ABSTRACT
Patient-derived xenograft tumors retain molecular and histopathological features of the originating tumor and are useful preclinical tools for drug discovery and assessment. We recently reported that ‘rapid’ engraftment of head and neck squamous cell carcinoma samples is highly prognostic and correlates with deregulation of the G1/S checkpoint. Tumors with genetic alterations in cyclinD1 (CCND1) and/or cyclin-dependent kinase inhibitor 2A (CDKN2A) are more likely to respond to abemaciclib.
KEYWORDS: Patient-derived xenografts, head and neck squamous cell carcinoma, engraftment, preclinical tool, biomarker discovery, abemaciclib
Head and neck squamous cell carcinomas (HNSCCs) arise in the mucosal lining of the upper aerodigestive tract. Classical risk factors for HNSCC are smoking, excessive alcohol consumption and infection with human papillomavirus (HPV). HPV(+) HNSCC is primarily localized to the oropharynx and exhibits distinct clinical responses; thus, our work focuses on HPV(-) HNSCC. Surgery is the first line of treatment and patients with more advanced disease also receive radiotherapy and/or chemotherapy. While current clinicopathological features are useful to identify patients that require more aggressive therapy, patient outcome remains poor with locoregional and distant failure. There is thus a need to develop more accurate methods for patient risk stratification and to enhance treatment options.
Next-generation sequencing technologies have revealed molecular heterogeneity within and across several cancer types, including HNSCC. Preclinical models with high translatability are essential to explore and dissect this heterogeneity. Patient-derived xenograft (PDX) models are well suited for such studies as they have been shown to recapitulate the molecular and histopathological features of the originating tumor and are able to predict drug responses.1–4 PDXs are the only model system that can incorporate the vast inter-patient and intra-tumor heterogeneity that is inherent to human cancers, making them a relevant preclinical tool for drug discovery.
From 2008 to 2015 we subcutaneously implanted fragments of surgically resected HNSCC tumor tissue into immune-compromised mice, establishing an extensive number of HNSCC-PDX models.5 Our xenografts consistently recapitulate histological and molecular features of the original human tumor and PDX tissues are viably banked for ongoing and future studies. In our recent study, the engraftment efficiency for 243 HNSCC implanted samples was ~60% and we found that engraftment associated significantly to poor patient overall survival, disease-free survival and distant metastasis. Poor patient outcome associated with engraftment has also been observed in other cancers.6,7
PDX growth rates were highly variable between patients. We, therefore, assessed outcomes of patients whose xenografts were palpable within 8 weeks of implantation and found that such “rapid engraftment” was associated more strongly to poor overall survival, disease-free survival and locoregional control. Currently, the only reliable predictor for patient outcome is nodal status. However, our findings show that engraftment ability, regardless of nodal status, is a strong predictor for patient outcome. The ability to acquire this result within 8 weeks fits well within the current HNSCC treatment timeline and could provide additional information to help devise or modify a patient’s treatment plan. For example, a patient presenting with clinical features that would not qualify for radiation therapy but does show rapid engraftment may be escalated to a more aggressive treatment plan to decrease the risk of locoregional failure. One patient in our study whose tumor demonstrated rapid engraftment was low risk according to clinical indications and was treated with surgery alone. The patient rapidly relapsed and died a few months later. With the knowledge that this tumor sample engrafted rapidly, additional treatment for this patient may have prevented recurrence. The clinical application of rapid engraftment as a biomarker for risk stratification could therefore potentially improve the outcome for HNSCC patients. In our opinion, a prospective study is clearly warranted.
While determining engraftment ability within an 8-week timeline is relatively inexpensive, it may prove challenging due to lack of expertise and facilities across hospitals. To identify underlying genetic alterations that could potentially serve as surrogate clinical biomarkers, we carried out next-generation sequencing and copy number analysis on a cohort of engrafting and non-engrafting patient samples. No single gene mutation was significantly associated with engraftment, but we found that loss of chromosome 3p and copy number gains at the 11q locus containing the cyclinD1 (CCND1) gene were significantly associated with engraftment. Amplification of this region of 11q has been previously reported in HNSCC patients and shown to be correlated with worse outcomes.8,9 Furthermore, alterations of CCND1 and/or cyclin-dependent kinase inhibitor 2A (CDKN2A) (both leading to deregulation of the G1/S cell cycle checkpoint pathway), were strongly associated with rapid engraftment.
PDXs have been shown to mimic human clinical trials and can be used to characterize drug efficacy, improve preclinical evaluation of treatment modalities and enhance the ability to predict clinical trial responses.3,4 CCND1 amplification and/or CDKN2A mutations are common in HNSCC (25% and 53%, respectively, in the Cancer Genome Atlas; and 27% and 44% in our study). Alterations in these genes enhance the activity of cyclin-dependent kinase-4 or −6 (CDK4/6), accelerating cancer cell progression through the G1/S cell cycle checkpoint. Thus, inhibition of CDK4/6 in these particular tumors may repress tumor growth. We used our HNSCC-PDX models to ascertain whether tumors containing CCND1 and/or CDKN2A alterations demonstrated selective sensitivity to the CDK4/6 inhibitor, abemaciclib. Ten HPV(-) HNSCC-PDX models were treated with abemaciclib: five of six CCND1 and/or CDKN2A mutated tumors and one of four of ‘wild type’ tumors showed a significant delay in tumor growth. Our results suggest that abemaciclib has anti-tumor activity in HNSCC, and CCND1 and/or CDKN2A alterations may be predictive of such a response.
CDK4/6 inhibitors are presently used for advanced estrogen receptor positive/human epidermal growth factor negative (ER+/HER2−) breast cancers, showing improved progression-free survival.10 Several clinical trials are in progress to evaluate CDK4/6 inhibitors in HNSCC, but only one is specifically taking into account CCND1 status. Since rapid engrafters are enriched for alterations in CCND1 and/or CDKN2A, these high-risk patients may benefit the most from the addition of abemaciclib to their treatment plan.
Our results support the assessment of abemaciclib in clinical trials and provide proof of principle for the use of PDXs for preclinical drug evaluation and predictive biomarker identification. The PDX model represents a patient ‘‘avatar’’ in which drug responses could be assessed within a relatively short time frame (Figure 1). Given that surgery and post-operative therapy can last several months, interrogations of candidate targeted treatments and predictive biomarkers could feasibly be determined before relapse. Over time, insight will be gained into the heterogeneous molecular landscape of HNSCCs and their specific responses to various drugs. With the help of PDXs the goal is to identify predictive biomarkers to be used directly and appropriately in the clinical setting.
Figure 1.
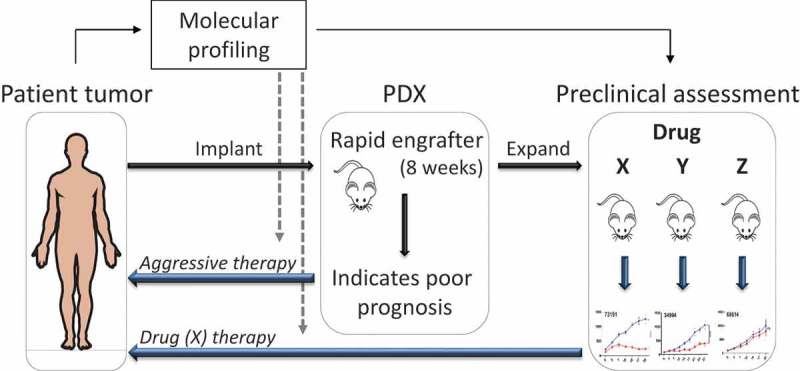
Patient-derived xenografts in personalized risk stratification and drug sensitivity screening. Head and neck squamous cell carcinoma (HNSCC) patient tumors that engraft into immuno-compromised mice by 8 weeks (rapid engrafters) have poor clinical outcome. Rapid engraftment can serve as a predictive biomarker to indicate the strong need for a patient to receive aggressive therapy (radiation and/or chemotherapy). Thus, engraftment status can be added to the existing pathological and clinical features used to decide the course of treatment. Given the rapid growth of these patient-derived xenografts (PDXs), they can be treated and tested quickly with approved drugs (X, Y, Z) to determine which one provides the highest tumor response and then applied to the patient. As extensive numbers of PDXs are tested, the heterogeneous landscape of mutations and their responses to various drugs will be captured with the goal that these molecular profiles can be used in the future, directly on patient tumors, to determine prognosis and predictive therapies (gray dashed arrows). Abbreviations: PDX = patient-derived xenograft.
Funding Statement
This work was supported by grants from the Canadian Institutes of Health Research (MOP126203), the Ontario Institute for Cancer Research (Investigator Award), the Princess Margaret Cancer Foundation, and the Joe and Cara Finley Centre for Head and Neck Cancer Translational Research, with additional philanthropic funds contributed by the Wharton Family and Gordon Tozer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Keysar SB, Astling DP, Anderson RT, Vogler BW, Bowles DW, Morton JJ, Paylor JJ, Glogowska MJ, Le PN, Eagles-Soukup JR, et al. A patient tumor transplant model of squamous cell cancer identifies PI3K inhibitors as candidate therapeutics in defined molecular bins. Mol Oncol. 2013;7(4):776–790. doi: 10.1016/j.molonc.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Wheeler S, Park Y, Ju Z, Thomas SM, Fichera M, Egloff AM, Lui VW, Duvvuri U, Bauman JE, et al. Proteomic characterization of head and neck cancer patient-derived xenografts. Mol Cancer Res. 2016;14(3):278–286. doi: 10.1158/1541-7786.MCR-15-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 4.Townsend EC, Murakami MA, Christodoulou A, Christie AL, Köster J, DeSouza TA, Morgan EA, Kallgren SP, Liu H, Wu S-C, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell. 2016;29(4):574–586. doi: 10.1016/j.ccell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karamboulas C, Bruce JP, Hope AJ, Meens J, Huang SH, Erdmann N, Hyatt E, Pereira K, Goldstein DP, Weinreb I, et al. Patient-derived xenografts for prognostication and personalized treatment for head and neck squamous cell carcinoma. Cell Rep. 2018;25(5):1318–1331 e4. doi: 10.1016/j.celrep.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 6.McAuliffe PF, Evans KW, Akcakanat A, Chen K, Zheng X, Zhao H, Eterovic AK, Sangai T, Holder AM, Sharma C, et al. Ability to generate patient-derived breast cancer xenografts is enhanced in chemoresistant disease and predicts poor patient outcomes. PLoS One. 2015;10(9):e0136851. doi: 10.1371/journal.pone.0136851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John T, Kohler D, Pintilie M, Yanagawa N, Pham N-A, Li M, Panchal D, Hui F, Meng F, Shepherd FA, et al. The ability to form primary tumor xenografts is predictive of increased risk of disease recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2011;17(1):134–141. doi: 10.1158/1078-0432.CCR-10-2224. [DOI] [PubMed] [Google Scholar]
- 8.Vincent-Chong VK, Salahshourifar I, Woo KM, Anwar A, Razali R, Gudimella R, Rahman ZAA, Ismail SM, Kallarakkal TG, Ramanathan A, et al. Genome wide profiling in oral squamous cell carcinoma identifies a four genetic marker signature of prognostic significance. PLoS One. 2017;12(4):e0174865. doi: 10.1371/journal.pone.0174865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Kempen PMW, Noorlag R, Braunius WW, Moelans CB, Rifi W, Savola S, Koole R, Grolman W, van Es RJJ, Willems SM.. Clinical relevance of copy number profiling in oral and oropharyngeal squamous cell carcinoma. Cancer Med. 2015;4(10):1525–1535. doi: 10.1002/cam4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Ettl J, Patel R, Pinter T, Schmidt M, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]


