ABSTRACT
The roles of RNA in the DNA damage response are emerging. We highlight findings from our recent study demonstrating the mechanism for transcription-associated homologous recombination repair (TA-HRR) of DNA double-strand breaks and the critical role of R-loops in TA-HRR.
Keywords: R-loop, DNA-RNA hybrid, DNA double-strand break, transcription-associated homologous recombination repair, RAD52, XPG
At transcribed regions of the genome, RNAs are enriched to regulate a range of cellular processes, including translation, replication, and DNA repair. R-loop, a three-stranded structure of nucleic acids comprising a DNA-RNA hybrid and displaced single-stranded DNA, has been shown to negatively regulate genome stability.1 However, recently in the DNA repair field, a positive role of RNA and R-loop in homologous recombination repair (HRR) of double-strand breaks (DSBs) has been revealed in S. pombe and S. cerevisiae.2,3 Previous studies have suggested that DNA damage signaling mediated by ATM or the DNA-dependent protein kinase inhibits the transcription upon DNA damage.4,5 However, the precise mechanisms of inhibiting transcription following DSB induction and whether R-loops are formed after DNA damage remain unclear. One of the technical barriers to the detection of R-loops following DSB induction has been that only the small amount of R-loop is associated with DSBs, i.e., the background levels of R-loop are too high to enable the detection of R-loop formed upon DSB induction.
In our recently published work, to address the technical barriers, we adopted two techniques to trace the formation and resolution of R-loops associated with DSB induction in real time.6 First, we employed a near infrared (730 nm) two-photon microbeam irradiation system, which, of the laser type systems frequently employed, induces mostly DSBs in a concentrated manner with minimum ultraviolet (UV) photoproducts.7 This is critical because UV irradiation can trigger transcription arrest and initiate transcription-coupled nucleotide excision repair, which is not what this study aimed to detect. The concentrated induction of DSBs enabled us to visualize accumulation of DNA/RNA structures and proteins, even if the amount of their accumulation to each DSB is very small. Second, we used a previously reported DNA-RNA hybrid indicator to examine the kinetics of DNA-RNA hybrid formation after DNA damage. This indicator consists of the fluorescent-tagged hybrid-binding domain of human RNaseH1 and detects DNA-RNA hybrids as effectively as the S9.6 antibody, a specific antibody that recognizes DNA-RNA hybrids.8 The combination of these techniques enabled us to trace DNA-RNA hybrid formation and resolution in real time.
Interestingly, our data revealed that DNA-RNA hybrids are rapidly formed upon DSB induction, peaking at 1–2 minutes, and most of them are resolved within 5 minutes after DSB induction. In combination with our analysis of the DSB repair pathway, we demonstrated that both DNA-RNA hybrid formation and resolution are critical for the initiation of transcription-associated HRR (TA-HRR) (Figure 1). Furthermore, the mechanism of TA-HRR is critical for the maintenance of genome stability in highly transcribed regions. Therefore, we have revealed that a very transient structure that has been difficult to detect in previous experimental systems can indeed protect our genome by regulating the DSB repair pathway.
Figure 1.
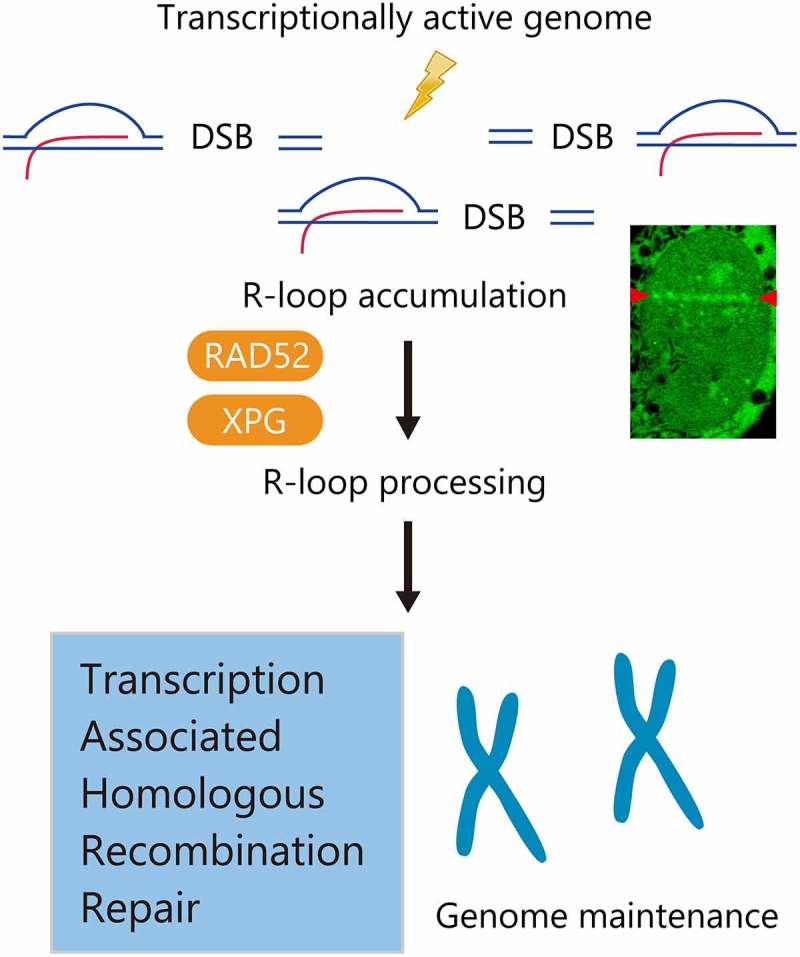
The role of R-loops in transcription-associated DNA double-strand breaks.
The model for the role of R-loops in transcriptionally active genome protection. R-loops formed upon DNA double-strand break (DSB) induction recruit RAD52 and XPG. These factors process R-loops to initiate transcription-associated homologous recombination repair (TA-HRR). The TA-HRR mechanism is critical for the maintenance of genome stability in transcriptionally active regions.
Using this system, we also identified the factors involved in the regulation of DNA-RNA hybrids formed upon DSB induction. First, RAD52 is recruited in a DNA-RNA hybrid-dependent manner upon DSB induction, which is critical for the resolution of DNA-RNA hybrids. Next, the nucleotide excision repair protein XPG (ERCC5, best known as XPG), not XPF (ERCC4, best known as XPF), is involved in the RAD52-dependent resolution of DNA-RNA hybrids. Considering that XPG is a structure-specific endonuclease, which recognizes and incises the 3ʹ junction of single- and double-strand DNA, XPG is assumed to incise the R-loop structure to facilitate the resolution of DNA-RNA hybrids.
Although we successfully visualized the R-loops formed upon DSB induction and demonstrated their significance in the promotion of TA-HRR, there are still numerous questions that remain to be resolved. One of the key issues is the dependence of R-loop regulation on the cell cycle. In the recent study, we focused solely on S/G2-phase cells to investigate R-loop functions in HRR. We also detected considerable amount of R-loops that did not seem to be involved in DSB repair. This observation suggests that among the R-loops formed upon DSB induction, only a limited fraction can participate in the regulation of the DSB repair pathway. Further analysis of R-loop regulation following DSB induction would enable us to understand this interesting observation.
Funding Statement
This work was supported by JSPS KAKENHI Grant Number JP15H06146 and JP18K18191 to T.Y., JP15H04902 and JP15K14376 to K.M.
Disclosure of interest
The authors report no conflict of interest.
References
- 1.Aguilera A, Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. PMID:22541554. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell. 2016;167:1001–1013e1007. PMID:27881299. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. PMID:25186730. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. PMID:20550933. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. PMID:22343725. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 6.Yasuhara T, Kato R, Hagiwara Y, Shiotani B, Yamauchi M, Nakada S, Shibata A, Miyagawa K. Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell. 2018;175:558–570. PMID:30245011. doi: 10.1016/j.cell.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds P, Botchway SW, Parker AW, O’Neill P. Spatiotemporal dynamics of DNA repair proteins following laser microbeam induced DNA damage – when is a DSB not a DSB?. Mutat Res/Genet Toxicol Environ Mutagen. 2013;756:14–20. PMID:23688615. doi: 10.1016/j.mrgentox.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. PMID:24896180. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]


