Abstract
Given the rapid advances in genomics, translating new genomic tests effectively into prenatal clinical practice remains challenging. We discuss emerging genetic tests, considerations for their use, how tests should ideally be validated prior to use in clinical practice, and the role of the Federal Drug Administration, Clinical Laboratory Improvement Amendments (CLIA) laboratories, commercial laboratories, insurers, and professional societies such as the American College of Obstetricians and Gynecologists (ACOG), and the Society for Maternal-Fetal Medicine (SMFM) in the introduction of new prenatal genetic tests. After the introduction of new tests into the prenatal clinic, it is critical to utilize shared databases with measured outcomes to improve clinical care as well as to advance science.
Keywords: prenatal, genetics, implementation, sequencing
Introduction
Fetal genetic testing by karyotype and chromosomal microarray analysis (CMA) using amniocytes or chorionic villi provides a diagnosis in only ~10% of pregnancies with suspected fetal genetic disease.1,2 Other technologies, such as whole genome sequencing (WGS) or whole exome sequencing (WES) can be performed on amniocytes and chorionic villi obtained via amniocentesis or chorionic villus sampling. Case reports and “proof of concept” reports have been published showing the ability to interrogate the fetal genome via cell-free DNA found in maternal plasma.3 Despite these advances, significant gaps in our knowledge regarding fetal genetic diagnosis remain and new tests are rapidly being developed and implemented into clinical practice to fill these gaps. As new tests are offered to obstetrical providers and to pregnant women, it is essential that efforts are made to educate patients and providers about best practices by incorporating available peer reviewed data and society recommendations to ensure that new tests are appropriately implemented into clinical practice.
What new tests are coming and what are the considerations for prenatal use?
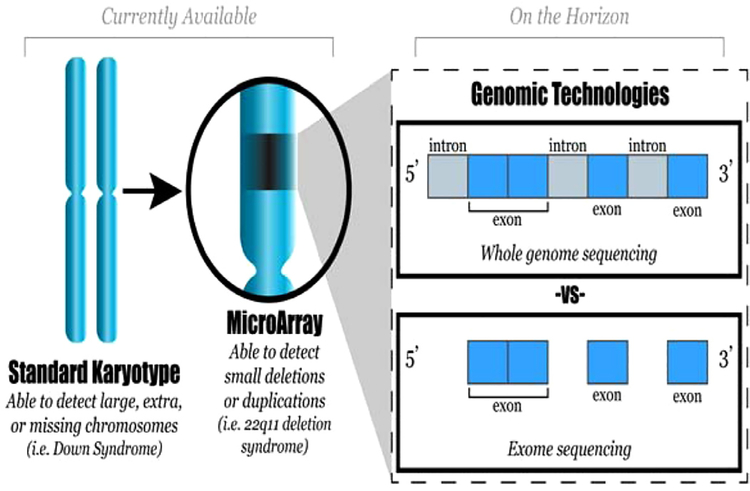
Genomic tests utilizing next generation sequencing technology can be performed directly on fetal samples obtained via amniocentesis or chorionic villus sampling. Current standard genetic testing includes karyotype and CMA. Whole exome sequencing (sequencing the exons or protein coding regions of the genome) is beginning to find a niche in clinical adult and pediatric medicine (Fig. 1). Limited case reports and case series regarding the use of WES prenatally are available and more studies are finding that WES can identify molecular etiologies in fetuses with multiple anomalies that do not receive a diagnosis with standard genetic testing.4–7 ACOG recommends that WES not be used routinely but states there are some clinical circumstances in which it may be appropriate to perform WES.8
Fig. 1 -.
Diagnostic capability of prenatal genetic tests.
Although there have been reports of prenatal use of whole genome sequencing (sequencing the entire genome, including the exons as well as the introns), use of WGS is limited by cost and the difficulty with interpreting intronic areas. Other tests under investigation make use of transcriptomics and epigenomics to shed light on multifactorial disorders, and to better understand the interaction of genetics and environmental exposures and, for example, why fetuses of women with poorly controlled diabetes are at higher risk of congenital anomalies. However, such tests are currently under investigation and not available for clinical prediction or use.
Cell-free DNA (cfDNA) screening for aneuploidy, introduced in 2011, has high sensitivity and specificity for common trisomies and has rapidly been integrated into clinical practice.9,10 Expansion of non-invasive cell free DNA based screening for copy number variants, such as 22q11.2 deletion syndrome (also known as velocardiofacial syndrome), has been limited by low positive predictive value (less than 10%) and small studies that generally have made use of in vitro generated samples.11–18 One study showed high sensitivity and specificity for genome wide analysis of copy number variants but this study was limited by small size and low PPV for these rare conditions.19 ACOG and SMFM does not advocate use of noninvasive cell free DNA screening for microdeletion syndromes, duplication syndromes, and genome wide copy number variants given limitations in available data on clinical performance, as well as a low positive predictive value, and significant concerns for high false positive and false negative rates.
Cell free DNA screening for single gene Mendelian disorders is just becoming clinically available, and although prospective clinical data are not yet available, proof of principle has been published regarding the possibility of detecting single gene disorders noninvasively.20,21 Techniques for analysis of cell-free DNA of single gene disorders include digital PCR, next-generation sequencing and relative haplotype dosage. Complexities of the technology, cost, and the need for validation samples for rare disorders have slowed development of the use of cell-free DNA for single gene disorders.22 Currently, these tests are limited by cost, low sensitivity and specificity, and the need for confirmatory diagnostic testing. Thus, the role for use of cell free DNA screening for single gene disorders is unclear at this time. In certain populations, such as, the Amish, who typically decline diagnostic tests and are at risk for autosomal recessive conditions that may benefit from immediate treatment in the neonatal period, such screening may ultimately found to be useful. However, broad population based application either for targeted testing or for universal screening using cell free DNA would have minimal advantage over diagnostic testing given the exceedingly low loss rate of amniocentesis and chorionic villus sampling.
Noninvasive WES and WGS have been reported, but at present there are multiple limitations including the cost of sequencing, the limitations of using cell free placental DNA, and the need for paternal blood draw to infer the fetal genome.23,24 There are numerous ethical and social implications of offering whole genome or whole exome screening noninvasively, which would likely lead to maternal anxiety especially if it is only a screening and not a diagnostic test. Until fetal variants are curated and we can categorize variants into known pathogenic and known benign, there does not seem to be a foreseeable future for introduction of noninvasive WES or WGS to prenatal practice.
Other tests that have been explored are utilizing the rare, much sought after, and promising intact fetal cell that can be found in maternal circulation. Intact fetal cells are not fragmented, represent pure fetal material, and can be analyzed using next generation sequencing technologies with higher sensitivity and specificity than cell-free placental DNA given that most of the utilized cells directly represent the fetus and not the placenta. However, to date, this technology is limited by the challenges of isolating fetal cells in sufficient quality and quantity to perform diagnostic testing. Most recently, trophoblast cells were identified via endocervical cells obtained via standard speculum exam with cytobrush and are being investigated as a possible additional option for noninvasive prenatal diagnosis.25,26
What are the requirements for test validation before introduction for prenatal use?
Standard components of test validation are needed before introduction into prenatal practice; these components include examination of analytical validity, clinical validity, and clinical utility. Thus, compliance with standards and guidelines for clinical genetics laboratories is recommended. These should include quality control and proficiency testing requirements similar to those used in clinical molecular laboratory tests. For instance, labs performing cell free DNA screening for aneuploidy should disclose assay validation, performance specifications for aneuploidy detection, risk interpretation in result reporting, and should make it clear on the report that confirmatory diagnostic testing is recommended for any positive results. Given that cell free DNA technologies make use of bioinformatics protocols that are proprietary, it is difficult to perform the much needed comparative effectiveness studies to determine the performance of different algorithms.27
What are the roles of commercial laboratories, insurers, and professional societies in the appropriate introduction of new tests?
Before any new tests are offered, large scale validation trials should be conducted followed by peer-reviewed publications demonstrating high sensitivity, specificity, and positive and negative predictive value. Currently available tests for sex chromosome abnormalities and microdeletion syndromes using cell free DNA did not undergo large-scale studies and the positive predictive value of these tests are low. As the tested conditions become rarer, large clinical validation studies become less feasible. Noninvasive prenatal screening and testing will inevitably expand; this will be driven in part by for-profit companies. Payers should systematically monitor society guidelines and track which tests are ordered as well as the outcomes of the testing, especially the costs of follow up testing due to false positive results.28 Professional and academic societies should make every effort to gather and systematically analyze data in order to provide optimal guidance for introduction of new tests. In addition, professional societies should work together and publish joint statements showing consensus rather than separate conflicting statements about best practices.
Should shared databases be required or recommended as tests are introduced?
Companies should voluntarily deposit data about results in a public database. This will enable the science to move forward at a faster pace. A research priority should be to obtain funding for coordination of a central data repository with clinical outcomes for clinically available tests.
Conclusions
The rapid pace at which genomic tests are being introduced into clinical practice necessitates close communication and collaboration between research laboratories, commercial laboratories, professional societies and clinicians. Every effort should be made to coordinate these groups to improve both the science and care of women.
Acknowledgments
Grant funding
Supported in part by NICHD BIRCWH award: 2K12HD00144116.
Footnotes
Disclosures
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Shaffer LG, Rosenfeld JA, Dabell MP, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenatal Diagnos. 2012;32(10):986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hillman SC, McMullan DJ, Hall G, et al. Use of prenatal chromosomal microarray: prospective cohort study and systematic review and meta-analysis. Ultrasound Obstet Gynecol Off J Int Soc Ultrasound Obstet Gynecol. 2013;41(6):610–620. [DOI] [PubMed] [Google Scholar]
- 3.Talkowski ME, Ordulu Z, Pillalamarri V, et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N. Engl J Med 2012;367(23):2226–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filges I, Nosova E, Bruder E, et al. Exome sequencing identifies mutations in KIF14 as a novel cause of an autosomal recessive lethal fetal ciliopathy phenotype. Clin Genet. 2014;86(3): 220–228. [DOI] [PubMed] [Google Scholar]
- 5.Hillman SC, Willams D, Carss KJ, McMullan DJ, Hurles ME, Kilby MD. Prenatal exome sequencing for fetuses with structural abnormalities: the next step. Ultrasound Obstet Gynecol Off J Int Soc Ultrasound Obstet Gynecol. 2015;45(1):4–9. [DOI] [PubMed] [Google Scholar]
- 6.Drury S, Williams H, Trump N, et al. Exome sequencing for prenatal diagnosis of fetuses with sonographic abnormalities. Prenat Diagn. 2015;35(10):1010–1017. [DOI] [PubMed] [Google Scholar]
- 7.Alamillo CL, Powis Z, Farwell K, et al. Exome sequencing positively identified relevant alterations in more than half of cases with an indication of prenatal ultrasound anomalies. Prenat Diagn. 2015;35(11):1073–1078. [DOI] [PubMed] [Google Scholar]
- 8.Vora NLRS, Ralston SJ, Dugoff L, Kuller JA. Microarrays and next-generation sequencing technology: the use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstet Gynecol. 2016;128(6):e262–e268. [DOI] [PubMed] [Google Scholar]
- 9.Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med Off J Am Coll Med Genet. 2011;13(11):913–920. [DOI] [PubMed] [Google Scholar]
- 10.Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. May 2012;119(5):890–901. [DOI] [PubMed] [Google Scholar]
- 11.Peters D, Chu T, Yatsenko SA, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. New Engl J Med. 2011;365(19):1847–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen TJ, Dzakula Z, Deciu C, van den Boom D, Ehrich M. Detection of microdeletion 22q11.2 in a fetus by next-generation sequencing of maternal plasma. Clin Chem. 2012;58(7):1148–1151. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Lau TK, Zhang C, et al. A method for noninvasive detection of fetal large deletions/duplications by low coverage massively parallel sequencing. Prenat Diagn. 2013;33(6):584–590. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013;92(2):167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vora NL, O’Brien BM. Noninvasive prenatal testing for micro-deletion syndromes and expanded trisomies: proceed with caution. Obstet Gynecol. 2014;123(5):1097–1099. [DOI] [PubMed] [Google Scholar]
- 16.Valderramos SG, Rao RR, Scibetta EW, Silverman NS, Han CS, Platt LD. Cell-free DNA screening in clinical practice: abnormal autosomal aneuploidy and microdeletion results. Am J Obstet Gynecol. November 2016;215(5): 626 e621–626 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wapner RJ, Babiarz JE, Levy B, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 2015;212(3): 332 e331–339. [DOI] [PubMed] [Google Scholar]
- 18.Yaron Y, Jani J, Schmid M, Oepkes D. Current Status of Testing for Microdeletion Syndromes and Rare Autosomal Trisomies Using Cell-Free DNA Technology. Obstet Gynecol. 2015;126(5):1095–1099. [DOI] [PubMed] [Google Scholar]
- 19.Lefkowitz RB, Tynan JA, Liu T, et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. Am J Obstet Gynecol. 2016;215(2): 227 e221–227 e216. [DOI] [PubMed] [Google Scholar]
- 20.Camunas-Soler J, Lee H, Hudgins L, et al. Noninvasive prenatal diagnosis of single-gene disorders by se if droplet digital PCR. Clin Chem. 2018;64:336–345. [DOI] [PubMed] [Google Scholar]
- 21.Vermeulen C, Geeven G, de Wit E, et al. Sensitive monogenic noninvasive prenatal diagnosis by targeted haplotyping. Am J Hum Genet. 2017;101:326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen S, Young E, Bowns B. Noninvasive prenatal diagnosis for single gene disorders. Curr Opin Obstetr Gynecol. April 2017;29(2):73–79. [DOI] [PubMed] [Google Scholar]
- 23.Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan HC, Gu W, Wang J, et al. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487:320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolnick AD, Fritz R, Jain C, et al. Trophoblast retrieval and isolation from the cervix for noninvasive, first trimester, fetal gender determination in a carrier of congenital adrenal hyperplasia. Reprod Sci. 2016;23(6):717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolnick JM, Kilburn BA, Bajpayee S, et al. Trophoblast retrieval and isolation from the cervix (TRIC) for noninvasive prenatal screening at 5 to 20 weeks of gestation. Fertil Steril. 2014;102(1):135–142: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregg AR, Skotko BG, Benkendorf JL, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med Off J Am Coll Med Genet. 2016;18(10):1056–1065. [DOI] [PubMed] [Google Scholar]
- 28.Allyse M, Chandrasekharan S. Too much, too soon?: Commercial provision of noninvasive prenatal screening for subchromosomal abnormalities and beyond. Genet Med Off J Am Coll Med Genet. 2015;17(12):958–961. [DOI] [PMC free article] [PubMed] [Google Scholar]