals where germline-competent embryonic stem cells (ESCs) are not readily available, generation of KO animals mainly relies on gene targeting in somatic cells followed by somatic cell nuclear transfer (SCNT) (Laible & Alonso-Gonzalez, 2009). The approach is challenging due to low efficiency of gene targeting in somatic cells, developmental problems associated with SCNT, and the high cost in large animal husbandry (Bacci, 2007; Niemann, Kues, & Carnwath, 2005). Although transgenesis through SSCs has been demonstrated in domestic animal species such as pigs and goats (Honaramooz et al., 2008; Zeng et al., 2012, 2013), random integration of transgenes into the genome did not allow specific and targeted genetic engineering. The recent advent of engineered nucleases such as Zinc-finger nucleases (ZFNs), Transcription Activator-like Effector Nucleases (TALENs), and Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated-9 (CRISPR/Cas-9), has greatly advanced the gene-specific genome editing in domestic animals (Cong et al., 2013; Joung & Sander, 2013; Porteus & Carroll, 2005). Guided either by fused DNA recognition domains (ZFNs and TALENs) or by interacting short RNAs (CRISPRs/Cas-9), the engineered nucleases are targeted to a specific genome locus to create double strand (ds) breaks. The induced ds breaks can be repaired either via non-homologous end joining (NHEJ) or via homologous recombination (HR). Compared to conventional gene targeting that relies on spontaneous events of HR, the efficiency of nucleases-facilitated mutagenesis is much higher with NHEJ-mediated mutations being detected in up to 50% of transfected cells (Urnov, Rebar, Holmes, Zhang, & Gregory, 2010). In several cell lines, targeting efficiency by nuclease-stimulated HR was >1,000 fold higher than that by spontaneous HR in conventional gene targeting (Hauschild-Quintern, Petersen, Cost, & Nieman, 2013).
So far, ZFNs, TALENs, and CRISPR/Cas-9 have been used to generate mono-allelic and bi-allelic knock-out pigs, cattle, and goats through the combination of gene targeting in somatic cells and SCNT (Bao et al., 2014; Carlson et al., 2012; Hauschild et al., 2011; Luo et al., 2014; Ni et al., 2014; Yang et al., 2011; Yu et al., 2011; Zhou et al., 2015). A locus-specific transgene knock-in pig model has also been generated by using CRISPR/Cas-9 and SCNT (Ruan et al., 2015). As a result of their high efficiency in mutagenesis, microinjection of TALENs, ZFNs, and CRISPRs/Cas-9 into pig zygotes resulted in production of live piglets with engineered mutations (Hai, Teng, Guo, Li, & Zhou, 2014; Lillico et al., 2013; Park et al., 2017; Wang et al., 2015). However, CRISPR/Cas9 mediated gene editing in zygotes can result in target allele mosaicism in animals due to independent multiple gene editing events at early embryonic cleavage stages (Niu et al., 2014; Yen et al., 2014). As a result, targeted alleles can differ between somatic tissues and the germline, requiring extensive outcrossing of mutants in order to generate non-mosaic germline of animals isogenic for specific targeted allele in all cells of their body.
To avoid generation of mosaic mutant progeny, direct germline editing using engineered nucleases has recently been implemented for targeting in rodent SSCs (Chapman et al., 2015; Sato et al., 2015; Wu et al., 2015). Both gene knockout and gene correction have been achieved in SSCs and sperm derived from those genome-edited SSCs were used by in vitro fertilization or natural breeding to produce offspring with desired genetic modifications. Similar to what has been observed in other cell types, nucleases-facilitated gene targeting in SSCs showed higher targeting efficiency compared to conventional gene targeting in SSCs (Fanslow et al., 2014; Kanatsu-Shinohara et al., 2006; Sato et al., 2015).
Improved techniques to enrich germ cell populations greatly facilitate other processes such as transfection or gene editing of germ cells, in vitro culture of germ cells, or germ cell transplantation. In the current study, we used a novel approach to sort germ cells by using light scatter to enrich the spermatogonia population which enabled us to optimize conditions for nucleofection of spermatogonia and then to demonstrate that gene targeting by TALENs can be achieved in porcine spermatogonia.
2 | RESULTS
2.1 | Enrichment of spermatogonia by flow cytometry using light scatter properties
We previously developed a differential plating protocol for enriching spermatogonia from prepubertal porcine testis (Luo et al., 2006). Here, we refined the protocol with additional rounds of plating and were able to obtain an enriched germ cell population with 40–60% cells being spermatogonia (UCH-L1+) without the intermediate STA-PUT velocity sedimentation process. This represented a ~8–12 fold of enrichment from the initial testicular cell suspension obtained from testis digestion. As a highly enriched population of spermatogonia is a prerequisite for establishing a culture system for porcine SSCs and for efficient genetic modification, we explored options for further enrichment of spermatogonia including Magnetic- and Flow-Activated Cell Sorting (MACS and FACS, respectively).
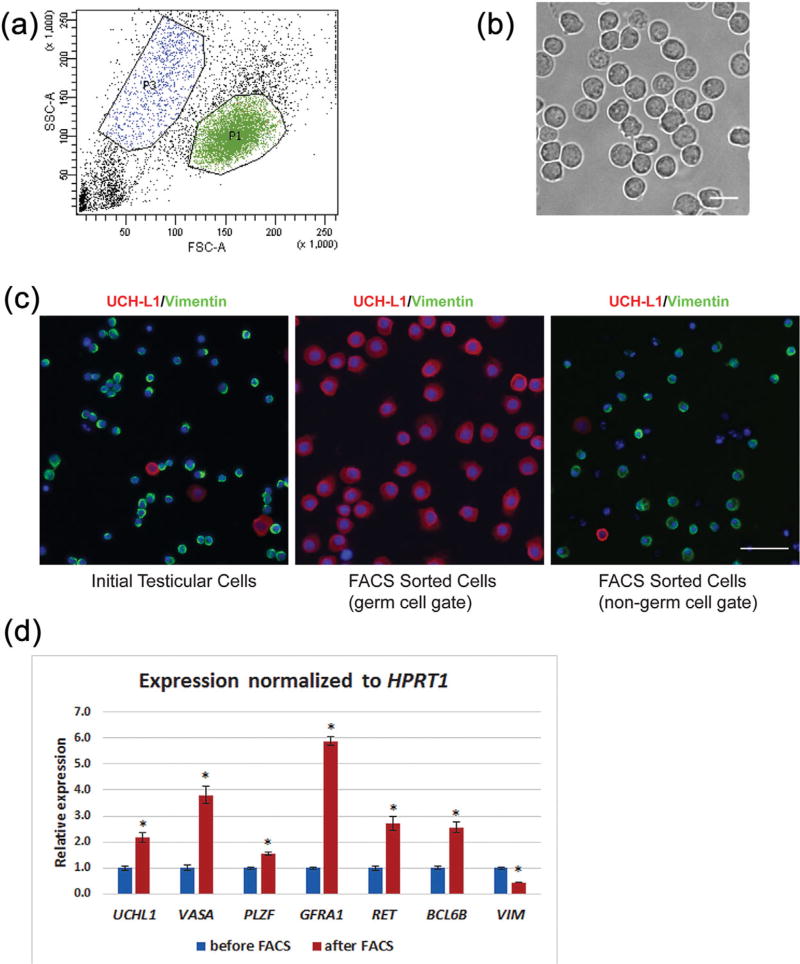
When relatively enriched spermatogonia obtained from differential plating were subjected to flow cytometry, a distinctive population of cells was evident on the light (forward and side) scatter dot plot (Figure 1a). We gated this population of cells and sorted cells within this gate. Cells within the gates of this distinctive population represented 21.68 ± 8.17% of the input cells, were relatively homogenous in morphology (Figure 1b) and had viability of over 95%. Immunocytochemical analysis of these cells for the spermatogonia marker UCH-L1 and the somatic cell marker Vimentin revealed that 92.27% ± 6.10% of the cells were UCH-L1-positive (UCH-L1+)/Vimentin negative (Vimentin−) spermatogonia (Figure 1c) (n = 7). We defined this gate as the germ cell gate. The proportion of Vimentin+ cells varied from 0.11% to 4.17% in the germ cell gate between different experiments (n = 7). Cells outside the germ cell gate (defined as the non-germ cell gate, Figure 1a) were mostly Vimentin+ somatic cells (50–75%), UCH-L1−/Vimentin− cells and cell fragments (Figures 1a and 1c).
FIGURE 1.
Isolation of a cell population highly enriched for undifferentiated spermatogonia by sorting for light scatter properties. (a) Representative scatter plot. P1 indicates the germ cell gate, P3 the non-germ cell gate. (b) Phase contrast microscopic image of cells sorted from the germ cell gate (P1) illustrating the homogenous morphology of sorted cells. Bar = 20 µm. (c) Immunofluorescence analysis of cell populations before and after sorting. UCH-L1 labels undifferentiated spermatogonia, vimentin is expressed in testicular somatic cells. Cells sorted from the germ cell gate are highly enriched in undifferentiated spermatogonia while the majority of cells in the non-germ cell gate are somatic cells. Bar = 50 µm. (d) Relative expression levels of genes specifically expressed in spermatogonia or somatic cells. Sorted cells had significantly higher expression of genes previously reported to be specifically expressed in germ cells and significantly lower expression of vimentin
Gene expression analysis by qRT-PCR revealed significantly higher expression levels of germ cell specific genes such as UCH-L1, VASA, PLZF, BCL6B, GFRα1, and RET. A significantly lower expression level of the somatic cell marker vimentin was observed in sorted cells compared to cells collected prior to sorting (Figure 1d). This finding confirmed that the sorted population was highly enriched for spermatogonia.
While the experiments reported above used cells from 10 weeks old donors (n = 12), the approach was applicable to sorting spermatogonia from 1, 4, and 14 week old donors (n = 6) with similar recovery of input cells (23.92 ± 2.52 %) and percentage of UCH-L1 + spermatogonia (90.57 ± 1.74) in the sorted cell population.
2.2 | Transfection of spermatogonia by nucleofection
Spermatogonia proliferate very slowly and are refractory to lipid-based transfection. We previously used viral vectors (AAV and Lentivirus) to deliver transgenes into goat and pig spermatogonia (Honaramooz et al., 2008; Zeng et al., 2013). Due to the safety concerns and limitations of viral vectors, we adopted nucleofection, an electroporation-based transfection method, and showed that it can be used to transfect goat spermatogonia (Zeng et al., 2012). Initially, we explored the application of different electroporation approaches such as Neon™ (Invitrogen, ThermoFisher Scientific, Burlington, ON, Canada), Gene Pulser MXcell™ (Biorad, Bio-Rad Laboratories, Mississauga, ON, Canada) and Nucleofector™ (Lonza, Lonz, Walkersville, MD) for transfection of pig spermatogonia and found that nucleofection gave the best balance between transfection efficiency and cell viability. We then tested a variety of combinations of nucleofection solutions and programs using a GFP reporter plasmid (pmaxGFP™, Lonza) and performed quantitative assessment on three solution/program combinations that yielded promising results (solution-V/ programX-005, solution-L/programX-001 and solution-B/programU-030). Out of three combinations evaluated, transfection with solution V/program X–005 resulted in the highest recovery of transfected spermatogonia with 49.7 ± 5.72% of cells being recovered after nucleofection and 46.5 ± 2.25% of the recovered germ cells being GFP + spermatogonia (GFP + UCH-L1 +; Table 1).
TABLE 1.
Nucleofection of enriched porcine germ cells
| Solution/ program |
% of cell recovery |
% of GFP + cells |
% of UCH- L1 + cells |
% of GFP + UCH-L1 + cells from recovered cells |
% of GFP + UCH-L1 + from recovered UCH-L1 + cells |
% overall yieldb |
|---|---|---|---|---|---|---|
| V/X-005 | 49.7 ± 5.72a,1 | 39.6 ± 2.161 | 52.9 ± 5.951 | 31.5 ± 7.671 | 46.5 ± 2.251 | 15.2 ± 1.991 |
| L/X-001 | 54.8± 5.571 | 25.9 ± 7.741 | 50.5 ± 3.051 | 19.6 ± 10.831,2 | 37.1 ± 8.291,2 | 10.5 ± 5.011,2 |
| B/U-030 | 33.1 ± 6.782 | 34.3 ± 2.871 | 43.6 ± 12.661 | 15.2 ± 7.622 | 39.1 ± 3.072 | 4.8 ± 2.172 |
Within each column, different numbers indicate statistically significant difference (p < 0.05).
mean ± SD; n = 3 for V/X-005 and B/U-030; n = 4 for L/X-001.
% of GFP+ UCH-L1+ cells from input cells subjected to nucleofection.
Therefore, we used solution-V/programX-005 for subsequent transfection experiments based on the number of cells recovered, the number of transfected spermatogonia recovered and transfection efficiency of germ cells.
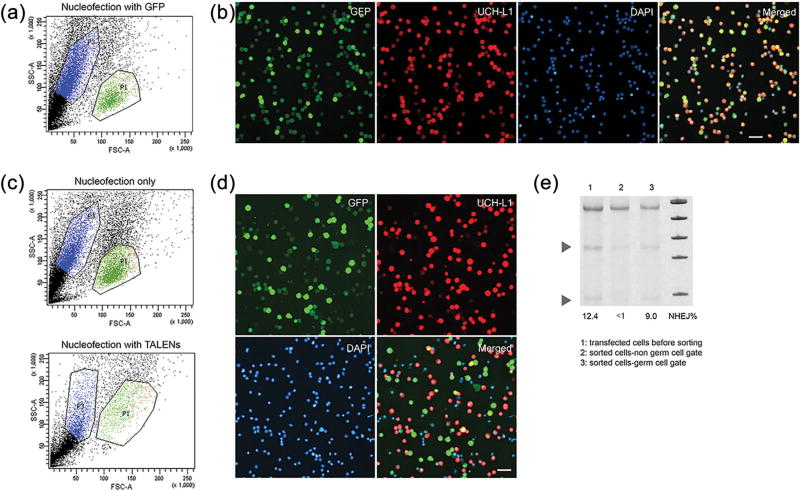
We used flow cytometry to isolate spermatogonia prior to nucleofection; however, the viability and recovery rate of sorted cells subsequently transfected by nucelofection were low (total recovery of input cells was 16.2 ± 6.36% with 1.2 ± 0.22% of input cells GFP+ UCH-L1+; n = 5) resulting in insufficient yield of live cells for applications that require a large number of cells such as germ cell transplantation. Therefore, we did not pursue this approach further, and rather than sorting germ cells prior to nucleofection, we investigated whether we could sort spermatogonia using light scatter properties after nucleofection. Cellular damage due to nucleofection was apparent as fewer cells could be gated in the germ cell gate after nucleofection compared to non-transfected cells (Figure 2a vs. 1b). Despite this, sorting of cells using light scatter properties after nucleofection substantially enriched the proportion of spermatogonia (88.67% ± 7.55% spermatogonia compared to 45.22% ± 6.86% spermatogonia prior to sorting; n = 3). By sorting of GFP+ cells within the germ cell gate we obtained over 80% UCH-L1+ GFP+ spermatogonia (Figures 2a, and 2b). The cell population outside of the germ cell gate (non-germ cell gate) contained only 2–15% spermatogonia (UCH-L1+) and had an overall transfection efficiency of approximately 30% (GFP+). While sorting based on light scatter properties was useful for enrichment of transfected spermatogonia, the percentage of cells recovered from the initial input cells after nucleofection and sorting was lower than after sorting of non-transfected cells (16.31 ± 7.37; n = 8 vs. 21.68 ± 8.17 %; n = 18).
FIGURE 2.
Evaluation of porcine testicular cells transfected with GFP and TALENs by nucleofection. (a) Representative scatter plot of GFP transfected cells. P1 indicates germ cell gate, P3 the non-germ cell gate. (b) Immunofluorescence analysis of the germ cell population after FACS-sorting of GFP transfected porcine testicular cells. The merged image illustrates >80% germ cell transfection efficiency. (c) Representative scatter plots of porcine testicular cells after sham nucleofection (top panel) and after nucleofection with TALENs (lower panel). Note lower recovery of cells in the germ cell gate after nucleofection with TALENs. (d) Immunofluorescence analysis of the germ cell population after TALENs + GFP nucleofection and FACS-sorting. (e) Evaluation of NHEJ by Surveyor assay in TALENs transfected FACS-sorted germ cells, confirming NHEJ occurred in germ cells
2.3 | Gene targeting in spermatogonia with TALENs
Targeted mutagenesis has not yet been achieved in germ cells of domestic animals. Having confirmed that pig spermatogonia could be effectively transfected by nucleofection and that spermatogonia could be isolated based on light scatter properties, we aimed to conduct gene targeting in pig spermatogonia. We used a pair of TALENs (DMD7.1) that specifically target the porcine Duchenne's muscular dystrophy (DMD) locus (Carlson et al., 2012). Previous targeting of the DMD gene in porcine fibroblast cells which resulted in 16–38% NHEJ in transfected cells at day 3 post-transfection (Carlson et al., 2012).
We performed a series of transfection experiments with TALENs to explore the effect of temperature and the amount of DNA on targeting efficiency (the percentage of NHEJ) in transfected cells. Cells used for transfection were enriched by the modified differential plating protocol to contain 40–60% spermatogonia. We used 5, 10, or 25 µg of DMD TALENs followed by incubation of cells at either 30 °C or 37 °C for 3–5 days before analysis. For each TALENs transfection, we also co-transfected 2 µg GFP plasmid to monitor transfection efficiency.
Although a higher amount of TALENs DNA (such as 25 µg) resulted in a higher percentage of NHEJ, this reduced cell viability compared to lower amounts of DNA (5 or 10 µg). A relatively high percentage of NHEJ (17.65 ± 0.92%) could be achieved with 25 µg TALENs but at the expense of low viability with only 17.6 ± 1.27% of nucleofected cells recovered following incubation at 30°C and a lower percentage of GFP+ spermatogonia (Table 2). In contrast, 34.7–50% of cells could be recovered from transfection with 5 or 10 µg TALENs. For later experiments, we chose 20 µg DNA to achieve a balance between cell recovery and NHEJ efficiency. Similar to what was reported previously for fibroblasts, TALENs activity was higher at a lower temperature of 30 °C, resulting in a higher percentage of NHEJ compared to 37 °C in each dose of TALENs tested (Table 2). Although the survival of spermatogonia was slightly lower at 30 °C, we used 30 °C for future experiments due to higher NHEJ efficiency.
TABLE 2.
Transfection of porcine spermatogonia with TALENs
| TALENs (µg) |
Incubation temperature (°C) |
% of cell recovery |
% of GFP + cells |
% of UCH- L1 + cells |
% of GFP+ UCH- L1 + cells |
% yield of GFP+UCH- L1 + cellsa |
% of NHEJ | % yield of cells with NHEJb |
|---|---|---|---|---|---|---|---|---|
| 5 | 37 | 49.9 ± 7.731 | 45.4 ± 3.211 | 62.7 ± 2.121 | 31.3 ± 4.251 | 16.1 ± 3.811 | 3.9 ± 1.101 | 2.0 ± 0.631 |
| 10 | 37 | 38.4 ± 0.711,2 | 36.5 ± 8.361,2 | 57.5 ± 1.352 | 27.7 ± 8.201,2 | 10.6 ± 2.961,2 | 11.5 ± 3.892,3 | 4.4 ± 1.412,3 |
| 25 | 37 | 17.6 ± 3.964 | 30.9 ± 9.102 | 53.7 ± 1.462 | 17.2 ± 2.062 | 3.0 ± 0.333 | 16.9 ± 0.493 | 3.0 ± 0.581,2 |
| 5 | 30 | 36.8 ± 4.642,3 | 50.1 ± 1.363 | 58.3 ± 0.492 | 30.9 ± 0.621 | 11.4 ± 1.341 | 8.1 ± 0.85 2 | 3.0 ± 0.461,2 |
| 10 | 30 | 34.7 ± 0.713 | 37.9 ± 2.642 | 54.7 ± 0.232 | 25.9 ± 0.081 | 9.0 ± 0.162 | 14.9 ± 1.703 | 5.2 ± 0.483 |
| 25 | 30 | 17.6 ± 1.274 | 41.2 ± 1.961,2 | 43.2 ± 0.113 | 23.3 ± 7.711,2 | 4.1 ± 1.073 | 17.7 ± 0.923 | 3.1 ± 0.062 |
| After sortingc: | In germ cell gate | % NEHJ in non-germ cell gate | ||||||
| 20 | 30 | 5.08 ± 2.51 | 55.94 ± 9.72 | 77.60 ± 11.99 | 42.19 ± 9.29 | 2.14 ± 1.06 | 5.76 ± 2.59 | 0.55 ± 1.10 |
Within each column, different numbers indicate statistically significant difference (p < 0.05), n = 4.
% of GFP+ UCH-L1+ cells from input cells subjected to nucleofection.
% of modified cells (NHEJ) from input cells subjected to nucleofection.
Separate experiment.
To evaluate if NHEJ occurred in spermatogonia, we sorted transfected cells on day 4 post-transfection using light scatter properties. Transfection with TALENs changed the distribution of germ cells on the dot plot. We no longer observed a very distinctive germ cell population. Although we defined a germ cell gate with cells treated with nucleofection only (without TALENs), we had to widen the gate to sort enough cells representing 5.08 ± 2.51 % (n = 4) of the input cells for subsequent analysis (Figure 2c). We were able to obtain a germ cell enriched fraction containing 66–89% spermatogonia (Figure 2d) with the percentage of NHEJ ranging from 2.80% to 9% (Figure 2e) with NHEJ 0–2.2% in the non-germ cell fraction (Table 2).
Analysis of a subset of samples after nucleofection by sequencing confirmed the percentage of targeted modification detected by Surveyor assay (Table 3).
TABLE 3.
Targeted modifications in porcine germ cells detected by surveyor assay and MiSeq targeted sequencing
| Samplesa | % NHEJ by surveyor assay | MiSeq total count | MiSeq indel count | % Indels by MiSeq |
|---|---|---|---|---|
| 1 | 6 | 186,635 | 11,780 | 6.31 |
| 2 | 1.7 | 194,437 | 508 | 0.26 |
| 3 | 9.2 | 93,557 | 7,470 | 7.98 |
| 4 | 0 | 337,179 | 296 | 0.08 |
Samples 1–3 were transfected with 20 µg TALEN constructs, sample 4 is sham transfected control.
3 | DISCUSSION
3.1 | Enrichment of spermatogonia based on light scatter properties
Our study represents the first report on using light scatter properties to isolate spermatogonia from the prepubertal pig testis. This flow cytometry based approach is different from other FACS-based live germ cell sorting in that it does not require the knowledge of surface proteins present on spermatogonia or the availability of a spermatogonia-specific transgenic reporter.
A highly pure population of spermatogonia will be useful for transcriptome and proteome profiling, for identifying unique surface markers for fractionating and isolating subsets of spermatogonia, and for establishing a long term culture system for porcine SSCs. In 10 week old pig testis, all the germ cells in the seminiferous tubules are spermatogonia. Some of these are residing at the basement membrane and others are still in the process of migrating toward the basement membrane. Spermatogonia at this developmental stage are relatively uniform in their size and granularity. We found they showed distinctive forward and side light scatter properties when subjected to flow cytometry. Taking advantage of these unique characteristics, we established a simple approach to isolate spermatogonia using a flow cytometer. Germ cells manifested as a distinct population on the light scatter plot, and through gating we were able to conveniently and consistently obtain a highly enriched spermatogonia faction (up to 98% purity). Similar results could be obtained with cells obtained from pigs from 1 to 14 weeks of age, making the approach applicable to a broader pre-pubertal age range. FACS sorting protocols for germ cells that rely on surface marker-mediated sorting require highly selective and robust antibodies and the presence of antigens on the cell surface that might be affected by protease digestion. Although germline specific transgenic lines (such as gc-Oct4-GFP, Vasa-Venus, Vasa-GFP, Id4-GFP) that can be used for sorting germ cells are available in species such as mice and fish (Chan et al., 2014; Hubner et al., 2003; Shiura et al., 2013; Sun, Xu, Zhao, & Chen, 2015; Yoshizaki, Takeuchi, Sakatani, & Takeuchi, 2000), they are not available for domestic animals. Sorting by light scatter properties does not require staining as described previously in studies where various dyes such as Hoechst 33,342 nucleic acid dye, rhodamine 123 mitochondrial dye and CDy1 stem cell dye were used to enrich spermatogonia and spermatogonial stem cells from fish or mouse testis (Falciatori et al., 2004; Hayashi et al., 2014; Kanatsu-Shinohara et al., 2016; Lo, Burg, Parker, & Lamb, 2005).
Isolating spermatogonia by flow cytometry using light scatter properties has been reported previously in teleost fish where a cell population highly enriched with undifferentiated type A spermatogonia (~93% VASA positive) could be obtained from immature rainbow trout testis (Kise et al., 2012). In the fish study, researchers used a transgenic rainbow trout line that carries a Vasa-GFP reporter to set a germ cell gate on the light scatter plot and sorted type A spermatogonia from immature non-transgenic males based on light scatter properties. They also showed that this gate can be readily used to enrich spermatogonia from various salmonid species with reduced purity (~75–80% spermatogonia). In our study, we did not have a germ cell-specific transgenic pig model for isolating or gating spermatogonia. Instead, we sorted spermatogonia from the prepubertal pig testis based on forward and side light scatter using a non-referenced gate. This gate was drawn around the dense population of cell events manifested on the light scatter dot plot. In the rainbow trout report, no enrichment of germ cells was required prior to sorting as the initial testicular suspension already contained ~36% type A spermatogonia. In our case, the initial testicular suspension only contained ~4–7% spermatogonia. We observed that a certain degree of enrichment of germ cells before sorting is preferred for spermatogonia to manifest as a distinctive cell population on the light scatter plot and for the sorting process to be time-efficient. Higher spermatogonia content in the input cells tended to result in higher purity in sorted cells and significantly reduced the sorting time required. To specifically demonstrate functionality of the sorted germ cells would require homologous transplantation which given the numbers of sorted cells needed was beyond the scope of the current study. However, we showed previously that germ cells enriched by differential plating, as performed here prior to sorting, retain their ability to colonize the pig testis and support spermatogenesis (Dores & Dobrinski, 2014), and as flow sorted mouse and fish SSCs retain their function after transplantation (Chan et al., 2014; Hayashi et al., 2014; Kise et al., 2012; Shinohara, Orwig, Avarbock & Brinster, 2000) we expect flow sorted porcine germ cells to remain functional. Use of flow sorted pig germ cells for homologous transplantation will become more feasible with improvements of in vitro expansion of cell numbers.
The enrichment approach reported here is largely independent of the transcriptomic and proteomic profile of spermatogonia and it was also applicable to enrichment of spermatogonia from pigs at the age of 1-, 4-, and 14-week old. Cells sorted based on physical properties are likely more heterogeneous in molecular signature compared to cells obtained by isolation methods relying on molecular or functional characteristics of spermatogonia and SSCs. Before the identification of authentic SSC marker(s), this heterogeneity may present an advantage in investigating in vitro culture conditions as it has been shown that murine SSCs form clusters in vitro within which cells are molecularly and functionally heterogeneous (Yeh, Zang, & Nagano, 2007, 2012). Sorting based on physical properties is likely also applicable to isolating spermatogonia from neonatal and prepubertal males from other non-rodent species.
3.2 | Targeted mutagenesis in spermatogonia
We have shown previously, that transgenes delivered to goat or pig SSCs by either viral vectors or by nucleofection can stably integrate into the genome and be detected in recipient sperm (Honaramooz et al., 2008; Zeng et al., 2012, 2013). Those studies established that SSCs can be a useful genetic carrier for transgene transmission. In the current study, we went one step further to demonstrate and confirm that gene targeting using engineered nucleases is feasible in pig SSCs.
The lack of a robust in vitro culture system to sustain, expand and screen pig SSCs made conventional gene targeting in those cells nearly impossible to achieve. In preliminary experiments, we tested both TALENs and CRISPR/Cas9 and editing was as good or better with the TALENs under the conditions tested. We therefore chose to use sequence-specific TALE nucleases for gene targeting in pig SSCs due to their demonstrated high targeting efficiency in a wide range of cell lines, model organisms as well as in porcine fibroblast cells and embryos (Carlson et al., 2012; Joung & Sander, 2013; Lillico et al., 2013; Yao et al., 2014). The targeting efficiency in primary germ cells achieved in the current study was similar to that reported for rodent SSCs (Chapman et al., 2015; Fanslow et al., 2014).
Precise genetic engineering via homologous recombination (gene targeting) in mouse ES cells and subsequent germline transmission of the mutations revolutionized the field of functional genomics and became the gold standard for creating mouse models for biomedical and pharmaceutical studies. Although SSC-mediated mutagenesis bypasses the steps of chimeric embryo formation and germline transmission of mutations carried in ES cells, the lengthy time required for isolating, and expanding targeted SSC clones precludes SSC-based gene targeting from being a standard tool for functional genomics in mice. However, genetic engineering through SSCs has tremendous potential in non-rodent species such as goats, pigs, and cattle where ES cell based transgenic technology is not yet available and currently practiced methods for producing transgenic animals are generally inefficient and costly (Gonzalez & Dobrinski, 2015).
Several developments in the field have paved the way for SSC-mediated genetic engineering in domestic animals. We and others showed that homologous germ cell transplantation (into unrelated same-species animals) is feasible in pigs, goats, sheep, and cattle without any apparent immune reaction (Honaramooz, Behboodi, Blash, Megee, & Dobrinski, 2003; Honaramooz, Megee, & Dobrinski, 2002; Izadyar et al., 2003; Rodriguez-Sosa, Silvertown, Foster, Medin, & Hahnel, 2009). Also, while depletion of endogenous SSCs is not absolutely required for germ cell transplantation into immature testis of large animals such as goats, pigs, sheep, and cattle (Herrid, Vignarajan, Davey, Dobrinski, & Hill, 2006; Honaramooz, Behboodi, & Megee, 2003; Rodriguez-Sosa et al., 2009; Zeng et al., 2013), genetically modified pigs that lack endogenous germ cells have recently been reported (Park et al., 2017; Tan et al., 2013). As in vitro expansion of genetically modified porcine germ cells is currently not possible, these animals are expected to serve as efficient recipient models for transplantation of primary, gene-edited donor cells.
Genetic mutations created by NHEJ are unpredictable due to the nature of the repair. This means that genetic changes (insertions or deletions) occurred in each targeted cell could be slightly different at the targeted locus, creating mutations that potentially result in a null allele or a hypomorphic allele, or silent mutations. As a culture system to isolate, expand, and screen each individual targeted clone is currently not available for porcine SSCs, transplantation of all the targeted cells into pig recipients would use the testis as an in vivo clonal expansion system. In a preliminary transplantation experiment using intact, prepubertal recipients, we were not able to conclusively detect the colonization, and spermatogenic potential of DMD-edited cells potentially due to dilution of sperm resulting from targeted spermatogonia by wild-type sperm from endogenous SSCs and non-targeted transplanted cells that made the detection of genetically modified sperm above background levels extremely challenging. Therefore, future work will make use of infertile recipients devoid of endogenous germ cells for transplantation of gene targeted SSCs.
In conclusion, we report here for the first time an approach to isolate highly enriched populations of undifferentiated porcine spermatogonia and demonstrate TALEN-mediated gene targeting after nucleofection. With the recent development of genetically modified sterile pigs as potentially superior recipients for transplantation of modified SSCs, and ongoing improvements to in vitro systems that will allow clonal expansion of gene targeted cells prior to transplantation, the approaches reported here will serve as a resource towards achieving germ line mediated targeted mutagenesis in a large animal model.
4 | MATERIALS AND METHODS
4.1 | Testis digestion
Single-cell suspensions were prepared from testes from 1-, 4-, 10-, and 14-week-old pigs by a sequential enzymatic digestion protocol (Honaramooz et al., 2002). Briefly, the tunica albuginea and visible connective tissue were dissected and removed. The exposed seminiferous tubules were dissociated with Type IV collagenase (2 mg/ml; Sigma, Millipore-Sigma, Oakville, ON, Canada) in Dulbecco modified Eagle medium (DMEM, Sigma) at 37 °C for 20–40 min with occasional agitation, followed by incubation at 37 °C for 30 min in DMEM with Type IV collagenase (2 mg/ml; Sigma) and hyaluronidase (1 mg/ml; Sigma). The digested tubules were rinsed three times in Dulbecco phosphate-buffered saline (DPBS, Ca2+and Mg2+ free) and further digested with 0.125% (w/v) trypsin and 0.5 mM ethyl-enediaminetetra-acetic acid (EDTA) at 37°C for 15–20 min. DNase I (7 mg/ml in DMEM; Sigma) was added during the digestion process as needed. After trypsin digestion, the cell suspension was filtered through 70 µm and 40 µm cell strainers sequentially (BD Biosciences, Oakville, ON, Canada). The single cells were then collected by centrifugation at 500g for 5 min at room temperature (RT) and the cell pellet was resuspended in DMEM/F-12 (Sigma) with 5% fetal bovine serum (FBS) for differential plating.
4.2 | Differential plating
Differential plating of pig testicular cells was performed as previously described (Luo et al., 2006) with the following modifications. Immediately after tissue digestion, 2.5 × 107 cells in 8 ml DMEM/F-12 with 5% FBS were plated onto 100 mm tissue culture plates and incubated at 37 °C in 5% CO2. Three sequential rounds of differential plating were performed with the first round for 1.5 hr, the second round for 1 hr and the third round overnight. At the 2nd and 3rd round of plating, cell suspensions from two plates of the previous round were combined and plated onto a new 100 mm culture plate. Attached cells were discarded.
After overnight incubation, some germ cells remained suspended in the culture medium, and some adhered loosely to the somatic cell monolayer at the bottom. To collect germ cells in suspension, supernatant from all the plates were pooled. To collect loosely adhered germ cells, 2–3 ml of diluted Trypsin/EDTA (1:5 or 1:20 dilution with PBS) was added to each plate. Plates were incubated at 37 °C for 2 min and then at RT for 3 min with constant agitation to release attached germ cells without disturbing somatic cells. The reaction was stopped by adding an equal volume of DMEM/F12 with 10% FBS. Cell suspensions were pooled from all plates, combined with cells collected from the supernatants, pelleted by centrifugation at 500g for 5 min and washed twice with PBS. After washing, cells were plated again onto 100 mm plates in DMEM/F12 with 5% FBS for 8 min at RT and cell suspensions were gently and slowly collected from the top. This extra plating step helped to further remove cell debris, red blood cells, and other small somatic cells.
4.3 | FACS sorting
Enriched cell fractions collected after differential plating were resuspended in PBS with 1% BSA (Sigma) and subjected to sorting on a FACSAria III (Becton Dickinson). A gate was drawn around the distinctive germ cell population on the forward and side light scatter dot plot and cells within this gate were sorted. An arbitrary gate was drawn away from the germ cell gate (the non-germ cell gate) and cells within this gate were sorted as the negative fraction. Sorted cells were pelleted and washed once with PBS. The viability of sorted cells was assessed by Trypan Blue staining. Cells were then fixed with paraformaldehyde (PFA) and assessed by immunocytochemistry with antibodies against UCH-L1 and Vimentin. The numbers of UCH-L1+ and Vimentin+ cells were scored and the percentage of UCH-L1+ and Vimentin+ cells within a particular fraction were calculated. UCH-L1 is a spermatogonia-specific marker that was used to assess the enrichment efficiency and to determine the percentage of germ cells present in a given cell population (Luo et al., 2006). Vimentin was used to label somatic cells. For each sorting experiment, 1,000–2,000 cells were evaluated for UCH-L1 and vimentin in each fraction.
4.4 | Immunocytochemistry
Cells from various preparations and fractions (unenriched, enriched, sorted, and transfected) were fixed in 2% PFA for 30 min at RT and washed twice with PBS. Cells were then transferred onto slides for immunostaining by cytospin centrifugation (800g for 5 min at RT) (Shandon Inc, ThermoFisher Scientific, Whitby, ON, Canada), permeabilized in PBS with 0.1% Triton-X and washed three times in PBS prior to 1 hr blocking with 3% BSA. Cells were incubated with the following primary antibodies overnight at 4 °C: rabbit-anti-human UCH-L1 (AbD Serotec) at 1:1000, mouse-anti-pig vimentin-Cy3 at 1:400 (Sigma Aldrich). Three washes were performed after overnight primary antibody incubation and secondary antibodies donkey-anti-rabbit IgG Alexa Fluor 594 (1:1000) were added onto samples. After 1 hr RT incubation, cells were washed three times, and mounted in VECTASHIELD Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA) for imaging.
4.5 | Quantitative RT-PCR
Total RNA was isolated from 0.5–2 × 106 cells using RNeasy Mini Kit (Qiagen, Toronto, ON, Canada). For reverse transcription 2 µg of total RNA was used in a final volume of 25 µl reaction containing 0.5 µg of Oligo d(T) 12–18, RT buffer (1×), 10 mM dithiothreitol, 0.5 mM of dNTP, 5 U of RNase-inhibitor, and 10 U of SuperScript II Reverse transcriptase (Invitrogen). Reverse transcription was carried out at 42 °C for 1 hr. Quantitative RT-PCR amplification was performed using SsoFast Eva Green SYBR Green Master (Bio-Rad) in 7,500 Fast Real Time PCR System (Applied Biosystems, ThermoFisher Scientific, Burlington, ON, Canada). The HPRT1 and RPL4 genes were amplified as internal controls for each Real-Time PCR. Primer sequences, length of amplified products, and annealing temperatures were as outlined in Supplementary Table S1. All amplified products were verified by High Resolution Melting Curve analysis, and relative levels of gene expression were analyzed by delta-dCt method using 7,500 Software (Applied Biosystems).
4.6 | Germ cell nucleofection
Nucleofection was performed with the Nucleofector II electoporator system according to the manufacturer's protocol (Lonza Inc.). For nucleofection optimization, 106 enriched cells were transfected with 2 µg of pmaxGFP™ (Lonza) and cells were harvested at 48 hr for viability assessment and fixation. The Amaxa® nucleofection solution kits V, L, and B from Lonza were used. The programs used were X–005, X–001, and U-030 for V-, L-, and B-solution, respectively. After nucleofection, cells were transferred onto six-well plates in a StemPro-based culture medium supplemented with 1% FBS, 0.1% BSA, 1× nonessential amino acids, 1 mM Sodium Pyruvate, 15 mM HEPES, 2 mM L-glutamine, 10 µM beta-mercaptoethanol, 100 U/ml Penicillin-100 µg/ml Streptomycin, and 10 ng/ml glial cell line-derived neurotrophic factor (GDNF) for cell recovery and short-term cell culture.
For nucleofection with TALENs, enriched cells were resuspended in solution V, and transfected with the program X–005. One million cells were used for each reaction with 20µg of DMD 7.1 TALENs DNA and 2µg of pmaxGFP™. After nucleofection, cells were transferred onto pre-warmed six-well plates in supplemented StemPro-based culture medium and allowed to recover overnight at 37 °C in 5% CO2. After overnight recovery, the medium was replaced with fresh Stem-Pro-based culture medium, and cells were incubated at 30 °C or 37 °C for 3–5 days depending on the experimental design. At the end of incubation, cells were harvested by gentle trypsinization (1:5 dilution of 0.25% Trypsin/EDTA). The number of cells collected was counted by hemocytometer and the viability was assessed by Trypan Blue staining. Collected cells were either used for FACS sorting or split into fractions to be used for immunocytochemistry and the surveyor assay. The percentage of UCH-L1 positive cells and NHEJ in cells collected after TALENs transfection and 3–5 days of cell incubation were scored.
4.7 | TALENs and NHEJ analysis (surveyor assay)
We used a pair of DMD TALENs (DMD7.1) which have previously been tested in porcine fibroblast cells (Carlson et al., 2012). PCR flanking the targeted sites was conducted using AccuStart™ Taq DNA Polymerase HiFi (Quanta Biosciences, QuantaBio, Beverly, MA) with 100 ng of template DNA according to the manufacturer's recommendations. The frequency of mutation in a population was analyzed with the Surveyor mutation detection kit (Transgenomic, Transgenomic, Inc., Omaha, NE) according to the manufacturer's recommendations using 10 ul of the PCR product as described above. Surveyor reactions were resolved on a 10% TBE polyacrylamide gels and visualized by ethidium bromide staining. Densitometry measurements of the bands were performed using ImageJ; and mutation rate of Surveyor reactions was calculated as described previously (Guschin et al., 2010). The primer sequence information used for detecting NHEJ was reported in Carlson et al. (2012).
4.8 | MiSeq targeted sequencing
Genomic DNA was extracted using the Gentra Puregene Cell Kit (QIAGEN) per the manufacturer's instructions. Primers were designed (ssDMD Sense: 5′-TGGGCATGTGTTGTCAGTCA-3′, ssDMD Anti-sense: 5′-TGGTAGTCCCAAAATGCACT-3′) to amplify the region of interest and a 6-nucleotide unique barcode was added to the 5′ end of the antisense primer. Polymerase chain reaction was performed using Accustart Taq DNA Polymerase HiFi (Quanta Biosciences) following the manufacturer's instructions. PCR products were purified using the QIAquick 96 PCR Purification Kit (QIAGEN), and then quantified. The products were then pooled at equimolar concentrations prior to Illumina MiSeq next generation sequencing. Paired end reads were stitched together to form a long complete read using FastqJoin (Aronesty, 2013) with a default 6 bp minimum overlap and 8% maximum difference. Merged FASTQ files were then trimmed, de-multiplexed, and assigned to each sample with corresponding barcodes. For each of the samples with a distinct barcode, count analysis was performed to calculate the ratio between wild type and indel/SNV mutations using a custom Perl script. All mutant sequences with the same barcode are aligned against the wild type using BLASTN (Camacho et al., 2008).
4.9 | Statistical analysis
Data were analyzed using GraphPad Prizm 4.0 (GraphPad Software Inc., La Jolla, CA). A Student's t-test and ANOVA were performed to compare groups. Data were expressed as means ± S.E.M and p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This study was supported by NIH/ORIP 9 R01 OD016575-12. We thank Dr. Jeff Biernaskie for use of the FACS facilities and Drs. Jonathan Hill and David Davidson for critical reading of the manuscript.
Abbreviations
- CRISPR/Cas9
Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated-9
- DMD
Duchenne Muscular Dystrophy
- DMEM
Dulbecco's Modified Eagle Medium
- DPBS
Dulbecco's PhoSphate Buffered Saline
- EDTA
ethylenediaminetetra acetic acid
- ESC
embryonic stem cell
- FACS
fluorescence activated cell sorting
- GFP
green fluorescent protein
- HR
homologous recombination
- iPSC
induced pluripotent stem cell
- MACS
magnetic activated cell sorting
- NHEJ
non-homologous end joining
- SCNT
somatic cell nuclear transfer
- SSC
spermatogonial stem cell
- TALEN
Transcription Activator-like Effector Nuclease
- UCH-L1
ubiquitin carboxyterminal hydrolase L-1
- ZFN
zinc finger nuclease.
Footnotes
CONFLICTS OF INTEREST
Drs. Carlson, Webster, and Fahrenkrug are employees of Recombinetics, Inc. Dr. Dobrinski is a member of the Scientific Advisory Board of Recombinetics, Inc.
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- Abbasi H, Tahmoorespur M, Hosseini SM, Nasiri Z, Bahadorani M, Hajian M, Nasr-Esfahani MH. THY1 as a reliable marker for enrichment of undifferentiated spermatogonia in the goat. Theriogenology. 2013;80:923–932. doi: 10.1016/j.theriogenology.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Aponte PM, Soda T, Teerds KJ, Mizrak SC, van de Kant HJ, de Rooij DG. Propagation of bovine spermatogonial stem cells in vitro. Reproduction. 2008;136:543–557. doi: 10.1530/REP-07-0419. [DOI] [PubMed] [Google Scholar]
- Aronesty E. Comparison of sequencing utility programs. The Open Bioinformatics Journal. 2013;7:1–8. [Google Scholar]
- Bacci ML. A brief overview of transgenic farm animals. Veterinary Research Communications. 2007;1:9–14. doi: 10.1007/s11259-007-0001-z. [DOI] [PubMed] [Google Scholar]
- Bao L, Chen H, Jong U, Rim C, Li W, Lin X, Huang H. Generation of GGTA1 biallelic knockout pigs via zinc-finger nucleases and somatic cell nuclear transfer. Science China Life Sciences. 2014;57:263–268. doi: 10.1007/s11427-013-4601-2. [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. The Journal of Cell Biology. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2008;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F, Oatley MJ, Kaucher AV, Yang Q-E, Bieberich CJ, Shashikant CS, Oatley JM. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes & Development. 2014;28:1351–1362. doi: 10.1101/gad.240465.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KM, Medrano GA, Jaichander P, Chaudhary J, Waits AE, Nobrega MA, Hamra FK. Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Reports. 2015;10:1828–1835. doi: 10.1016/j.celrep.2015.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores C, Dobrinski I. De novo morphogenesis of testis tissue to investigate the role of VEGF-165 during testis formation. Reproduction. 2014;148:109–117. doi: 10.1530/REP-13-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebata KT, Zhang X, Nagano MC. Expression patterns of cell-surface molecules on male germ line stem cells during postnatal mouse development. Molecular Reproduction and Development. 2005;72:171–181. doi: 10.1002/mrd.20324. [DOI] [PubMed] [Google Scholar]
- Falciatori I, Borsellino G, Haliassos N, Boitani C, Corallini S, Battistini L, Vicini E. Identification and enrichment of spermatogonial stem cells displaying side-population phenotype in immature mouse testis. FASEB Journal. 2004;18:376–378. doi: 10.1096/fj.03-0744fje. [DOI] [PubMed] [Google Scholar]
- Fanslow DA, Wirt SE, Barker JC, Connelly JP, Porteus MH, Dann CT. Genome editing in mouse spermatogonial stem/progenitor cells using engineered nucleases. PLoS ONE. 2014;9:e112652. doi: 10.1371/journal.pone.0112652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara M, Kim SM, Minami N, Yamada M, Imai H. Characterization and in vitro culture of male germ cells from developing bovine testis. Journal of Reproduction and Development. 2011;57:355–364. doi: 10.1262/jrd.10-185m. [DOI] [PubMed] [Google Scholar]
- Goel S, Sugimoto M, Minami N, Yamada M, Kume S, Imai H. Identification, isolation, and in vitro culture of porcine gonocytes. Biology of Reproduction. 2007;77:127–137. doi: 10.1095/biolreprod.106.056879. [DOI] [PubMed] [Google Scholar]
- Gonzalez R, Dobrinski I. Beyond the mouse monopoly: Studying the male germ line in domestic animal models. ILAR Journal. 2015;56:83–98. doi: 10.1093/ilar/ilv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods in Molecular Biology. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Research. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamra FK, Gatlin J, Chapman KM, Grellhesl DM, Garcia JV, Hammer RE, Garbers DL. Production of transgenic rats by lentiviral transduction of male germ-line stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14931–14936. doi: 10.1073/pnas.222561399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild-Quintern J, Petersen B, Cost GJ, Niemann H. Gene knockout and knockin by zinc-finger nucleases: Current status and perspectives. Cellular and Molecular Life Sciences. 2013;70:2969–2983. doi: 10.1007/s00018-012-1204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Schwinzer R. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Sato M, Nagasaka Y, Sadaie S, Kobayashi S, Yoshizaki G. Enrichment of spermatogonial stem cells using side population in teleost. Biology of Reproduction. 2014;91(1):23, 1–8. doi: 10.1095/biolreprod.113.114140. [DOI] [PubMed] [Google Scholar]
- Herrid M, Vignarajan S, Davey R, Dobrinski I, Hill JR. Successful transplantation of bovine testicular cells to heterologous recipients. Reproduction. 2006;132:617–624. doi: 10.1530/rep.1.01125. [DOI] [PubMed] [Google Scholar]
- Honaramooz A, Behboodi E, Blash S, Megee SO, Dobrinski I. Germ cell transplantation in goats. Molecular Reproduction and Development. 2003;64:422–428. doi: 10.1002/mrd.10205. [DOI] [PubMed] [Google Scholar]
- Honaramooz A, Behboodi E, Megee SO, Overton SA, Galantino-Homer H, Echelard Y, Dobrinski I. Fertility and germline transmission of donor haplotype following germ cell transplantation in immunocompetent goats. Biology of Reproduction. 2003;69:1260–1264. doi: 10.1095/biolreprod.103.018788. [DOI] [PubMed] [Google Scholar]
- Honaramooz A, Megee S, Zeng W, Destrempes MM, Overton SA, Luo J, Blash S. Adeno-associated virus (AAV)-mediated transduction of male germ line stem cells results in transgene transmission after germ cell transplantation. FASEB Journal. 2008;22:374–382. doi: 10.1096/fj.07-8935com. [DOI] [PubMed] [Google Scholar]
- Honaramooz A, Megee SO, Dobrinski I. Germ cell transplantation in pigs. Biology of Reproduction. 2002;66:21–28. doi: 10.1095/biolreprod66.1.21. [DOI] [PubMed] [Google Scholar]
- Hubner K, Fuhrmann G, Christenson LK, Kehler J, Reinbold R, De La Fuente R, Scholer HR. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- Iwamori N, Iwamori T, Matzuk MM. Characterization of spermatogonial stem cells lacking intercellular bridges and genetic replacement of a mutation in spermatogonial stem cells. PLoS ONE. 2012;7:e38914. doi: 10.1371/journal.pone.0038914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izadyar F, Den Ouden K, Stout TA, Stout J, Coret J, Lankveld DP, Van der Ploeg KD. Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction. 2003;126:765–774. [PubMed] [Google Scholar]
- Izsvak Z, Frohlich J, Grabundzija I, Shirley JR, Powell HM, Chapman KM, Hamra FK. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nature Methods. 2010;7:443–445. doi: 10.1038/nmeth.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: A widely applicable technology for targeted genome editing. Nature Rev. Mol. Cell Biology. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ikawa M, Takehashi M, Ogonuki N, Miki H, Inoue K, Oshimura M. Production of knockout mice by random or targeted mutagenesis in spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8018–8023. doi: 10.1073/pnas.0601139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Morimoto H, Shinohara T. Enrichment of mouse spermatogonial stem cells by a stem cell dye CDy1. Biology of Reproduction. 2016;94(1):13, 1–10. doi: 10.1095/biolreprod.115.135707. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Toyokuni S, Shinohara T. CD9 is a surface marker on mouse and rat male germline stem cells. Biology of Reproduction. 2004;70:70–75. doi: 10.1095/biolreprod.103.020867. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Toyokuni S, Shinohara T. Genetic selection of mouse male germline stem cells in vitro: Offspring from single stem cells. Biology of Reproduction. 2005;72:236–240. doi: 10.1095/biolreprod.104.035659. [DOI] [PubMed] [Google Scholar]
- Kise K, Yoshikawa H, Sato M, Tashiro M, Yazawa R, Nagasaka Y, Yoshizaki G. Flow-cytometric isolation and enrichment of teleost type A spermatogonia based on light-scattering properties. Biology of Reproduction. 2012;86:107, 1–12. doi: 10.1095/biolreprod.111.093161. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biology of Reproduction. 2004;71:722–731. doi: 10.1095/biolreprod.104.029207. [DOI] [PubMed] [Google Scholar]
- Laible G, Alonso-Gonzalez L. Gene targeting from laboratory to livestock: Current status and emerging concepts. Biotechnology Journal. 2009;4:1278–1292. doi: 10.1002/biot.200900006. [DOI] [PubMed] [Google Scholar]
- Lillico SG, Proudfoot C, Carlson DF, Stverakova D, Neil C, Blain C, Mileham AJ. Live pigs produced from genome edited zygotes. Scientific Reports. 2013;3:2847. doi: 10.1038/srep02847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KC, Brugh VM, Parker M, 3rd, Lamb DJ. Isolation and enrichment of murine spermatogonial stem cells using rhodamine 123 mitochondrial dye. Biology of Reproduction. 2005;72:767–771. doi: 10.1095/biolreprod.104.033464. [DOI] [PubMed] [Google Scholar]
- Luo J, Megee S, Rathi R, Dobrinski I. Protein gene product 9.5 is a spermatogonia-specific marker in the pig testis: Application to enrichment and culture of porcine spermatogonia. Molecular Reproduction and Development. 2006;73:1531–1540. doi: 10.1002/mrd.20529. [DOI] [PubMed] [Google Scholar]
- Luo J, Song Z, Yu S, Cui D, Wang B, Ding F, Li N. Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PLoS ONE. 2014;9:e95225. doi: 10.1371/journal.pone.0095225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DJ, Russell LD, Griswold MD. Biological activity and enrichment of spermatogonial stem cells in vitamin A-deficient and hyperthermia-exposed testes from mice based on colonization following germ cell transplantation. Biology of Reproduction. 2002;66:1374–1379. doi: 10.1095/biolreprod66.5.1374. [DOI] [PubMed] [Google Scholar]
- Morena AR, Boitani C, Pesce M, De Felici M, Stefanini M. Isolation of highly purified type A spermatogonia from prepubertal rat testis. Journal of Andrology. 1996;17:708–717. [PubMed] [Google Scholar]
- Nagano M, Brinster CJ, Orwig KE, Ryu BY, Avarbock MR, Brinster RL. Transgenic mice produced by retroviral transduction of male germ-line stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Shinohara T, Avarbock MR, Brinster RL. Retrovirus-mediated gene delivery into male germ line stem cells. FEBS Letters. 2000;475:7–10. doi: 10.1016/s0014-5793(00)01606-9. [DOI] [PubMed] [Google Scholar]
- Ni W, Qiao J, Hu S, Zhao X, Regouski M, Yang M, Chen C. Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS ONE. 2014;9:e106718. doi: 10.1371/journal.pone.0106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Li W. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Niemann H, Kues W, Carnwath JW. Transgenic farm animals: Present and future. Revue Scientifique Et Technique. 2005;24:285–298. [PubMed] [Google Scholar]
- Park K-E, Kaucher AV, Powell A, Waqas MS, Sandmaier SES, Oatley MJ, Blomberg LA. Generation of germline ablated male pigs by CRISPR/Cas9 editing of the NANOS2 gene. Scientific Reports. 2017;7:1–9. doi: 10.1038/srep40176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nature Biotechnology. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Reding SC, Stepnoski AL, Cloninger EW, Oatley JM. THY1 is a conserved marker of undifferentiated spermatogonia in the pre-pubertal bull testis. Reproduction. 2010;139:893–903. doi: 10.1530/REP-09-0513. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Sosa JR, Silvertown JD, Foster RA, Medin JA, Hahnel A. Transduction and transplantation of spermatogonia into the testis of ram lambs through the extra-testicular rete. Reproduction in Domestic Animals. 2009;44:612–620. doi: 10.1111/j.1439-0531.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H, Xu K, Wu T, Wei J, Zhou R, Ouyang H. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Scientific Reports. 2015;5:142–153. doi: 10.1038/srep14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Orwig KE, Oatley JM, Lin CC, Chang LJ, Avarbock MR, Brinster RL. Efficient generation of transgenic rats through the male germline using lentiviral transduction and transplantation of spermatogonial stem cells. Journal of Andrology. 2007;28:353–360. doi: 10.2164/jandrol.106.001511. [DOI] [PubMed] [Google Scholar]
- Sato T, Sakuma T, Yokonishi T, Katagiri K, Kamimura S, Ogonuki N, Ogawa T. Genome editing in mouse spermatogonial stem cell lines using TALEN and double-Nicking CRISPR/Cas9. Stem Cell Reports. 2015;5:75–82. doi: 10.1016/j.stemcr.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T, Avarbock MR, Brinster RL. Functional analysis of spermatogonial stem cells in Steel and cryptorchid infertile mouse models. Developments in Biologicals. 2000;220:401–411. doi: 10.1006/dbio.2000.9655. [DOI] [PubMed] [Google Scholar]
- Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8346–8351. doi: 10.1073/pnas.97.15.8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiura H, Ikeda R, Lee J, Sato T, Ogonuki N, Hirose M, Abe K. Generation of a novel germline stem cell line expressing a germline-specific reporter in the mouse. Genesis. 2013;51:498–505. doi: 10.1002/dvg.22391. [DOI] [PubMed] [Google Scholar]
- Sun F, Xu Q, Zhao D, Chen CD. Id4 marks spermatogonial stem cells in the mouse testis. Scientific Reports. 2015;5:17594. doi: 10.1038/srep17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehashi M, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Shinohara T. Adenovirus-mediated gene delivery into mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2596–2601. doi: 10.1073/pnas.0609282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Carlson DF, Lancto CA, Garbe CA, Webster DA, Hackett PB, Fahrenkrug SC. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutation Research. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- van Dissel-Emiliani FM, de Rooij DG, Meistrich ML. Isolation of rat gonocytes by velocity sedimentation at unit gravity. Journal of Reproduction and Fertility. 1989;86:759–766. doi: 10.1530/jrf.0.0860759. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhou J, Cao C, Huang J, Hai T, Wang Y, Miao X. Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Scientific Reports. 2015;5:13348. doi: 10.1038/srep13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou H, Fan X, Zhang Y, Zhang M, Wang Y, Liang D. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Research. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Yang H, Li W, Zhao B, Ouyang Z, Liu Z, Tian J. Generation of PPARgamma mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Research. 2011;21:979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Huang J, Hai T, Wang X, Qin G, Zhang H, Yuan Z. Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs. Scientific Reports. 2014;4:6926. doi: 10.1038/srep06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JR, Zhang X, Nagano MC. Establishment of a short-term in vitro assay for mouse spermatogonial stem cells. Biology of Reproduction. 2007;77:897–904. doi: 10.1095/biolreprod.107.063057. [DOI] [PubMed] [Google Scholar]
- Yeh JR, Zhang X, Nagano MC. Indirect effects of Wnt3a/beta-catenin signalling support mouse spermatogonial stem cells in vitro. PLoS ONE. 2012;7:e40002. doi: 10.1371/journal.pone.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen S-T, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, Behringer RR. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Developments in Biologicals. 2014;393:3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki G, Takeuchi Y, Sakatani S, Takeuchi T. Germ cell-specific expression of green fluorescent protein in transgenic rainbow trout under control of the rainbow trout vasa-like gene promoter. International Journal of Developmental Biology. 2000;44:323–326. [PubMed] [Google Scholar]
- Yu S, Luo J, Song Z, Ding F, Dai Y, Li N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Research. 2011;21:1638–1640. doi: 10.1038/cr.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Tang L, Bondareva A, Honaramooz A, Tanco V, Dores C, Paczkowski M. Viral transduction of male germline stem cells results in transgene transmission after germ cell transplantation in pigs. Biology of Reproduction. 2013;88(1):27, 1–9. doi: 10.1095/biolreprod.112.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Tang L, Bondareva A, Luo J, Megee SO, Modelski M, Overton SA. Non-viral transfection of goat germline stem cells by nucleofection results in production of transgenic spermafter germ cell transplantation. Molecular Reproduction and Development. 2012;79:255–261. doi: 10.1002/mrd.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, He Y, An J, Qin J, Wang Y, Zhang Y, Zeng W. THY1 is a surface marker of porcine gonocytes. Reproduction, Fertility and Development. 2014;26:533–539. doi: 10.1071/RD13075. [DOI] [PubMed] [Google Scholar]
- Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Lai S. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cellular and Molecular Life Sciences. 2015;72:1175–1184. doi: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.