ABSTRACT
More than 90% of thyroid cancer belongs to the papillary and follicular thyroid carcinomas based on pathological subtypes. Papillary and follicular thyroid carcinoma are generally associated with a good prognosis. In contrast, other pathological subtypes such as poorly-differentiated and anaplastic thyroid carcinoma (PDTC and ATC) have a poor clinical outcome with a short life expectancy. To identify the genetic variations and biomarkers that may potentially distinguish the aggressive form of thyroid cancer, we performed a retrospective analysis of the formalin-fixed paraffin-embedded tumor samples from 50 patients who mainly displayed aggressive thyroid cancer using next-generation sequencing of 416 solid tumor-related genes. We adopted extensive bioinformatic analysis to vigorously remove germline single-nucleotide polymorphism and systematic sequencing errors, and report here that mutation in DNMT3A gene was significantly enriched in patients with PDTC or ATC.
Keywords: thyroid carcinoma, gene mutation, next-generation sequencing
Introduction
Thyroid cancer is a common endocrine malignancy, and it has been divided into several pathological subgroups. Papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC) account for approximately 90% of all thyroid cancer cases.1 PTC and FTC are collectively considered as well-differentiated thyroid cancer (WDTC), and are generally associated with good prognosis with over 10-year survival rate in 90% of patients.1 Medullary thyroid carcinoma (MTC) is the next dominant subtype, accounting for approximately 3%-5% of all thyroid cancer cases. MTC can be associated with inherited predisposition depending on the mutational status of the rearranged with introduction of (RET) proto-oncogene.1 Although rare, poorly-differentiated thyroid carcinoma (PDTC) and anaplastic (also called undifferentiated) thyroid carcinoma (ATC), show the most aggressive progression with frequently uncontrolled local disease or distant metastasis, and with an extremely short life expectation.1
With the recent advancement of DNA sequencing technology, biomarkers have been vigorously evaluated in order to improve disease stratification and prognostic implication in the treatment of thyroid cancer.2 Aberrant activation of the mitogen activated protein kinase (MAPK) pathway is a frequently mutated signaling pathway in PTC and FTC, with mutations in BRAF and RAS genes showing the highest prevalence.3 BRAFp.V600E mutation accounts for approximately half of all PTC and FTC patients, and has been demonstrated its prognostic value of recurrence.4,5 RET rearrangement and point mutation are also common in WDTC and MTC, respectively.3 However, the exact molecular mechanism and genetic characteristics related to PDTC and ATC remain poorly understood.
To better understand the thyroid cancer and identify biomarkers with clinical implication, we retrospectively analyzed the formalin-fixed paraffin-embedded tumor samples of a cohort of 50 thyroid carcinoma patients using next-generation sequencing. As EGFR mutation was detected in FTC with lung metastasis by the amplification refractory mutation system (ARMS) method in our hospital (unpublished), we focused on FTC and those deadly or uncontrolled thyroid carcinoma. We interrogated the exonic regions of 416 genes and the intronic regions of a selected subset of genes that are frequently mutated in solid tumors. Due to lack of matched whole blood sample as a negative control, we deployed a series of bioinformatic algorithms to remove germline single nucleotide polymorphisms (SNP) and sequencing errors. Unfortunately, the EGFR exon 19 in-frame deletion case which was identified by ARMS failed to pass the examination of next-generation sequencing because of the biopsied tumor sample was too small. The EGFR mutations were also reported by other groups.6 We confirmed that NRASp.Q61 and BRAFp.V600E is a recurrent mutation in this cohort, but with much less lower prevalence. We also found that DNMT3A mutation was significantly enriched in PDTC and ATC, suggesting its potential value in informing personalized treatment for patients with thyroid cancer.
Methods
Patient recruitment
This study was approved by the ethics committee of the First Affiliated Hospital of Soochow University, China. A total of 50 thyroid cancer patients diagnosed and treated in our hospital were recruited to this study from Aug 2012 to Dec 2016. Unstained formalin-fixed paraffin-embedded (FFPE) slides or FFPE scrolls were sectioned from archived FFPE tissue blocks. These cases were reviewed by a pathologist (LCG) to confirm the diagnosis and select areas of tumors for DNA extraction.
DNA extraction and library preparation
All FFPE samples were examined by Nanjing Geneseeq Technology Inc., Nanjing, China for testing following previous described procedures.7 Briefly, FFPE samples were first de-paraffinized with xylene and DNA was extracted using QIAamp DNA FFPE Tissue Kit (Qiagen), according to the manufacturer’s protocols. Resulting DNA sample was evaluated using NanoDrop 2000 (Thermo Fisher Scientific) for potential organic solvent contamination, and quantified using Qubit 2.0 dsDNA HS Assay (Life Technologies) following manufacturer’s instructions.
Sequencing libraries were prepared using KAPA Hyper Prep Kit (KAPA Biosystems) following manufacturer’s recommended protocols. Briefly, 1 μg of DNA, if necessary, mechanically sheared into 350bp fragments using Covaris M220 instrument, was processed sequentially by end-repairing, A-tailing, adaptor ligation, and PCR amplification. Purification and size selection was performed using AMPure XP beads.
Hybridization capture and sequencing
Different libraries with unique sample indices were pooled together in desirable ratios for up to 2 μg of total library input. Human cot-1 DNA (Life Technologies) and xGen Universal blocking oligos (Integrated DNA Technologies) were added as blocking reagents. Customized xGen lock down probes (Integrated DNA Technologies) targeting the exonic regions of 416 cancer-relevant genes and selected intronic regions.7 The capture reaction was performed with the Nimble GenSeqCap EZ Hybridization and Wash Kit (Roche) and Dynabeads M-270 (Life Technologies). Captured libraries were on-beads amplified with Illumina p5 (5′ AAT GAT ACG GCG ACC ACC GA 3′) and p7 primers (5′ CAA GCA GAA GAC GGC ATA CGA GAT 3′) in KAPA HiFiHotStartReadyMix (KAPA Biosystems). The post-capture amplified library was purified by AMPure XP beads and quantified by qPCR using the NEB Next Library Quantification kit (New England Biolabs). The target-enriched library was then sequenced on HiSeq 4000 NGS platforms (Illumina) according to the manufacturer’s instructions with an anticipated average depth of coverage of approximately 700X.
Sequence data processing
After performing demultiplexing by bcl2fastq, Trimmomatic was used for FASTQ file quality control.8 Leading/trailing low quality (quality reading below 15) or N bases were removed. Reads from each sample were mapped to the human reference genome hg19 using Burrows-Wheeler Aligner (BWA-mem, v0.7.12).9,10 Local realignment around indels and base quality score recalibration were applied with the Genome Analysis Toolkit (GATK 3.4.0).11 PCR duplicates were removed by Picard (available at: https://broadinstitute.github.io/picard/). VarScan2 was employed for detection of single nucleotide variants12 and insertion/deletion mutations with the following settings: minimum read depth = 20, minimum base quality = 15, minimum variant allele frequency = 0.01, minimum variant supporting reads = 5, variant supporting reads mapped to both strands, and strand bias no greater than 15%. CNVkit was used for identification of copy number variants,13 with a segment log2 ratio cutoff of 0.32 for copy number gain and −0.41 for copy number loss.
Variant filtering and annotation
The vcf files generated by VarScan were annotated using ANNOVAR14 against the following database: dbSNP (v138),15 1000Genome16, ESP6500 (available at: http://evs.gs.washington.edu/evs/), ExAC,17 CG46, COSMIC (v70),18 ClinVAR,19 and PolyPhen2.20 Mutations that did not result in non-synonymous or frameshift, or mutations that was not documented in ClinVAR or COSMIC were remove. Mutations showing 1% or higher frequency in the 1000 Genome, ESP6500, ExAC, or CG46 were removed as well. The result mutation lists were filtered through an internally maintained list of recurrent sequencing errors. Mutations occurred within the repeat masked regions while not annotated by COSMIC or ClinVAR were also removed. Remaining mutations were reverse-referenced against the other samples (non-thyroid cancer samples and whole blood samples) sequenced within the same flow-cell. We removed variant that is detected within more than 20% of the non-thyroid cancer samples or within more than 10% of the whole blood samples.
Statistical analysis
Statistical analysis was performed using the R Programming Language.21 When comparing our PTC and FTC patient data with TCGA results, Fisher’s exact test was performed. When comparing patient demographics, Freeman-Halton extension of Fisher’s exact test and Cox proportional hazards model was performed. For genes whose mutation was detected in more than 3 samples (43 genes), Freeman-Halton extension of Fisher’s exact test was performed to identify significantly mutated genes within subgroups of the patients. Multiple test correction was performed using false discovery rate.
Results
Patient demographics
Fifty thyroid cancer patients diagnosed and treated in our hospital between Aug 2012 and Dec 2016 were included in this study. Each patient was diagnosed with either PTC with distant metastasis during initial assessment, or had FTC, MTC, PDTC, ATC at initial diagnosis. Because of the selection process, half of the cohort was MTC, PDTC and ATC, deviating from the reported statistics. Consistent with the literature, females have a higher incidence of thyroid cancer than males (Table 1). However, no significant difference was detected in the disease pathological type distribution (Fisher’s exact test, p = 0.610) and disease stage (WHO 7th edition) distribution (Fisher’s exact test, p = 0.073) between genders. Most of (45/50) patients received partial or totalthyroidectomy as the standard treatment protocol. Seven patients with metastatic tumor to the lung (2), brain (2), thoracic vertebra (2) and skull (1) were diagnosed based on the biopsies. Following surgery or biopsy, four patients received radioactive iodine therapy and two patients received chemotherapy while two patients received both radioactive iodine and chemotherapy treatment. Samples including 43 from thyroid tissue and 7 from distant metastases were meeted the criteriaused for analysis by next-generation sequencing.
Table 1.
Summary of patient demographics, thyroid carcinoma subtypes and clinical staging.
| Cases | Significance (Cox Proportional Hazards model) | ||
|---|---|---|---|
| Gender | Female | 31 | p = 0.039 * |
| Male | 19 | ||
| Age | <60 | 24 | p = 0.139 |
| ≥ 60 | 26 | ||
| Stage | I | 11 | |
| II | 9 | p = 1.000 | |
| III | 9 | p = 0.998 | |
| IVA | 9 | p = 0.998 | |
| IVB | 2 | p = 0.998 | |
| IVC | 10 | p = 0.998 | |
| Pathology | Papillary TC | 2 | |
| Follicular TC | 23 | p = 0.130 | |
| Medullary TC | 14 | p = 0.193 | |
| Poorly Differentiated TC | 4 | p = 0.214 | |
| Anaplastic TC | 7 | p = 0.749 |
* p-value <0.05. TC: Thyroid Carcinoma
Mutation profile
We performed targeted sequencing of the exonic regions of 416 solid tumor related genes and the intronic regions of a subset of the genes using Illumina sequencing platform on the archived FFPE samples of our cohort. Regrettably, due to those patients who failed to thrive, we could not collect the matched whole blood samples as a negative control to filter patient specific germline mutations. To circumvent this shortcoming, we screened the detected single nucleotide variants (SNV) and small insertion/deletion (Indel) through the following criteria: 1) the mutations may not demonstrate more than 1% population frequency within the 1000 Genome, ESP6500, ExAC, and CG46 databases; 2) the mutations may not be detected within more than 20% of the non-thyroid cancer samples or within more than 10% of the whole blood samples sequenced within the same flow cell; 3) remove any mutation if all detected incidents were below 2% variant allelic frequency; 4) rescue any mutations removed in the previous steps if it is documented within the COSMIC or ClinVAR databases. For copy number variants (CNV), we set the cutoff of Log2 ratio for calling CNV gain and loss to be 0.32 (Log2 of 1.25) and −0.41 (Log2 of 0.75), respectively. We also removed CNV fragments longer than 5,000,000 base pairs to focus our analysis on focal copy number aberrations.
We obtained a total of 305 SNV, 78 Indel, and 167 CNV after applying the aforementioned filters. Figure 1 displayed the mutational profile of the analyzed cohort. Due to limited sample sizes, we grouped PTC and FTC as WDTC, and PDTC and ATC as LDTC for downstream analysis. NRAS (13), RET (10), TP53 (9), BRAF (6), and HRAS (6) genes remain the most frequently mutated genes, with number in brackets indicating the number of patients carrying the mutations. Three of the four incidences of BRAF SNV were c.1799T>A, p.(V600E) mutations, while the other one was a c.1790T>A, p.(L597Q) mutation. Among the 21 mutational events occurred on NRAS, HRAS, and KRAS, 19 involves the glutamine at codon 61 and 2 involves the glycine at codon 13, consistent with previously reported bias. 22
Figure 1.
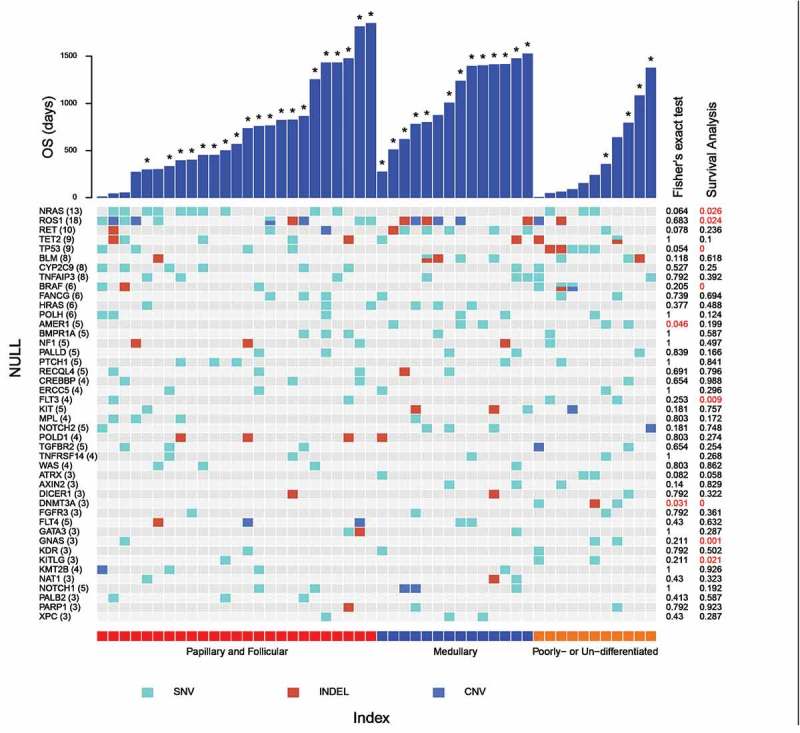
Mutational profile of aggressive thyroid cancer patients. Each column represents an individual patient. Only genes whose mutations were detected in at least 3 patients were shown. The numbers in brackets following each gene name indicates the number of patients carrying the mutation. Overall survival of patients was plotted as bar graph, with asterisk marking the patients who still thrive at last revisit. The p-value of Freeman-Halton extension of Fisher’s exact test and survival analysis were listed on the right side with red color highlighting p-values < 0.05.
We cross-referenced our mutational profiles against the Cancer Genome Atlas (TCGA) database. Interestingly, among the 504 available thyroid cancer (THCA) patients documented in TCGA, 235 of 241 BRAF mutations occurred at p.V600E, among which only 12 were detected in FTC patients. None of the mutations within NRAS, TP53, and BRAF genes were detected in MTC patients, further supporting that MTC display a distinctive mutational profile from the other subtypes. By applying a 2 × 3 extension of Fisher’s exact test to the genes whose mutations were recurrently detected in at least 3 patients, we identified that AMER1 mutation was significantly enriched in MTC and LDTC patients, while DNMT3A mutations were significantly enriched in LDTC patients.
Survival analysis
Survival analysis of the patient overall survival (OS) was then applied to our dataset. Consistent with established statistic data, three groups of patients displayed significant difference in survival pattern (p = 0.002), with PDTC and ATC being the group showing worst outcome (Figure 2A). We then extended the analysis for each gene by comparing the OS of the patients with mutations within the gene to that of the patients with wild type. We found that NRAS mutations were significantly associated with a poor OS, but the significance was lost after multiple test correction (Figure 2B). We also found that mutations in TP53, BRAF, DNMT3A, and GNAS were significantly associated with the poor survival (p < 0.05, FDR<0.1) (Figure 2C-F).
Figure 2.
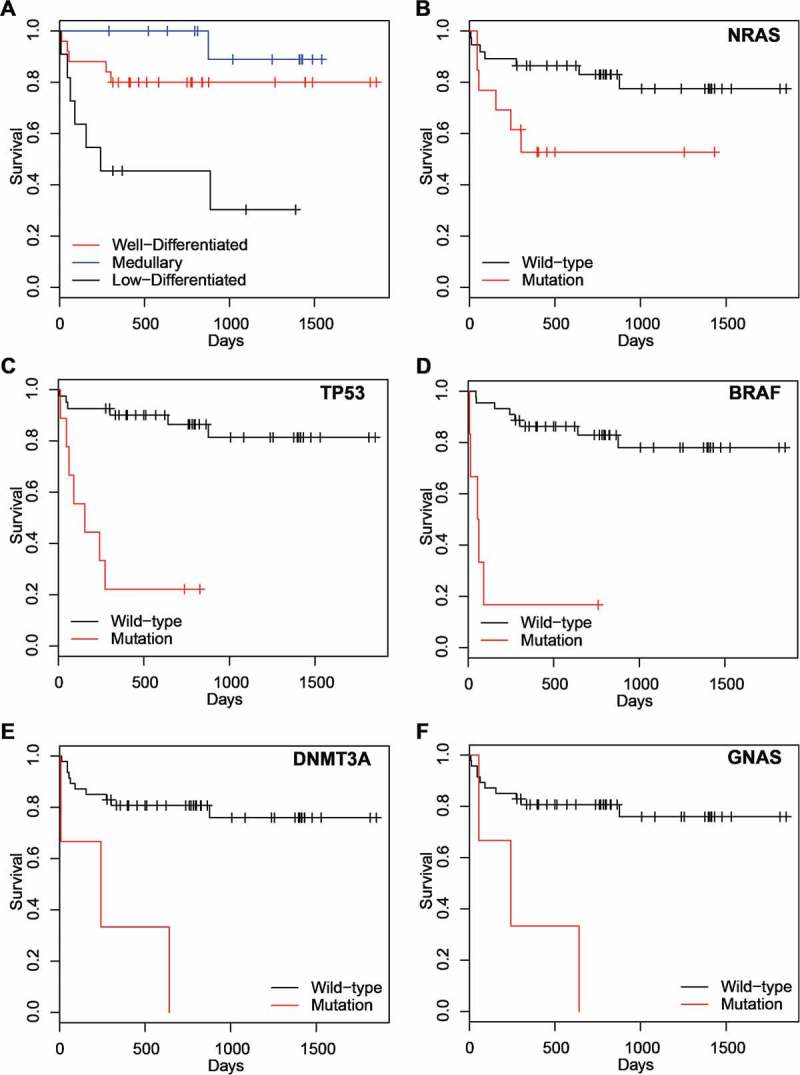
Survival Plot. Patient grouping is based on A) pathology subtypes and mutational status, B) the NRAS gene, C) the TP53 gene, D) the BRAF gene, E) the DNMT3A gene, and F) the GNAS gene.
Discussions
Thyroid cancer is the most prevalent type of endocrine neoplasm. Molecular pathology has greatly been developed in recent years, and it has been applied in the diagnosis and guideline for the treatment.23 The diagnosis of thyroid cancer has been heavily relied on genetic analysis as an extra adjunct of morphology and immuno-histochemical staining results. For instance, genetic alterations defining subtypes in thyroid cancer include RET and NTRK mutation in subsets of PTC24,25 and PAX8-PPARγ in FTC.26 In general, molecular transcripts have been respectively associated with thyroid cancer, such as BRAF p.V600E, RAS, DNMT3 mutation, RET, NTRK1, NTRK3, ALK, PAX8 gene rearrangement, and other related oncogenes.27–30 Besides diagnostic and genetic values, the detection of genetic changes in thyroid cancer has been becoming an effective way to search for new therapeutic targets. Recently, the RET inhibitors, including vandetanib31 and cabozantinib,32 have been approved by the US Food and Drug Administration (FDA) to treat metastatic MTC because they have anti-proliferative role in RET-mutant MTC. Also, the sensitivity of vandetanib has been observed in PTC with RET mutation.33 But little is known about oncogene driven mutation in aggressive thyroid cancer. Therefore, efforts to target several other molecular alterations are still under way.
EGFR 19del was first detected in a FTC female patient with ARMS method in our hospital in 2015. EGFR mutation often occurred in lung adenocarcinoma, especially in Asian female without smoking.34 Target therapy with small molecular EGFR tyrosine kinase inhibitor has achieved amazing outcomes in these EGFR mutation non-small cell lung cancer patients, but little is reported in thyroid carcinoma.6,35 It is interesting that the PFS is nearly 11 months in this patient treated with EGFR tyrosine kinase inhibitor. As far as we know, this is the first reported clinical efficacy of EGFR TKI in FTC with EGFR mutation. Even only one case with EGFR mutation was found in our study, it is also suggested that EGFR mutation should be detected especially in uncontrolled thyroid cancer in clinical practice as only 50 thyroid cancer patients were recruited in our study. In this study, we retrospectively analyzed the archived FFPE tumor samples from 50 thyroid cancer patients with next-generation sequencing. Despite lacking the matched whole blood sample, which may constitute aminor setback to the analysis, we performed extensive bioinformatic algorithms and vigorously removed contamination from patient specific germline mutations. The filtering process is also capable of suppressing systematic sequencing error unique to the flow cell, as it evaluates each mutation against the samples of other cancer types and the unrelated whole blood control samples within the same sequencing run. Nevertheless, it should be pointed out that bioinformatic filtering is not capable of replacing matched normal sample control at removing germline mutations. Indeed, 201 of the 305 SNV mutations had a variant allelic frequency between 40% and 60% or greater than 98%, highly reminiscent of that of a germline mutation. The AMER1 mutations enriched in MTC and LDTC were an example, all 5 incidences of AMER1 mutation were c.85G>A and were documented in dbSNP as rs138399473. The three GNAS mutations identified through survival analysis also falls into this category.
We noticed that although BRAFp.V600E was a frequently mutated gene in WDTC and LDTC, its recurrence frequency was much lower than the reported frequency in TCGA (Fisher’s exact test, p < 0.001). This is likely a consequence of the fact that FTC and MTC are the principal subtypes of our cohort. Indeed, a closer examination of TCGA data showed that BRAFp.V600E has a much lower prevalence in FTC (12 incidences in 102 cases) than PTC (192 incidences in 358 cases) (Fisher’s exact test, p < 0.001), while several published work collectively supported that BRAF p.V600E is rare in MTC.36–38 Among the three RAS gene family members, most of the mutations occurred at codon Q61 instead of the classical codon G12 or G13. This observation supports the previous report, and suggests a mutational profile unique to the cancer type.22
By dividing our cohort into WDTC, MTC, and LDTC and applying Freeman-Halton extension of Fisher’s exact test, we also identified DNMT3A mutation to be significantly enriched in our LDTC patients. Regrettably, the statistical significance is lost (FDR = 0.6) after multiple test correction, and survival analysis suggests that mutation in DNMT3A is significantly associated with short life expectancy in LDTC patients. This gene encodes a DNA methyltranserase that is essential to maintain the methylation status of the genome. Aberrant methylation of promoter region of the DNA mismatch repair gene hMLH1 has been associated with the development of BRAFp.V600E mutation in PTC.39 To our knowledge, DNMT3A mutation was only previously reported as a recurrently mutated gene in a Saudi Arabia cohort of 886 PTC patients (1.5% recurrence) and was associated with adverse parameters such as extra-thyroidal extension and late-stage in PTC. Given the limited PTC patients in our cohort, it is not surprising that DNMT3A mutations were only detected in our LDTC patients. Our findings suggest that DNMT3A can be explored as a potential predictive biomarker or therapeutic target for prognosis and treatment of thyroid cancer.
Funding Statement
This study was funded by grants from National Natural Sciences Foundation of China (81473240, 81773749) to Yi Zhang; National Natural Sciences Foundation of China (81472191) to Shundong Ji; Natural Science Foundation of Jiangsu Province of China (BK20151209) to Cheng Ji; and sponsored by Qing Lan Project to Yi Zhang.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Ethical approval
All procedures performed in studies involving human participants were in accordance with ethics committee of the First Affiliated Hospital of Soochow University and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
References
- 1.Fahiminiya S, de Kock L, Foulkes WD.. Biologic and clinical perspectives on thyroid cancer. N Engl J Med. 2016;375:2306–2307. doi: 10.1056/NEJMc1613118. [DOI] [PubMed] [Google Scholar]
- 2.Cha YJ, Koo JS, Cos X, Solé C, Mestre S, Canela M, Boquet A, Cabré -J-J, Barrio F, Flores-Mateo G, et al. Next-generation sequencing in thyroid cancer. J Transl Med. 2016;14:322. doi: 10.1186/s12967-016-0867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikiforov YE, Nikiforova MN.. Molecular genetics and diagnosis of thyroid cancer. Nat Reviews Endocrinol. 2011;7:569–580. doi: 10.1038/nrendo.2011.142. [DOI] [PubMed] [Google Scholar]
- 4.Xing M, Alzahrani AS, Carson KA, Viola D, Elisei R, Bendlova B, Yip L, Mian C, Vianello F, Tuttle RM, et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. Jama. 2013;309:1493–1501. doi: 10.1001/jama.2013.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xing M, Alzahrani AS, Carson KA, Shong YK, Kim TY, Viola D, Elisei R, Bendlová B, Yip L, Mian C, et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clinical Oncology: Official Journal Am Soc Clin Oncol. 2015;33:42–50. doi: 10.1200/JCO.2014.56.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masago K, Asato R, Fujita S, Hirano S, Tamura Y, Kanda T, Mio T, Katakami N, Mishima M, Ito J. Epidermal growth factor receptor gene mutations in papillary thyroid carcinoma. Inter J Can. 2009;124:2744–2749. [DOI] [PubMed] [Google Scholar]
- 7.Shu Y, Wu X, Tong X, Wang X, Chang Z, Mao Y, Chen X, Sun J, Wang Z, Hong Z, et al. Circulating Tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017;7:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30;2014:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-Wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29;2001:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D83. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics 2013; Chapter 7: Unit720.doi: 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed]
- 21.R Development Core Team R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2008. [Google Scholar]
- 22.Nikiforov YE. Molecular diagnostics of thyroid tumors. Arch Pathol Lab Med. 135;2011:569–577. [DOI] [PubMed] [Google Scholar]
- 23.Baloch ZW, LiVolsi VA. Special types of thyroid carcinoma. Histopathology. 2018;72:40–52. doi: 10.1111/his.13348. [DOI] [PubMed] [Google Scholar]
- 24.Jargin SV. RET/PTC3 rearrangement in papillary thyroid carcinoma: possible marker of tumor progression. Ann Surg. 2017;266:e120–e1. doi: 10.1097/SLA.0000000000002031. [DOI] [PubMed] [Google Scholar]
- 25.Prasad ML, Vyas M, Horne MJ, Virk RK, Morotti R, Liu Z, Tallini G, Nikiforova MN, Christison-Lagay ER, Udelsman R, et al. NTRK fusion oncogenes in pediatric papillary thyroid carcinoma in northeast United States. Cancer. 2016;122:1097–1107. doi: 10.1002/cncr.29887. [DOI] [PubMed] [Google Scholar]
- 26.Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn GW 2nd, Tallini G, Kroll TG, Nikiforov YE. RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88:2318–2326. doi: 10.1210/jc.2002-021907. [DOI] [PubMed] [Google Scholar]
- 27.Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129. doi: 10.1186/s13073-015-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Z, Xie WJ, Zhao M, Cheng GP, Wu MJ. DDR2 facilitates papillary thyroid carcinoma epithelial mesenchymal transition by activating ERK2/Snail1 pathway. Oncol Lett. 2017;14:8114–8121. doi: 10.3892/ol.2017.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrari SM, Fallahi P, Politti U, Materazzi G, Baldini E, Ulisse S, Miccoli P, Antonelli A. Molecular targeted therapies of aggressive thyroid cancer. Front Endocrinol (Lausanne). 2015;6:176. doi: 10.3389/fendo.2015.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siraj AK, Masoodi T, Bu R, Beg S, Al-Sobhi SS, Al-Dayel F, Al-Dawish M, Alkuraya FS, Al-Kuraya KS. Genomic profiling of thyroid cancer reveals a role for thyroglobulin in metastasis. Am J Hum Genet. 2016;98:1170–1180. doi: 10.1016/j.ajhg.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells SA Jr., Robinson BG, Gagel RF, Dralle H, Fagin JA, Santoro M, Baudin E, Elisei R, Jarzab B, Vasselli JR, et al. Vandetanib in patients with locally advanced or metastatic medullary thyroid cancer: a randomized, double-blind phase III trial. J Clinical Oncology: Official Journal Am Soc Clin Oncol. 2012;30:134–141. doi: 10.1200/JCO.2011.35.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldenberg MM. Pharmaceutical approval update. P & T: a Peer-Reviewed J Formulary Manage. 38;2013:86–95. [PMC free article] [PubMed] [Google Scholar]
- 33.Vitagliano D, De Falco V, Tamburrino A, Coluzzi S, Troncone G, Chiappetta G, Ciardiello F, Tortora G, Fagin JA, Ryan AJ, et al. The tyrosine kinase inhibitor ZD6474 blocks proliferation of RET mutant medullary thyroid carcinoma cells. Endocr Relat Cancer. 2011;18:1–11. doi: 10.1677/ERC-09-0292. [DOI] [PubMed] [Google Scholar]
- 34.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 35.Hogan T, Jing Jie Y, Williams HJ, Altaha R, Xiaobing L, Qi H. Oncocytic, focally anaplastic, thyroid cancer responding to erlotinib. J Oncology Pharmacy Practice: Official Publication Int Soc Oncol Pharm Pract. 2009;15:111–117. doi: 10.1177/1078155208101212. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal N, Jiao Y, Sausen M, Leary R, Bettegowda C, Roberts NJ, Bhan S, Ho AS, Khan Z, Bishop J, et al. Exomic sequencing of medullary thyroid cancer reveals dominant and mutually exclusive oncogenic mutations in RET and RAS. J Clin Endocrinol Metab. 2013;98:E364–9. doi: 10.1210/jc.2012-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boichard A, Croux L, Al Ghuzlan A, Broutin S, Dupuy C, Leboulleux S, Schlumberger M, Bidart JM, Lacroix L. Somatic RAS mutations occur in a large proportion of sporadic RET-negative medullary thyroid carcinomas and extend to a previously unidentified exon. J Clin Endocrinol Metab. 2012;97:E2031–5. doi: 10.1210/jc.2012-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moura MM, Cavaco BM, Pinto AE, Leite V. High prevalence of RAS mutations in RET-negative sporadic medullary thyroid carcinomas. J Clin Endocrinol Metab. 2011;96:E863–8. doi: 10.1210/jc.2010-1921. [DOI] [PubMed] [Google Scholar]
- 39.Guan H, Ji M, Hou P, Liu Z, Wang C, Shan Z, Teng W, Xing M. Hypermethylation of the DNA mismatch repair gene hMLH1 and its association with lymph node metastasis and T1799A BRAF mutation in patients with papillary thyroid cancer. Cancer. 2008;113:247–255. doi: 10.1002/cncr.23548. [DOI] [PubMed] [Google Scholar]


