Abstract
Given the alarming rise in instances of antibiotic resistance displayed by disease-causing microorganisms, it is necessary to accelerate efforts to find new antibiotic agents. One prominent approach is to identify potent inhibitors of receptors that are indispensable for the microorganism’s survival. Dihydrofolate reductase, DHFR, is one such target1,2 in the gram negative bacteria Escherichia coli that is indispensable for the microorganism’s survival3,4. Traditional drug discovery approaches rely exclusively on quantitative structure activity relationships based elaboration of core scaffolds to discover new and potent inhibitors for this enzyme. However, the advent of Next Generation Virtual Ligand Screening methodologies that rely on evolution-based ligand-binding information, which utilize the principles of both structure-based and ligand similarity-based approaches, have significantly changed the pace with which new inhibitors have been discovered for E. coli DHFR (EcDHFR). Moreover, while efforts at targeting alternative pockets to overcome drug-resistant variants of the enzyme have failed miserably in the past, recent work has been very promising. This review summarizes recent efforts at the effective interfacing of computational and experimental efforts to discover novel classes of inhibitors against both drug-sensitive and drug-resistant variants of EcDHFR. Furthermore, we posit that targeting multiple pockets on an enzyme by both active-site and alternative-site binding inhibitors has the potential to significantly overcome drug resistance in target enzymes.
Keywords: Drug-resistance, virtual ligand screening, Dihydrofolate reductase, Escherichia coli, QSAR
2.1. Escherichia coli and prominent protein targets currently exploited by antibiotics
Rapid acquisition of resistance to available antibiotics in Gram negative bacteria is an increasing cause for concern for human health5. The evolutionary selection pressure exerted by widespread antibiotic abuse has contributed to the selection of drug-resistant variants of target enzymes, which, in turn, have contributed to the drug-resistant phenotype displayed by the organisms. However, the trend has been alarming in recent years with multiple deaths attributed to drug resistant bacterial strains6.
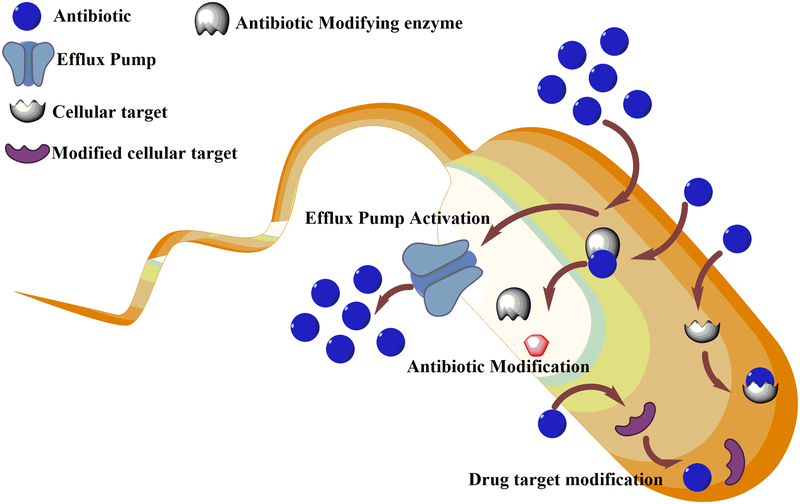
Escherichia coli is a normal commensal of the human colon. However, in immunocompromised hosts, certain strains are known to cause infections like gastroenteritis, peritonitis, thrombocytopenia, septicemia, bloody diarrhea and hemolytic uremic syndrome (HUS)7. E. coli acquires drug resistance in the shortest possible time span by means of several different mechanisms8. Prominent among these are mutations and modulation of drug efflux pumps, acquisition of plasmids encoding antibiotic-resistance genes and acquisition of mutations in a biological target making it refractory to the action of the drug9 (see Fig 1). A previous study has presented statistics showing that among antibiotics against gram-positive bacteria, approximately 90% showed no cytotoxicity for Escherichia coli10. Given the alarming rise in instances of hospital-acquired infections caused by drug-resistant gram-negative bacteria11, it becomes imperative to search for novel targets for antibiotic agents against these organisms.
Figure 1.
Schematic view depicting the mechanisms by which bacteria acquire antibiotic resistance.
The most common targets that have been exploited to date are those that interfere with either DNA replication, transcription or translation, those that interfere with peptidoglycan synthesis and those that alter the microbial cytoplasmic membrane by causing increased permeability12. The fluoroquinolone class of antibiotics blocks topoisomerases and gyrases that are pivotal for DNA replication13, while rifampicin binds to bacterial RNA polymerase and blocks transcription14. However, one of the most prominent class of antibiotics are those that block translation by either binding to the 30S ribosomal subunit and out compete tRNA binding (aminoglycosides and tetracyclins)15, or the 50S ribosomal subunit and obstruct the exit tunnel (macrolide antibiotics, e.g., Erythromycin)16. Peptidoglycan interfering antibiotics operate by either blocking the transport of peptidoglycan monomers across the cytoplasmic membrane, or inhibiting transpeptidases and transglycosidases that are pivotal for formation of peptide cross-links or formation of glycosidic bonds, respectively9. Antibiotics that disrupt bacterial membranes are cationic cyclic peptides with fatty acid chains (Polymixins). They intercalate with the bacterial membrane to modulate its permeability and thereby cause membrane disruption17. However, discovery of additional targets is essential to counteract the increasing drug resistance displayed by several validated targets against which drugs are available.
2.2. Dihydrofolate reductase
Dihydrofolate reductase, DHFR, is an important enzyme in the de novo pathway of purine and thymidine synthesis. Small-molecules targeting this enzyme have demonstrated utility as potential antibiotics2. There are two main variants of EcDHFR that are either chromosomally-encoded or plasmid-encoded. Plasmid-encoded Type II R67 EcDHFR from trimethoprim-resistant bacteria are especially interesting since they are genetically unrelated to chromosomal EcDHFR. R67 EcDHFR is a homotetramer and is structurally distinct, both at the overall protein-fold level and at the active site, from chromosomal EcDHFR. The episomally encoded EcDHFR is also fascinating from the perspective of multiple levels of regulation demonstrating positive cooperativity in binding the substrate dihydrofolate and negative cooperativity in binding the cofactor NADPH18 that could be potentially harnessed in an inhibitor-discovery project. While, it would be desirable to discuss small-molecules that target both chromosomal and plasmid-encoded DHFR from the perspective of antibiotic discovery, the current review exclusively focuses on chromosomal DHFR as a model system. This is because of the availability of a large amount of structural and mechanistic data for the latter and its indispensable nature for the survival of the microorganism, making it an ideal target for drug discovery. The enzyme converts dihydrofolate to tetrahydrofolate by hydride transfer from the cofactor NADPH to the C6 atom of the pterin ring and an additional concomitant protonation at N5. At cellular concentrations of the cofactor and substrate and under steady-state conditions, the catalytic cycle of EcDHFR goes through 5 kinetic intermediates: E: NADPH (holoenzyme), E:DHF:NADPH (Michaelis complex) and E:THF:NADP+, E:THF, and E:THF:NADPH (the product complexes)19. DHFR is the sole source of cellular tetrahydrofolate and thus plays an important role in the maintenance of tetrahydrofolate pools. Tetrahydrofolate is an important precursor of purine and thymidine synthesis, and thus is critical for growth and proliferation of cells. Consequently, targeting DHFR is lethal for rapidly proliferating cells like cancer or bacterial cells. Several classes of compounds have been explored for their potential anti-folate activity, among the most prominent are diaminoquinazoline20, diaminopyrimidine21,22, diaminopteridine23 and diaminotriazines24. DHFR inhibitors that have found widespread application in therapy are methotrexate (used in chemotherapy against cancer cells and rheumatoid arthritis), trimethoprim, (for bacterial DHFR) and pyrimethamine (against Plasmodium falciparum DHFR).
However, in spite of continuous efforts to discover novel small-molecule inhibitors of this enzyme, most studies exclusively rely on QSAR-based elaboration of known antifolates to discover novel small-molecules25. Moreover, the rapid acquisition of drug resistance by the enzyme compounds the challenges associated with drug discovery. It was recently demonstrated that the laboratory based selection for E. coli cells resistant to trimethoprim showed step-wise acquisition of resistance phenotype mainly localized on either the promoter or the substrate binding site of the enzyme DHFR26. Mutations in the DHFR amino-acid coding region were P21L, A26T, A26V, A26S, L28R, W30C, W30G, W30R and I94L, respectively and have been either shown or predicted to affect DHFR enzymatic activity26. Three of these mutations (c-35t, P21L and W30R) have also been reported from clinical isolates26, four (P21L, A26T, W30R and I94L) have been reported in laboratory selection and four (L28R and W30C in the coding region and −35C>T and −9G>A in promoter) appeared in independent selection experiments performed on agar plates. Furthermore, there has been extensive documentation of mutations in DHFR leading to drug resistance in pathogenic organisms like Plasmodium falciparum, Streptococcus pneumoniae, etc27–29. Hence, it is necessary to keep discovering novel small molecule inhibitors for this enzyme.
2.3. Next generation fold-based and pocket-based virtual ligand screening
In virtual ligand screening (VLS), computer algorithms predict the likelihood of a particular small-molecule interacting with the protein target of interest30. Subsequently, these predictions are assessed by high-throughput experimental screening of the predicted ligands for binding to their protein target. Thus, VLS can reduce the number and kinds of molecules that have to be screened experimentally, thereby saving both time and cost. Recently, the introduction of various statistical, filtering and informatics protocols has fostered the efficient integration of experimental and in silico screening methodologies resulting in enhancing their importance in drug discovery31.
There are two distinct types of virtual ligand screening protocols to identify potential lead molecules: structure-based and ligand-based virtual screening (VS). Traditional structure-based approaches rely on the presence of a high-resolution structure for the target protein32. Then, molecular docking of the ligand to the protein target is often employed as it does not require a priori knowledge of known binders33. Furthermore, they can also target a specific binding pocket of interest. In practice, molecular docking employs empirical force fields to compute the free energy of interaction of the small-molecule with its protein target. However, to a large extent, the accuracy of its predictions depend upon the quality of the receptor’s structure, accessory information about its dynamics34 and the availability of a uniform high-quality validation set35. It has been demonstrated that docking accuracy may be reduced by almost 90 % if the structure employed has a root-mean square deviation of greater than ~1.5 Å from the native state36,37. Thus, it is very sensitive to rather minor structural distortions. Furthermore, the reliability of the docked poses depends upon aspects like water molecule locations, the small-molecule conformational ensemble and the accuracy of the force-field38. A variant of structure-based VLS is fragment based drug discovery whereby weakly binding fragments to the protein target of interest are fused together in silico in order to arrive at a lead molecule for subsequent organic synthesis and assessment39–42. However, implementing in silico approaches to fragment discovery has remained challenging because of the low binding affinity of the fragments and the inability of existing force fields to differentiate binders from non-binders43. In some cases, the absence of high-resolution structures has been compensated for by the use of homology models that have been refined and manually cross-checked for accuracy44–46. Some examples of structure-based VS approaches include AutoDock, Dock, FlexX, Glide, Gold Surflex, ICM, LigandFit, and eHiTS33.
As pointed out above, the major rate-limiting step associated with structure-based drug discovery is the presence of either a high resolution protein structure or a confidently predicted protein model. However, not all protein targets are amenable to x-ray crystallography-based structure determination or high resolution structure prediction (due to the lack of appropriate template structures). The problem becomes all the more acute for membrane anchored proteins or large macromolecular complexes. To circumvent this limitation, ligand-based VS approaches have been developed47. Though ligand-based VS approaches are robust, they require at least one known small-molecule compound that binds to the protein target of interest. These methods focus exclusively on the comparative molecular similarity analysis of the ligand demonstrated to bind to a particular protein target with molecules in a database. Some ligand-based VS approaches rely on Tanimoto coefficient 2-D fingerprint48, pharmacophore49, or 3-D based shape similarities50 between the known binder and database molecules. Ligand-based VS does not provide information about the site of binding in the protein and requires an experimentally determined bioactive compound. Thus, it is clear that both traditional structure-based and ligand-based VS, though advantageous, possess their fair share of limitations, especially vis-à-vis therapeutically relevant proteins, many of which are either membrane proteins lacking substantial structural information51–53 or lacking known binding ligands.
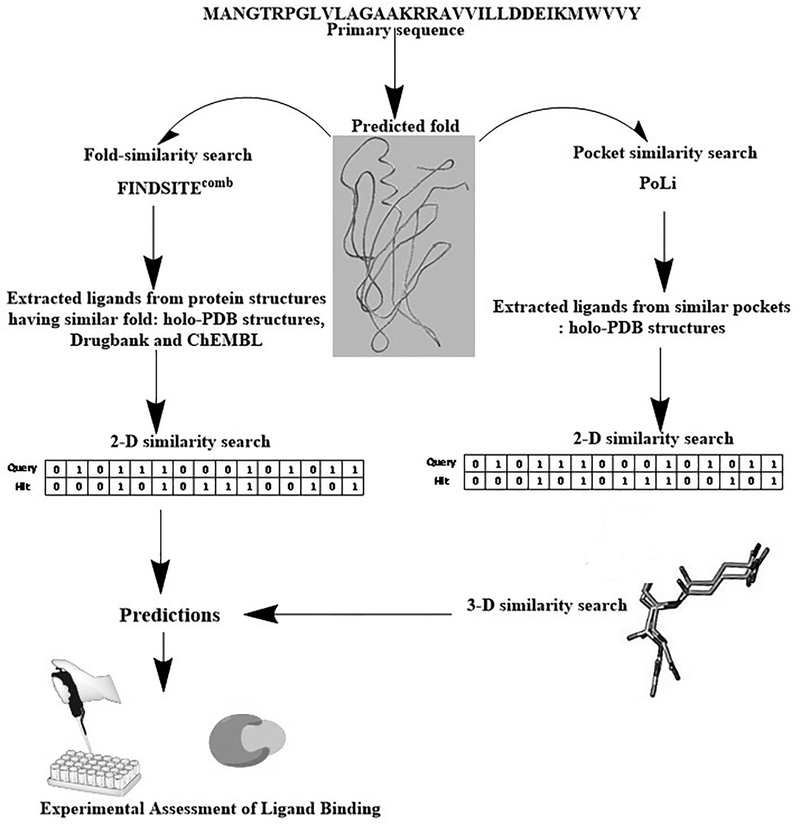
To overcome the limitations of the above two classical approaches, hybrid methods that rely on structural and ligand similarity combined with evolution-based ligand binding information have been pioneered by our group54–56 (Fig. 2). These approaches encompass both global structural similarity and pocket similarity. The first approach is called FINDSITEfilt 54 and can use either an experimental structure or a low-resolution predicted protein structure to find similar template proteins in the PDB holo-template library (PDB holo-templates are protein structures bound to either their prosthetic groups, to the substrate/product and/or their respective analogues or inhibitors). Subsequently, it employs 2D fingerprint similarity to screen for database molecules that are similar to template ligands excised from the selected holo-structures. FINDSITEcomb extends FINDSITEfilt for proteins having holo protein template structures to target proteins without holo-templates, by generating an artificial library of predicted holo-structures using known template ligand binding information from the ChEMBL57 and DrugBank58 databases (Fig. 2). Since predicted models can be employed with minor diminution in accuracy, these methods neither need high-resolution structures nor known binders. These methods also possess the advantage of speed and are capable of predicting diverse small-molecular structural scaffolds as compared to conventional structure and ligand-based methods or traditional quantitative structure-activity relationship based approaches. In practice, the predictions from FINDSITEcomb have been experimentally assessed on a significant number of medically relevant target proteins belonging to different fold-classes and coming from several different organisms55 and achieves good enrichment in identifying active small-molecules. The methods predicted low nanomolar binders for the enzyme dihydrofolate reductase from Escherichia coli and also predicted micromolar binders for several different protein targets such as the phosphatase domain of protein tyrosine phosphatase delta (from rat) and omega (from Homo sapiens), tryptophanyl tRNA synthetase from H. sapiens, ubiquitin-conjugating enzyme from P. falciparum, nucleosome assembly protein 1 from P. knowlesi, thioredoxin peroxidase 2 from P. falciparum, the catalytic domain of cAMP dependent protein kinase from H. sapiens55 and N-glycanase 159.
Figure 2.
Schematic representation of the virtual ligand screening pipelines discussed in this review.
However, the above approaches, though successful at predicting small-molecule binders for several different medically relevant targets55, are not capable of a priori selecting a particular ligand binding site in the protein of interest. Rather, whatever binding sites are occupied by template ligands in the PDB are then inferred to bind to the target protein in a similar pocket. Moreover, they are constrained by target similarity at the global fold level, rely on ligand similarity at the 2D-level and cannot recognize the stereochemical similarity of ligands that adopt a similar geometric shape with similar functional groups located at equivalent positions when the functional moieties of the ligands are substantially different.
To address these issues, PoLi, a new pocket centric approach capable of targeting specific binding pockets in holo-protein templates, was developed60 (Fig. 2). This method takes advantage of our recent demonstration that the number of stereochemically distinct ligand binding pockets is small and likely complete61,62. PoLi can target specific ligand binding pockets in the target protein, does not rely on the similarity between the template and the target at the global fold-level and implements both 2-D and 3-D small-molecule similarity approaches to identify ligands from holo-templates60. More specifically, the method models the target protein, predicts their ligand-binding pockets, aligns the predicted pockets to database of holo-pockets, copies and prunes the ligands from the holo-pocket to weight the binding pharmacophore, and then undertakes ligand-based VS approaches with both 2D and 3D similarity metrics to come up with a ranked prediction for experimental assessment. This method was benchmarked extensively in silico followed by high-throughput experimental validation on EcDHFR. As expected, the experimentally obtained hits not only belonged to those that were already obtained by FINDSITEcomb but also included ligands excised from evolutionarily and structurally unrelated protein scaffolds60. Finally, apart from these above mentioned approaches, we have also successfully developed a new iterative combined pocket detection with an interaction-weighted ligand-similarity search-based approach to obtain high affinity binders for the olfactomedin domain of human myocilin implicated in glaucoma63.
2.4. Classification of DHFR Inhibitors
2.4.1. Substrate and Cofactor Analogues
Conventional classes of DHFR inhibitors are heterocyclic with one to three nitrogen atoms in the ring and two amino groups arranged in the para position. They are mostly analogues of the substrate dihydrofolate64 and all have been characterized for their potency of DHFR inhibition4,20,24,55,60. A few classes have been kinetically characterized in detail to understand their site and order of binding20,24. Results from these studies indicate that all bind to the dihydrofolate binding site in the enzyme, and hence, competitively displace the substrate. Unlike the substrate dihydrofolate that can bind to the enzyme with or without the cofactor NADPH, the binding of these inhibitors is ordered in nature and conditional upon NADPH binding. As mentioned earlier, prominent classes of conventional DHFR inhibitors are diaminotriazines, diaminopteridines, diaminoquinazolines and diaminopyrroloquinazolines20,24,55,60.
Analogues of the NADPH cofactor have also been explored as potential inhibitors of DHFR65. Pyridine nucleotides NADP, NHDP, ε-NADP, APADP and NAD function as analogues of NADPH and inhibit the enzyme in a linear competitive fashion vis-à-vis NADPH and linear noncompetitive fashion vis-a-vis dihydrofolate, DHF65. However, the reduced and oxidized forms of thionicotinamide adenine dinucleotide phosphate inhibit the enzyme such that it shows linear noncompetitive inhibition with respect to both NADPH and DHF. Furthermore, adenosine 5’-phosphate, adenosine 2’-phosphate, ADP-ribose and NAD all preferably bind to the free enzyme to form the E.I binary complex compared to their affinity for the Enzyme-DHFR complex. Analogues such as adenosine 2’,5’-diphosphate, ATP-ribose, APADP, NHDP, ε-NADP and NADP show increased affinity for the enzyme-dihydrofolate form to make the ternary complex over the free enzyme. However, NADPH analogues may give rise to extensive cross-reactivity with other NADPH binding proteins and proteins containing nucleotide-binding pockets, and hence, they might not be appropriate for clinical applications. Interestingly, it has also been demonstrated that inhibition of NAD kinase by thionicotinamide adenine dinucleotide phosphate (NADPS) led to cofactor NADPH depletion which, in turn, led to DHFR degradation in neoplastic cells66. This seems to be a novel route to inhibit DHFR and could be tested in bacteria provided that the bacterial homologue is as unstable as the mammalian one in the absence of the cofactor.
2.4.2. Classical and Non-Classical inhibitors
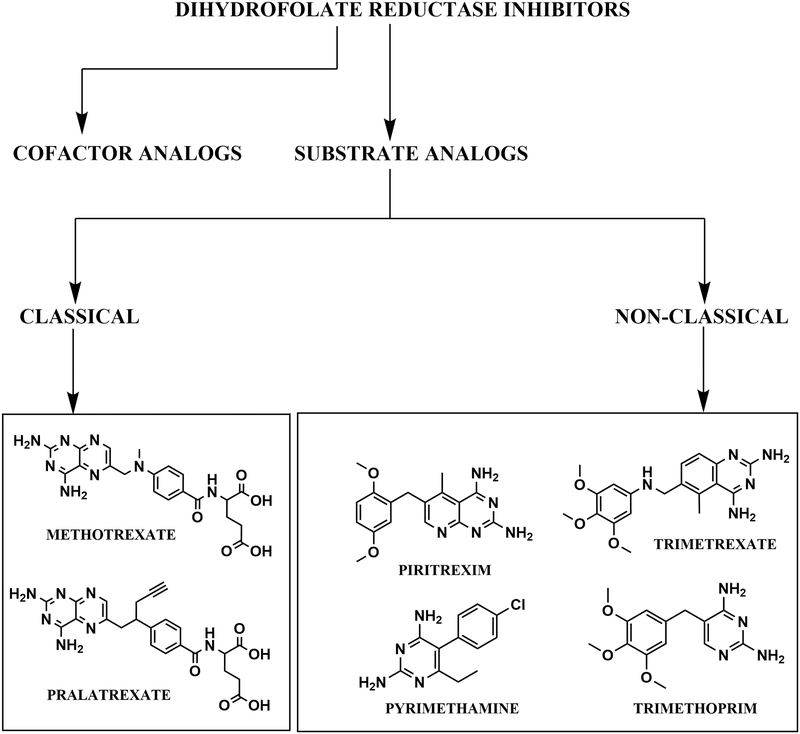
Yet another classification of DHFR inhibitors is based on their chemical structures. According to structural differences, inhibitors have been classified as either classical or non-classical67 (Fig. 3). Classical inhibitors are folate analogues that possess a heterocyclic ring (most often a pteridine) that is linked to an aryl group and a glutamate tail. For this group to inhibit DHFR, they need to be imported into the cell through folate transporters (RFC-1’s in Eukaryotes68) and need to be polyglutamylated by folylpolyglutamyl synthetase (FPS)69,70. E. coli possesses an FPS analogue (folC) that functions both in polyglutamylation of tetrahydrofolate (or its analogues) and in the synthesis of dihydrofolate by addition of a glutamate residue to dihydropteroate71. However, E. coli has a de novo folate synthesis pathway and lacks a functional homologue of the RFC-1 transporter. Recently, it has been demonstrated that the gram negative bacteria have abgT transporters that can uptake p-aminobenzoate to facilitate the biosynthesis of folate within cells72,73 . It can be speculated that the abgT transporters can import folate analogues within bacterial cells. However, we have not come across any examples in the literature supporting this conviction (see section 2.4.3). Some examples of classical folate inhibitors include methotrexate (MTX) and pralatrexate (PDX).
Figure 3.
Classification of DHFR inhibitors based on their site of binding and chemical structure.
On the contrary, non-classical inhibitors do not possess the glutamate tail. This confers both desirable and undesirable properties on them to be employed as drugs. The desirable properties include uptake by passive diffusion through the membrane, and thus, they do not require any transporters. However, the undesirable effects include reduced water solubility, and, not being the substrate for FPS, they are incapable of being polyglutamylated, resulting in reduced retention inside the cell subsequent to uptake. A few examples of this class of inhibitors include trimethoprim (TMP), pyrimethamine (PYR), trimetrexate (TMQ) and piritrexim (PTX) (Fig. 3).
2.4.3. Membrane permeability as a factor in DHFR inhibitor discovery
Antibiotic development faces two fold challenges in terms of affinity of the small-molecule for its intended target to bring about inhibition and bioavailability. The latter indicates the amount of the small-molecule that can cross the cytoplasmic membrane and outer membrane (in the case of Gram negative bacteria), either actively or passively, for it to be available to interact with its target74. With regard to uptake, aqueous porins on the outer membrane of Gram negative bacteria facilitate the passive uptake of selected small molecules that are subsequently taken up by the cells (traversing the inner membrane) by either passive diffusion or active transport. This process of uptake should essentially be faster than possible efflux mechanisms operational at any given time74. The success or failure of an antibiotic-discovery initiative, to a large extent, depends on determining the membrane permeability of the small-molecule apart from studies that throw light on its interaction with the target of interest. A judicious combination of the above two factors in determining the structure-activity relationship based medicinal chemistry synthesis of derivatives is ideal for successful antibiotic discovery.
EcDHFR is a cytosolic enzyme. As such, the high negative charge on classical DHFR inhibitors at physiological pH makes them unsuitable for passive diffusion through the membrane, thereby making bioavailability issues for this class of inhibitors a major issue. As briefly discussed in section 2.4.2, this problem is compounded by a lack of conventional folic acid transporters due to the reliance of E. coli on de novo folate synthesis. Thus, classical DHFR inhibitors like methotrexate (MTX), which is extensively used in chemotherapy in mammalian cells, have a poor MIC above 1 mM for E. coli despite of their high affinity for the purified bacterial enzyme. Mutations in acrA or tolC, resulting in inactivation of the TolC-dependent AcrAB multidrug resistance efflux pump, result in an approximately 10 fold reduction in MIC (the MIC drops from 1 mM to about 0.064 mM) indicating that efflux plays a major part in the methotrexate resistance of E. coli75. Having said that, the MIC still does not correlate well with the low nanomolar affinity of MTX for the in vitro enzyme.
On the contrary, non-classical inhibitors can diffuse passively through cell membranes. A prominent example includes trimethoprim, which is weakly basic at physiological pH and shows potent cytotoxicity for E. coli (Its MIC is approximately 6.9 μM76) in spite of showing poorer affinity (vis-à-vis methotrexate) for the recombinantly expressed bacterial enzyme (IC50 is ~20.4 ± 2.3 nM77).
Recently, it has also been demonstrated that zwitterionic compounds such as propargyl-linked antifolates are DHFR inhibitors of Gram-negative bacteria and can diffuse passively across the cell membrane. These hybrid antifolates, according to the authors, conserve the features made by negatively charged glutamate tails while being permeable across the bacterial cell membrane78.
Our studies that employ fold-based hybrid virtual ligand screening approaches (section 2.5.) were successful in picking analogues with both acidic tails and no-tails20,24,55,79. However, the best inhibitors had long tails with localized negative charges. These group of inhibitors, we posit, would face the same kind of troubles discussed above in terms of cell permeability. For instance, the top nine best compounds tested from our studies (along with appropriate positive and negative controls) against a panel of seven organisms belonging to the gram positive, gram negative and yeast cells returned non-significant inhibition at a concentration of 20 μM (unpublished results). However, our pocket based approaches (section 2.6), apart from picking the classical and non-classical inhibitors60,80,81, were also successful in predicting a unique set of inhibitors targeting a novel allosteric pocket. These compounds had MIC values very similar to their IC50 values for the enzyme strongly suggesting that they can efficiently diffuse across gram negative bacterial cell membranes80,81.
2.5. Conventional classes of inhibitors targeting the active site of DHFR: Capturing novel analogues of known DHFR inhibitors by fold-based VLS approach:
2.5.1. The 2,4-diaminopteridine and diaminopyrimidine
templates are known DHFR inhibitors. As pointed out above, the main representative of the family diaminopteridine is methotrexate, a potent inhibitor of DHFR82,83. Our work spanning both fold-based FINDSITEcomb54,55 and pocket-based PoLi60 was successful in recapturing methotrexate. Previously, methotrexate has been shown to be a slow-onset, tight-binding inhibitor of the E. coli enzyme20,83. Inhibition by methotrexate obeys a mechanism where there is a rapid initial formation of an enzyme-NADPH-inhibitor complex followed by its slow isomerization to trap the inhibitor84. Further, it has also been shown that methotrexate binds to the folate binding site (competitive with respect to folate) and its binding on the enzyme is conditional upon NADPH binding85. Trimethoprim, the most successful inhibitor against bacterial DHFR as far as the antibacterial effect is concerned, belongs to the diaminopyrimidine class. Our studies with fold-based virtual ligand screening were always successful in recapturing this group of compounds as positive controls.
2.5.2. 2,4-diaminoquinazoline and diaminopyrroloquinazoline
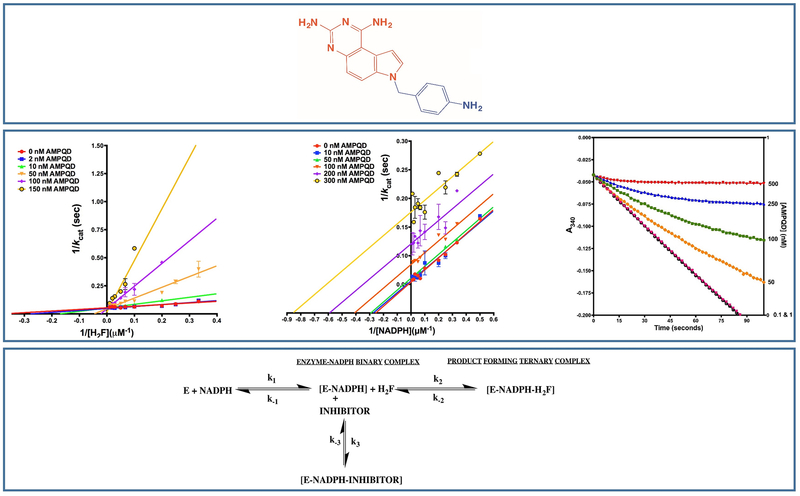
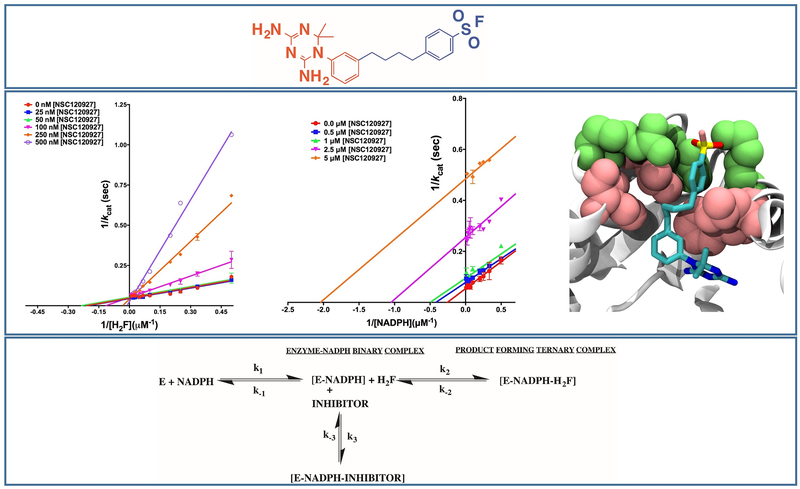
scaffolds are well known for their high potency on bacterial DHFR variants and have been extensively studied as part of several optimization programs86–92. Most of these efforts have focused on 6-substituted derivatives. The best hit from our study is a diaminopyrroloquinazoline, NSC309401, a compound with an aminophenyl methyl substitution on the 7th position of the diaminopyrroloquinazoline ring20,55,79 (Fig. 4). We have carried out the detailed kinetic characterization of diaminopyrroloquinazoline group of compounds vis-à-vis EcDHFR. Our studies show that the presence of aminophenyl methyl substitution in the core diaminopyrroloquinazoline moiety leads to inhibition of EcDHFR by a slow-onset, tight binding mechanism indicating to possible non-equilibrium effects (E.I to E.I*) subsequent to the initial E.I complex formation or to very slow rates of association of or dissociation from E.I* vis-à-vis the first order dissociation rates for EI to E + I (Fig. 4). Diaminopyrroloquinazolines binding to EcDHFR revealed clear non-linearity in reaction progress curves (transitioning from vinitial to vsteady-state) indicating a time-dependent establishment of enzyme-inhibitor equilibrium. The principal advantage of slow onset, tight binding inhibition is that the inhibitor binds with high affinity to the target enzyme and the residence time of the inhibitor on the enzyme is long because of low koff values. An approximate estimate for the drug residence time of NSC309401 on the enzyme was 8.5 minutes (a koff of 0.118 min−1)20 comparable to that of Trimethoprim93. Further, the next best hit, NSC339578, did not show the slow onset behavior20. It should be pointed out here that bulk of the drugs available in market exhibit a slow-onset of inhibition forming a very tight [E.I] complex. This confers the advantage of longer desired inhibition compared to the rate of pharmacokinetic clearance of the compound. One prominent example of slow onset inhibitor is the COX2 inhibitor DuP697.
Figure 4.
Inhibition of EcDHFR by diaminopyrroloquinazoline group of inhibitors. Top most panel shows the structure of the best hit with an aminophenyl methyl substitution. The central panel shows Lineweaver-Burk plots of the inhibitor titrated against substrate and cofactor (leftmost two panels) and slow-onset tight-binding mechanism (rightmost panel). The bottom most panel shows the kinetic scheme for the mechanism of inhibition. Adapted from20
Order of binding studies with respect to substrate and cofactor demonstrated that the inhibitors showed binding to the dihydrofolate binding site conditional upon NADPH binding (Fig. 4). For any small-molecule to be considered as a lead for potential antibiotic development, it is imperative to demonstrate that it possess higher affinity for the bacterial homologue of the enzyme as compared to the human one. The inhibition potency (IC50) of the best hit and the second best hit was approximately 3-fold and 30-fold less for the human homologue vis-à-vis the bacterial variant20.
2.5.3. Diaminotriazines,
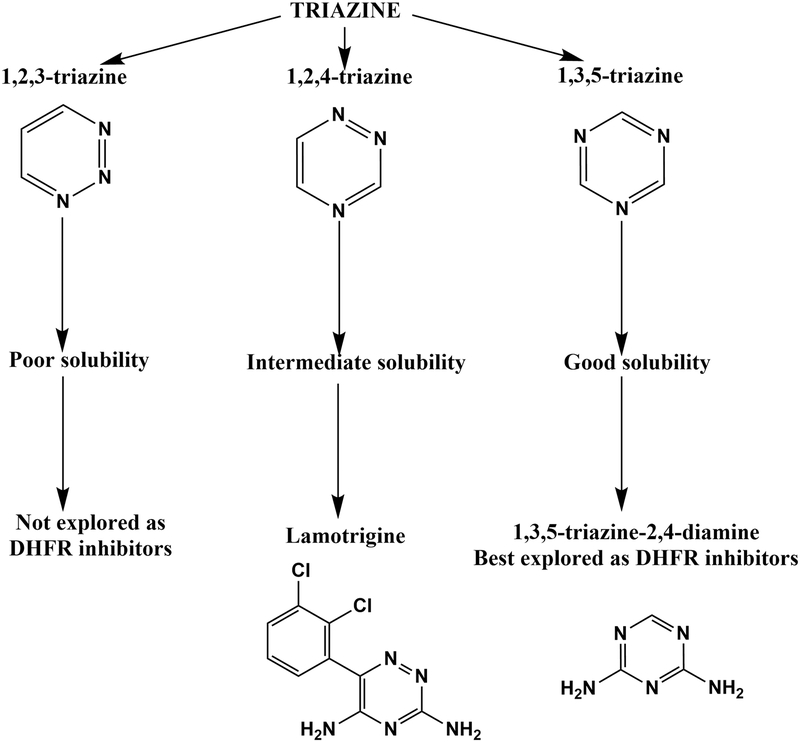
Triazines are organic heterocycles containing nitrogen. This group of compounds is classified into three different types based on the separation of the nitrogen atoms on the ring: 1,2,3-triazines, 1,2,4-triazines and 1,3,5-triazines (Fig. 5). 1,3,5-triazine are the best studied and 1,2,3-triazines compounds are the least studied among the three isomers, respectively (Fig. 5). The latter is because 1,2,3-triazine compounds show poor solubility. 1,2,4-triazines, with intermediate solubility properties, have been reasonably well studied with the most prominent example being lamotrigine, a sodium-channel blocker class of anti-epileptic drug. Compounds containing a 1,2-dihydro substitution on the 1,3,5-triazine 2,4-diamino core have been explored extensively for their potential as inhibitors of eukaryotic DHFRs24,94. Cycloguanil, a derivative of linear aliphatic proguanil, is the most prominent example as it is a potent inhibitor of P. falciparum DHFR95,96. Extensive QSAR analysis of the inhibition by the diaminotriazine series of compounds has been carried out on DHFR analogues from several different organisms24,25,97–103. Finally, hybrids of triazines also demonstrate inhibitory activity on DHFRs104.
Figure 5.
Classification and assessment of triazine compounds as inhibitors of dihydrofolate reductase.
Most studies have focused on understanding the inhibitory effect of diaminotriazines or their hybrids on DHFRs from eukaryotic sources, mainly P. falciparum, since they show poor affinities for the prokaryotic enzyme. To address this issue, we employed systematic QSAR analysis and insights obtained from docking studies to design appropriate inhibitors employing analogs of 1-phenyl-6,6-dimethyl-1,3,5-triazine-2,4-diamine (PDTD), as potent inhibitors of EcDHFR (Fig. 6) Our study was the first attempt at detailed mechanistic characterization of the diaminotriazine family of compounds by inhibition kinetics to assess their effect on EcDHFR24. Fifteen analogs of PDTD showed binding to EcDHFR as assessed by differential scanning fluorimetry, and subsequently showed inhibition of the enzyme. NSC120927 was the best hit obtained from this study of 1,3,5-triazine-2,4-diamine class of molecules and is the first ever to show potent inhibition of a DHFR isoform from gram-negative prokaryotes. We also explored the kinetic mechanism of inhibition by 1,2,4-triazine-3,5-diamines on EcDHFR. Detailed kinetic characterization demonstrated that, like diaminopyrroloquinazolines, this class of compounds also bind to the active site of the enzyme and their binding is conditional upon NADPH binding. However, the best hits obtained from this study failed to show slow-onset of tight binding inhibition indicating that the koff rates are not as slow as for the best hit from the diaminopyrroloquinazoline group of compounds. This is indicative of the short residence time on the enzyme, and hence, would require further organic synthesis efforts to design better inhibitors. Having said that, this study has opened up the possibility of exploring a new class of molecules that could potentially yield novel antibiotic candidates against gram-negative bacteria.
Figure 6.
Inhibition of EcDHFR by the diaminotriazine group of inhibitors. Top most panel shows the structure of the best hit. The central panel shows the Lineweaver-Burk plots of the inhibitor titrated against substrate and cofactor (leftmost two panels) and the docked pose of the small molecule inside the active site cavity of the enzyme (rightmost panel) (Adapted from24). The bottom most panel shows the kinetic scheme for the mechanism of inhibition. Adapted from20 .
2.6. Atypical classes of inhibitors targeting the EcDHFR allosteric site and the pocket based VLS approach: Deoxybenzoin, Stilbene and Chalcones
PoLi, the pocket based virtual ligand screening algorithm, was used to perform virtual ligand screening on EcDHFR. This yielded a set of small molecule predictions that were assessed by high-throughput experimental screening employing differential scanning fluorimetry. Most of the hits belonged to the conventional classes of DHFR inhibitors as elaborated in section 2.5. However, a few weak binders were novel, small-molecule scaffolds with no similarity to known classes of DHFR inhibitors (see next paragraph) and with no previous report of them interacting with DHFR from any organism whatsoever60.
Using conventional QSAR and systematic scaffold hopping, we assessed a series of small-molecule chalcones, stilbenoids, and other chemically similar scaffolds for their EcDHFR binding/ inhibition potential80,81. Six stilbenoid compounds (resveratrol, oxyresveratrol, SITS, DIDS, flavonic acid and DNDS), three chalcone derivatives and ononetin showed binding and inhibition of EcDHFR. This demonstrated that the general requirement for this class of molecules to inhibit EcDHFR involves small molecules possessing 3–4 degrees of freedom connecting the two benzene moieties, with appropriate hydrogen bonding acceptors or donors on the ring80,81. However, no information was available on the site or order of binding for this novel class of inhibitors. To address these questions, we performed detailed competition assays with substrate and inhibitor of the small-molecules. The resultant kinetic patterns demonstrated that the compounds, under concentrations that might be physiologically relevant, showed uncompetitive or linear mixed-type inhibition with respect to substrate dihydrofolate indicating that they do not bind to the substrate binding site as is the case with other EcDHFR inhibitors80 (Fig. 7). Furthermore, in a behavior reminiscent of other inhibitors, their binding is conditional upon NADPH binding. This implies that the inhibitors bind to a unique site distinct from either the substrate or the cofactor binding site, and hence, reports on a cryptic105 site on EcDHFR that is formed in the fully ligated ternary form of the enzyme. It has to be stated here that targeting cryptic sites is projected as one of the main challenges in designing small molecule drugs against target proteins and our demonstration opens up the avenue for discovery of such cryptic sites in other drug targets106. However, we would like to point out that there have been a few previous investigations that have tried to understand allosteric binding in DHFRs. One study, investigating nanobody binding in EcDHFR, showed that there are two epitopes to which the nanobody binds107. They predict that epitope α is a new allosteric site that is over 10 Å away from the active site, and nanobody binding to that site results in conformational restraints and alterations of protein dynamics in EcDHFR, causing either activation or inhibition107. Another study has pointed to the role of M42, a residue distal to the active site in EcDHFR, as being a allosteric site that regulates protein dynamics and thus turnover at the active site108. Employing a sequence based approach, yet another study has tried to demonstrate the network of residues that are involved in facilitating the conformational transition from the closed state to occluded state and vice-versa109. The resveratrol binding site predicted by us (residues I2, P105, K106, A107 and Q108)80,81 is distinct from the sites in the above mentioned previous studies. While the allosteric pocket in the nanobody study is comprised of residues V10, D11, H114, I115, D116, E118, F140, S150, Y151, C152107, the dynamics study points to M42 as a crucial residue that impacts the dynamics of the enzyme108.
Figure 7.
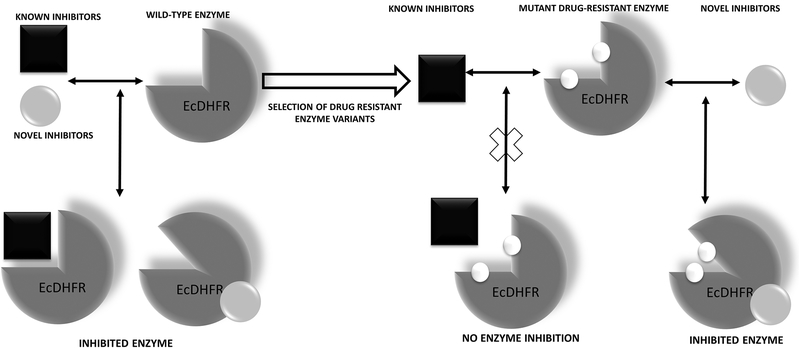
Design of allosteric site binders for EcDHFR capable of inhibiting both the wild-type and the drug resistant variants of the enzyme.
The class of inhibitors that were discovered, apart from their ability to inhibit wild-type EcDHFR, are also capable of inhibiting drug resistant rescue variants of the enzyme (A drug-resistant rescue variant of DHFR is defined as the form of DHFR that is enriched due to natural selection during persistent drug challenge and mostly possesses mutations at the inhibitor binding site, making it refractory to inhibitor binding). We assessed their behavior on three drug resistant variants of EcDHFR (the L28R single mutant and the A26T/L28R & P21L/L28R double mutants)80. They inhibited these variants with as much potency as for the wild-type enzyme (Fig. 7). Further, the inhibitors exhibited toxicity against E. coli strains that harbored the drug resistant variants. It must be emphasized that none of the conventional classes of DHFR inhibitors were capable of inhibiting either the drug-resistant variants of EcDHFR nor did they display cytotoxic effects against the microorganisms that harbored such drug resistant variants. Thus, these new molecules represent interesting antibiotic hits that are worthy of future development80,81.
To appreciate the significance of the above finding vis-à-vis discovering novel inhibitors for the drug resistant variant of EcDHFR, it is essential to understand the literature on the types of ligand protein interactions and their advantages and disadvantages, respectively. The below section, in brief, summarizes the various types of ligand-protein interactions that are known.
2.7. Monotherapy/polytherapy and monovalency/bivalency/multivalency
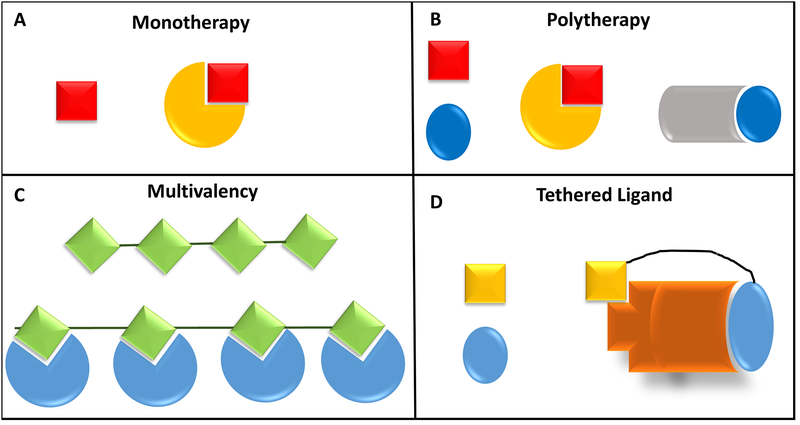
There are various modes whereby ligands (small-molecules or protein) interact with their target of interest in order to bring about the desired physiological outcome (Fig. 8). The most common kind of interactions are monovalency and/or monotherapy whereby a single small-molecule is designed to interact with one target of critical importance for the physiological outcome110. Polytherapy is the utilization of more than one small-molecule to target different receptor molecules or target pathways to achieve the desired outcome111. Both monotherapy and polytherapy are widely employed in clinical practices to counteract conditions such as epilepsy, psoriasis, depression and cancer, but their advantages and disadvantages remain a topic of debate110.
Figure 8.
Different modalities of ligands interacting with proteins. The simplest schemes are depicted here.
Multi/Polyvalency is another emerging concept in the ligand-protein interaction field, whereby a multivalent ligand comprised of multiple copies of ligands conjugated to scaffolds, allows the simultaneous binding of multivalent ligands to multiple binding sites or receptors112. Polyvalency has properties that are distinct from monovalent interactions in terms of conferring higher specificity and affinity. A few representative differences include achieving higher affinity of interactions for ligands with less surface area, signal amplification by non-linear graduation in biological response through possible induction of positive cooperativity, induction of oligomerization as a means of regulating the outcome; and inhibiting or suppressing undesirable interactions between ligands and non-specific targets113,114. Bivalency is a minor modification on the concept of multivalency whereby bivalent ligands, which are composed of two similar/ distinct functional pharmacophores linked by a spacer, can interact with either similar/distinct pockets on target protein/proteins115,116. A typical example of a bivalent ligand interacting with distinct pockets on a target protein is bitopic orthosteric/allosteric ligands of G protein-coupled receptors117. Another prominent example, especially vis-à-vis folate metabolism, is the discovery and synthesis of dual inhibitors that target both dihydrofolate reductase and thymidylate synthase in humans118. However, to the best of our knowledge, none of the studies to date has explored in considerable detail the application of two distinct untethered small-molecules targeting two distinct pockets on the same protein’s surface. The section below expands on the idea of targeting the allosteric pocket and the orthosteric pocket on the enzyme EcDHFR as a means of designing potent antibiotics which could have possible roles in killing drug resistant E. coli.
2.8. Combinatorial therapy: Targeting allosteric and active sites simultaneously
Why do we need novel classes of molecules targeting unique allosteric pockets on the enzyme dihydrofolate reductase? Will this new strategy prevent acquisition of drug resistance? Are we attempting to suggest that allosteric sites, somehow, are less prone to mutation induced resistance acquisition? The answer, of course, is no. Mutations at the active site on dihydrofolate reductases that confer drug resistance impose fitness costs on the organism119 that may, to some extent, impose stringent conditions upon the acquisition of such mutations. In other words, there is more selective pressure on the active site and hence more severe penalties in terms of fitness lost due to mutations on the active site. However, allosteric sites are organism specific120–123 and, to the best of our knowledge, are almost unknown in EcDHFR indicating that they may be either dispensable or are under less evolutionary pressure and hence, likely to be more mutable.
Having said that, it is well documented in the literature that small-molecules targeting the substrate and cofactor binding pockets, the usual targets for development of novel drugs in EcDHFR, have a tendency to evolve resistance by acquisition of mutations26. This is because of the selection of variants that can confer an evolutionary survival advantage by having either reduced or no binding for the small-molecule drug. However, novel allosteric pockets, which have not been previously targeted for small-molecule inhibitor discovery are a sterile niche for inhibitor discovery. These pockets, at the least, represent repositories of cavities that could be exploited for overcoming the drug resistance acquired by the original substrate binding site. Moreover, combined administration of folate binding-site targeting small-molecules and molecules that bind to the novel pocket may represent a stringent conditional probability that demands the presence of mutations in both the pockets for resistance acquisition against both small-molecules. Even if we assume that such mutations exist in both the pockets, the probability of such an event happening simultaneously will be rarer than a unique mutation in just one of the pockets. Moreover, simultaneous mutations in both pockets may constrain the loop dynamics of the enzyme in such a way that might not be beneficial for the fitness of the organisms harboring such double mutants. Moreover, it might be difficult to acquire an array of compensatory mutations to restore the fitness that the organism lost124 in selecting for mutations at both the active and allosteric sites to become refractory to an antibiotic. Not only is this a far lower probability event, but the acquisition of two mutations might result in significant destabilization of the native protein structure which will increase the population of unfolded molecules. By implication, its ability to generate the requisite levels of the enzyme product will therefore be reduced.
Furthermore, there is an opinion in the literature that allosteric inhibitors are more selective and less toxic than those that target orthosteric sites125–127.
Additional arguments that supports the design of inhibitors for allosteric sites rather than the active site include the lack of homologue level resolution which would likely happen if the active site were targeted. Further, active site binding molecules are all inhibitors rather than modulators of the enzyme activity. Modulation is a more desirable property than inhibition since the latter has the disadvantage of shutting down the enzyme activity, basal levels of which might be pivotal for survival. Furthermore, active site binding small-molecules will be competitive inhibitors of the enzyme. Assessment of the IC50 for the competitive model of inhibition is trickier than that for non-competitive inhibition128. This is because of the substrate concentration dependence of the former’s potency. Under equilibrium conditions, an increase in substrate concentration can effectively displace the competitive inhibitor and shift the IC50 rightwards. Hence, competitive inhibition is reversible by an increase in substrate concentration that likely happens in the absence of substrate turnover in the proximity of the enzyme128. Assuming that the rate-limiting step in an enzyme catalytic cycle is product release (and not the chemical step) (which is the case with chromosomally encoded EcDHFR), most of the enzyme species under steady state condition would be product bound. This might also hold true for the physiological form of the enzyme. Hence, designing small-molecules that can target the product-bound holo-enzyme form (with either non-competitive or uncompetitive inhibitors) might yield a better outcome as compared to targeting the apo-form with competitive inhibitors129.
However, targeting allosteric sites, as in the case of uncompetitive or non-competitive inhibition, makes more sense. In the case of uncompetitive inhibition, the inhibitor can trap the substrate bound complex that may be evident as an increase in the affinity of the inhibitor for the enzyme resulting in leftward IC50 shift129. This leftward shift is because an increase in substrate concentration in the absence of substrate turnover will push the equilibrium towards the substrate or product bound form of the enzyme that, in turn, is the preferable receptor for the small-molecule inhibitor. In a recent paper from our group, we introduced the concept of COmposite protein LIGands (COLIG) whereby more than one ligand binds to a pocket on the protein’s surface which interact with each other as well as the protein within a single ligand binding pocket130. We have also demonstrated, by a systematic analysis of the structures deposited in the Protein data bank (PDB), how uncompetitive kinetics of EcDHFR paves the way for exploration of further cases of uncompetitive inhibition as potential targets of drug discovery130. These arguments, coupled with the wide resurgence of interest in targeting allosteric sites for drug discovery131,132, support the justification for selection of allosteric pockets for drug discovery.
2.9. Conclusions and Future Perspectives
Development of resistance due to mutations is a persistent problem in dihydrofolate reductase, in particular, and one of the mechanisms of antibiotic resistance, in general. Given the important role that this enzyme plays and that it is indispensable to the survival of the microorganisms that harbor it, it is important that continuous efforts be invested in discovering new and improved inhibitors for this enzyme. Our work with Next Generation Virtual Ligand Screening approaches has shown that we can predict a handful of candidates to be screened as compared to traditional approaches55, and yet, we have not only been successful in predicting novel small-molecule inhibitors belonging to the traditional inhibitor scaffold but also in predicting novel allosteric inhibitors for the enzyme. Though extensive follow-up work is required to translate the discoveries from the lab to conferring benefits in human health and well-being, our approaches show the potential power of the application of these novel VLS methodologies for discovering small-molecule binders and inhibitors for both very well studied, and hence saturated, and novel refractory targets implicated in many human diseases.
Acknowledgement
This research was supported in part by grant No. 1R35GM-118039 of the Division of General Medical Sciences of the National Institutes of Health
Abbreviations:
- QSAR
Quantitative Structure Activity Relationship
- VLS
Virtual ligand Screening
- EcDHFR
E. coli Dihydrofolate Reductase
Biographies
Bharath Srinivasan
Bharath Srinivasan is a Marie Skłodowska-Curie Actions Fellow at the Instituto Gulbenkian de Ciência, Portugal. He attended graduate school in Biochemistry at the Jawaharlal Nehru Centre for Advanced Scientific Research, India, receiving a PhD in Biochemistry studying the structure-function relationship in Haloacid Dehalogenase superfamily of enzymes. He then held a postdoctoral fellowship at the Center for the Study of Systems Biology in the School of Biological Sciences at the Georgia Institute of Technology. Among his awards and fellowships are the Marie Skłodowska-Curie Individual Fellowship, Fundação para a Ciência e a Tecnologia Postdoctoral Fellowship, The Gordon Research Seminar travel award, The World Academy of Sciences travel award, Senior Research Fellowship, Junior Research Fellowship and Eligibility for Lectureship awarded by the Council of Scientific & Industrial Research (CSIR) and University Grants Commission Govt. of India, Senior Research Fellowship awarded by Indian Council for Agricultural Research, Govt. of India, and International symposium on cyanobacterial biotechnology commemorative medal. His research interests are in the areas of Enzymology, Drug-discovery and Medicinal Chemistry. He is the author of 24 publications, 1 patent awarded by USPTO and has an h-index of 9.
Sam-Tonddast-Navaei
Sam is a Data Scientist at SysteMatrix Solutions in Atlanta. He received his PhD in computational chemistry from University of Cincinnati, during which he studies protein unfolding assisted by molecular chaperones using Molecular Dynamics (MD) simulations. Afterwards and for the postdoctoral appointment, he joined Prof. Skolnick’s group at the Center for the Study of Systems Biology, Georgia Institute of Technology to study the interplay between promiscuity and specificity in biological systems with applications to drug discovery. His research resulted in more than 10 publications.
Ambrish Roy
Ambrish is a Research Scientist at Vertex Pharmaceuticals, Boston. He received his PhD in Bioinformatics from the University of Kansas, where his research focused on developing algorithms for protein structure and function prediction. He later did his post-doctoral training at the Center for the Study of Systems Biology, Georgia Institute of Technology developing algorithms in computational chemistry. He has authored 23 publications and has an h-index of 16. His research interests are in the areas of computational chemistry, computational systems biology and development of algorithms for modeling protein 3D structure and protein-ligand interaction.
Hongyi Zhou
Hongyi Zhou is a Research Scientist II at the Center for the Study of Systems Biology in the School of Biological Sciences at the Georgia Institute of Technology. He received his Ph.D. in theoretical physics in Department of Physics, Tsinghua University, China. He then joined the faculty member of Department of Physics, Tsinghua University, China. He conducted his postdoctoral research in Biophysics at the Germany Cancer Research Center and Department of Physiology and Biophysics, University of Buffalo. He received the Alexander von Humboldt Research Fellowship Award, Germany to conduct research in the Institute of Theoretical Physics, Heidelberg University. He also received First, Second class Science and Technology Achievement Awards from the Ministry of Education, China. He published more than 60 peer reviewed journal articles and one of them was cited more than 800 times. He is also one of the inventors of a US patent.
Jeffrey Skolnick
Jeffrey Skolnick is the Director of the Center for the Study of Systems Biology in the School of Biological Sciences at the Georgia Institute of Technology. He is also the Mary and Maisie Gibson Chair in Computational Systems Biology and a Georgia Research Alliance Eminent Scholar in Computational Systems Biology. He attended graduate school in Chemistry at Yale University, receiving a Ph.D. in Chemistry in polymer statistical mechanics. He then held a postdoctoral fellowship at Bell Laboratories. Next, he joined the faculty of the Chemistry Department at Louisiana State University, Baton Rouge. Next, he moved to Washington University, where he was subsequently appointed Professor of Chemistry. Following his expanding interest in biology, he joined the Department of Molecular Biology of the Scripps Research Institute, where he held the rank of Professor. Among his awards are the Sigma Xi Sustained Research Award, Southeastern Universities Research Association (SURA), Distinguished Scientist Award and an Alfred P. Sloan Research Fellowship. He is a Fellow of the American Association for the Advancement of Science, the Biophysical Society, and the St. Louis Academy of Science. His research interests are in the areas of Computational Systems Biology and Bioinformatics including the development of algorithms and their application to proteomes for the prediction of protein structure and function, the prediction of small molecule ligand-protein interactions with applications to drug discovery and the prediction of off-target uses of existing drugs with applications to aging, cancer and chronic fatigue syndrome, cancer metabolomics, precision medicine, fundamental studies on the nature and completeness of protein structure space and the exploration of the interplay between protein physics and evolution in determining protein structure and function, prediction of protein-protein and protein-DNA interactions, and molecular simulations of subcellular processes. He is the author of over 375 publications, has an h-index of 83, and has served on numerous editorial boards including the Scientific Reports, Peer J, Biology Direct, Biophysical Journal, Biopolymers, Proteins, and the Journal of Chemical Physics. He is also a cofounder of an early stage drug discovery company, PanXome and a structural proteomics company, Geneformatics, and his software has been commercialized by Tripos
References
- 1.Rengarajan J, Sassetti CM, Naroditskaya V, Sloutsky A, Bloom BR, Rubin EJ. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol Microbiol. 2004;53(1):275–282. [DOI] [PubMed] [Google Scholar]
- 2.Schweitzer BI, Dicker AP, Bertino JR. Dihydrofolate reductase as a therapeutic target. FASEB J. 1990;4(8):2441–2452. [DOI] [PubMed] [Google Scholar]
- 3.Chopra I, Hesse L, O’Neill AJ. Exploiting current understanding of antibiotic action for discovery of new drugs. Symp Ser Soc Appl Microbiol. 2002(31):4S–15S. [PubMed] [Google Scholar]
- 4.Hawser S, Lociuro S, Islam K. Dihydrofolate reductase inhibitors as antibacterial agents. Biochem Pharmacol. 2006;71(7):941–948. [DOI] [PubMed] [Google Scholar]
- 5.Exner M, Bhattacharya S, Christiansen B, et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg Infect Control. 2017;12:Doc05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luepke KH, Suda KJ, Boucher H, et al. Past, Present, and Future of Antibacterial Economics: Increasing Bacterial Resistance, Limited Antibiotic Pipeline, and Societal Implications. Pharmacotherapy. 2017;37(1):71–84. [DOI] [PubMed] [Google Scholar]
- 7.Papaconstantinou HT, Thomas JS. Bacterial colitis. Clin Colon Rectal Surg. 2007;20(1):18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R, Swick MC, Ledesma KR, et al. Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 2012;56(4):1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole K Mechanisms of bacterial biocide and antibiotic resistance. Symp Ser Soc Appl Microbiol. 2002(31):55S–64S. [PubMed] [Google Scholar]
- 10.Nikaido H Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178(20):5853–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362(19):1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakheet TM, Doig AJ. Properties and identification of antibiotic drug targets. BMC Bioinformatics. 2010;11:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31 Suppl 2:S24–28. [DOI] [PubMed] [Google Scholar]
- 14.Campbell EA, Korzheva N, Mustaev A, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104(6):901–912. [DOI] [PubMed] [Google Scholar]
- 15.Brodersen DE, Clemons WM Jr., Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103(7):1143–1154. [DOI] [PubMed] [Google Scholar]
- 16.Chittum HS, Champney WS. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr Microbiol. 1995;30(5):273–279. [DOI] [PubMed] [Google Scholar]
- 17.HsuChen CC, Feingold DS. The mechanism of polymyxin B action and selectivity toward biologic membranes. Biochemistry. 1973;12(11):2105–2111. [DOI] [PubMed] [Google Scholar]
- 18.Bradrick TD, Beechem JM, Howell EE. Unusual binding stoichiometries and cooperativity are observed during binary and ternary complex formation in the single active pore of R67 dihydrofolate reductase, a D2 symmetric protein. Biochemistry. 1996;35(35):11414–11424. [DOI] [PubMed] [Google Scholar]
- 19.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313(5793):1638–1642. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan B, Skolnick J. Insights into the slow-onset tight-binding inhibition of Escherichia coli dihydrofolate reductase: detailed mechanistic characterization of pyrrolo [3,2-f] quinazoline-1,3-diamine and its derivatives as novel tight-binding inhibitors. FEBS J. 2015;282(10):1922–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nammalwar B, Bourne CR, Wakeham N, et al. Modified 2,4-diaminopyrimidine-based dihydrofolate reductase inhibitors as potential drug scaffolds against Bacillus anthracis. Bioorg Med Chem. 2015;23(1):203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson RG, Rosowsky A. Dicyclic and tricyclic diaminopyrimidine derivatives as potent inhibitors of Cryptosporidium parvum dihydrofolate reductase: structure-activity and structure-selectivity correlations. Antimicrob Agents Chemother. 2001;45(12):3293–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson HC, Biggadike K, McKilligin E, et al. 6,7-disubstituted 2,4-diaminopteridines: novel inhibitors of Pneumocystis carinii and Toxoplasma gondii dihydrofolate reductase. Antimicrob Agents Chemother. 1996;40(6):1371–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan B, Tonddast-Navaei S, Skolnick J. Ligand binding studies, preliminary structure-activity relationship and detailed mechanistic characterization of 1-phenyl-6,6-dimethyl-1,3,5-triazine-2,4-diamine derivatives as inhibitors of Escherichia coli dihydrofolate reductase. Eur J Med Chem. 2015;103:600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Booth RG, Selassie CD, Hansch C, Santi DV. Quantitative structure-activity relationship of triazine-antifolate inhibition of Leishmania dihydrofolate reductase and cell growth. J Med Chem. 1987;30(7):1218–1224. [DOI] [PubMed] [Google Scholar]
- 26.Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2011;44(1):101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maskell JP, Sefton AM, Hall LM. Multiple mutations modulate the function of dihydrofolate reductase in trimethoprim-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1104–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch C, Pearce R, Pota H, et al. Emergence of a dhfr mutation conferring high-level drug resistance in Plasmodium falciparum populations from southwest Uganda. J Infect Dis. 2008;197(11):1598–1604. [DOI] [PubMed] [Google Scholar]
- 29.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A. 1988;85(23):9114–9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klebe G Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today. 2006;11(13–14):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bajorath J Integration of virtual and high-throughput screening. Nat Rev Drug Discov. 2002;1(11):882–894. [DOI] [PubMed] [Google Scholar]
- 32.Tang YT, Marshall GR. PHOENIX: a scoring function for affinity prediction derived using high-resolution crystal structures and calorimetry measurements. J Chem Inf Model. 2011;51(2):214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng T, Li Q, Zhou Z, Wang Y, Bryant SH. Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J. 2012;14(1):133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Vivo M, Masetti M, Bottegoni G, Cavalli A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J Med Chem. 2016;59(9):4035–4061. [DOI] [PubMed] [Google Scholar]
- 35.Hartshorn MJ, Verdonk ML, Chessari G, et al. Diverse, high-quality test set for the validation of protein-ligand docking performance. J Med Chem. 2007;50(4):726–741. [DOI] [PubMed] [Google Scholar]
- 36.Bordogna A, Pandini A, Bonati L. Predicting the accuracy of protein-ligand docking on homology models. J Comput Chem. 2011;32(1):81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem. 2004;47(1):45–55. [DOI] [PubMed] [Google Scholar]
- 38.Lionta E, Spyrou G, Vassilatis DK, Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem. 2014;14(16):1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat Chem. 2009;1(3):187–192. [DOI] [PubMed] [Google Scholar]
- 40.Davis BJ, Erlanson DA. Learning from our mistakes: the ‘unknown knowns’ in fragment screening. Bioorg Med Chem Lett. 2013;23(10):2844–2852. [DOI] [PubMed] [Google Scholar]
- 41.Erlanson DA, McDowell RS, O’Brien T. Fragment-based drug discovery. J Med Chem. 2004;47(14):3463–3482. [DOI] [PubMed] [Google Scholar]
- 42.Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. [DOI] [PubMed] [Google Scholar]
- 43.Sheng C, Zhang W. Fragment informatics and computational fragment-based drug design: an overview and update. Med Res Rev. 2013;33(3):554–598. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt T, Bergner A, Schwede T. Modelling three-dimensional protein structures for applications in drug design. Drug Discov Today. 2014;19(7):890–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evers A, Klabunde T. Structure-based drug discovery using GPCR homology modeling: successful virtual screening for antagonists of the alpha1A adrenergic receptor. J Med Chem. 2005;48(4):1088–1097. [DOI] [PubMed] [Google Scholar]
- 46.Dahl SG, Sylte I. Molecular modelling of drug targets: the past, the present and the future. Basic Clin Pharmacol Toxicol. 2005;96(3):151–155. [DOI] [PubMed] [Google Scholar]
- 47.Acharya C, Coop A, Polli JE, Mackerell AD Jr. Recent advances in ligand-based drug design: relevance and utility of the conformationally sampled pharmacophore approach. Curr Comput Aided Drug Des. 2011;7(1):10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willett P Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11(23–24):1046–1053. [DOI] [PubMed] [Google Scholar]
- 49.Taminau J, Thijs G, De Winter H. Pharao: pharmacophore alignment and optimization. J Mol Graph Model. 2008;27(2):161–169. [DOI] [PubMed] [Google Scholar]
- 50.Nicholls A, McGaughey GB, Sheridan RP, et al. Molecular shape and medicinal chemistry: a perspective. J Med Chem. 2010;53(10):3862–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeagle PL, Albert AD. G-protein coupled receptor structure. Biochim Biophys Acta. 2007;1768(4):808–824. [DOI] [PubMed] [Google Scholar]
- 52.Makley LN, Gestwicki JE. Expanding the number of ‘druggable’ targets: non-enzymes and protein-protein interactions. Chem Biol Drug Des. 2013;81(1):22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ngo T, Kufareva I, Coleman J, Graham RM, Abagyan R, Smith NJ. Identifying ligands at orphan GPCRs: current status using structure-based approaches. Br J Pharmacol. 2016;173(20):2934–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H, Skolnick J. FINDSITE(comb): a threading/structure-based, proteomic-scale virtual ligand screening approach. J Chem Inf Model. 2013;53(1):230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srinivasan B, Zhou H, Kubanek J, Skolnick J. Experimental validation of FINDSITE(comb) virtual ligand screening results for eight proteins yields novel nanomolar and micromolar binders. J Cheminform. 2014;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy A, Skolnick J. LIGSIFT: an open-source tool for ligand structural alignment and virtual screening. Bioinformatics. 2015;31(4):539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srinivasan B, Zhou H, Mitra S, Skolnick J. Novel small molecule binders of human N-glycanase 1, a key player in the endoplasmic reticulum associated degradation pathway. Bioorg Med Chem. 2016;24(19):4750–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy A, Srinivasan B, Skolnick J. PoLi: A Virtual Screening Pipeline Based on Template Pocket and Ligand Similarity. J Chem Inf Model. 2015;55(8):1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skolnick J, Gao M. Interplay of physics and evolution in the likely origin of protein biochemical function. Proc Natl Acad Sci U S A. 2013;110(23):9344–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skolnick J, Gao M, Roy A, Srinivasan B, Zhou H. Implications of the small number of distinct ligand binding pockets in proteins for drug discovery, evolution and biochemical function. Bioorg Med Chem Lett. 2015;25(6):1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srinivasan B, Tonddast-Navaei S, Skolnick J. Pocket detection and interaction-weighted ligand-similarity search yields novel high-affinity binders for Myocilin-OLF, a protein implicated in glaucoma. Bioorg Med Chem Lett. 2017;27(17):4133–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams JW, Duggleby RG, Cutler R, Morrison JF. The inhibition of dihydrofolate reductase by folate analogues: structural requirements for slow- and tight-binding inhibition. Biochem Pharmacol. 1980;29(4):589–595. [DOI] [PubMed] [Google Scholar]
- 65.Stone SR MA, Morrison JF. Interaction of analogs of nicotinamide adenine dinucleotide phosphate with dihydrofolate reductase from Escherichia coli. Biochemistry. 1984;23(19):4340–4346. [Google Scholar]
- 66.Hsieh YC, Tedeschi P, Adebisi Lawal R, et al. Enhanced degradation of dihydrofolate reductase through inhibition of NAD kinase by nicotinamide analogs. Mol Pharmacol. 2013;83(2):339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gangjee A, Mavandadi F, Kisliuk RL, Queener SF. Synthesis of classical and a nonclassical 2-amino-4-oxo-6-methyl-5-substituted pyrrolo[2,3-d]pyrimidine antifolate inhibitors of thymidylate synthase. J Med Chem. 1999;42(12):2272–2279. [DOI] [PubMed] [Google Scholar]
- 68.Matherly LH, Hou Z. Structure and function of the reduced folate carrier a paradigm of a major facilitator superfamily mammalian nutrient transporter. Vitam Horm. 2008;79:145–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.El Fadili A, Kundig C, Ouellette M. Characterization of the folylpolyglutamate synthetase gene and polyglutamylation of folates in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2002;124(1–2):63–71. [DOI] [PubMed] [Google Scholar]
- 70.Rots MG, Pieters R, Peters GJ, et al. Role of folylpolyglutamate synthetase and folylpolyglutamate hydrolase in methotrexate accumulation and polyglutamylation in childhood leukemia. Blood. 1999;93(5):1677–1683. [PubMed] [Google Scholar]
- 71.Mathieu M, Debousker G, Vincent S, Viviani F, Bamas-Jacques N, Mikol V. Escherichia coli FolC structure reveals an unexpected dihydrofolate binding site providing an attractive target for anti-microbial therapy. J Biol Chem. 2005;280(19):18916–18922. [DOI] [PubMed] [Google Scholar]
- 72.Carter EL, Jager L, Gardner L, Hall CC, Willis S, Green JM. Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate. J Bacteriol. 2007;189(9):3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hussein MJ, Green JM, Nichols BP. Characterization of mutations that allow p-aminobenzoyl-glutamate utilization by Escherichia coli. J Bacteriol. 1998;180(23):6260–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hong S, Moritz TJ, Rath CM, et al. Assessing Antibiotic Permeability of Gram-Negative Bacteria via Nanofluidics. ACS Nano. 2017;11(7):6959–6967. [DOI] [PubMed] [Google Scholar]
- 75.Kopytek SJ, Dyer JC, Knapp GS, Hu JC. Resistance to methotrexate due to AcrAB-dependent export from Escherichia coli. Antimicrob Agents Chemother. 2000;44(11):3210–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson M, Liu JW, Ollis D. Directed evolution of trimethoprim resistance in Escherichia coli. FEBS J. 2007;274(10):2661–2671. [DOI] [PubMed] [Google Scholar]
- 77.Cammarata M, Thyer R, Lombardo M, et al. Characterization of trimethoprim resistant E. coli dihydrofolate reductase mutants by mass spectrometry and inhibition by propargyl-linked antifolates. Chem Sci. 2017;8(5):4062–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scocchera E, Reeve SM, Keshipeddy S, et al. Charged Nonclassical Antifolates with Activity Against Gram-Positive and Gram-Negative Pathogens. ACS Med Chem Lett. 2016;7(7):692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Srinivasan B, Skolnick J, Zhou H, Inventor; Georgia Tech Research Corp assignee. Molecules with potent DHFR binding affinity and antibacterial activity. 2018. [Google Scholar]
- 80.Srinivasan B, Rodrigues JV, Tonddast-Navaei S, Shakhnovich E, Skolnick J. Rational Design of Novel Allosteric Dihydrofolate Reductase Inhibitors Showing Antibacterial Effects on Drug-Resistant Escherichia coli Escape Variants. ACS Chem Biol. 2017;12(7):1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Srinivasan B, Rodrigues JV, Tonddast-Navaei S, Shakhnovich E, Skolnick J. Correction to Rational Design of Novel Allosteric Dihydrofolate Reductase Inhibitors Showing Antibacterial Effects on Drug-Resistant Escherichia coli Escape Variants. ACS Chem Biol. 2018;13(5):1407. [DOI] [PubMed] [Google Scholar]
- 82.Huennekens FM. The methotrexate story: a paradigm for development of cancer chemotherapeutic agents. Adv Enzyme Regul. 1994;34:397–419. [DOI] [PubMed] [Google Scholar]
- 83.Williams JW, Morrison JF, Duggleby RG. Methotrexate, a high-affinity pseudosubstrate of dihydrofolate reductase. Biochemistry. 1979;18(12):2567–2573. [DOI] [PubMed] [Google Scholar]
- 84.Stone SR, Montgomery JA, Morrison JF. Inhibition of dihydrofolate reductase from bacterial and vertebrate sources by folate, aminopterin, methotrexate and their 5-deaza analogues. Biochem Pharmacol. 1984;33(2):175–179. [DOI] [PubMed] [Google Scholar]
- 85.Stone SR, Morrison JF. Mechanism of inhibition of dihydrofolate reductases from bacterial and vertebrate sources by various classes of folate analogues. Biochim Biophys Acta. 1986;869(3):275–285. [DOI] [PubMed] [Google Scholar]
- 86.Lam T, Hilgers MT, Cunningham ML, et al. Structure-based design of new dihydrofolate reductase antibacterial agents: 7-(benzimidazol-1-yl)-2,4-diaminoquinazolines. J Med Chem. 2014;57(3):651–668. [DOI] [PubMed] [Google Scholar]
- 87.Chan JH, Hong JS, Kuyper LF, et al. Selective inhibitors of Candida albicans dihydrofolate reductase: activity and selectivity of 5-(arylthio)-2,4-diaminoquinazolines. J Med Chem. 1995;38(18):3608–3616. [DOI] [PubMed] [Google Scholar]
- 88.Gangjee A, Adair OO, Pagley M, Queener SF. N9-substituted 2,4-diaminoquinazolines: synthesis and biological evaluation of lipophilic inhibitors of pneumocystis carinii and toxoplasma gondii dihydrofolate reductase. J Med Chem. 2008;51(19):6195–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gokhale VM, Kulkarni VM. Selectivity analysis of 5-(arylthio)-2,4-diaminoquinazolines as inhibitors of Candida albicans dihydrofolate reductase by molecular dynamics simulations. J Comput Aided Mol Des. 2000;14(5):495–506. [DOI] [PubMed] [Google Scholar]
- 90.Khabnadideh S, Pez D, Musso A, et al. Design, synthesis and evaluation of 2,4-diaminoquinazolines as inhibitors of trypanosomal and leishmanial dihydrofolate reductase. Bioorg Med Chem. 2005;13(7):2637–2649. [DOI] [PubMed] [Google Scholar]
- 91.Susten SS, Hynes JB, Kumar A, Freisheim JH. Inhibition of dihydrofolate reductase, methotrexate transport, and growth of methotrexate-sensitive and -resistant L1210 leukemia cells in vitro by 5-substituted 2,4-diaminoquinazolines. Biochem Pharmacol. 1985;34(12):2163–2167. [DOI] [PubMed] [Google Scholar]
- 92.Fukunaga JY, Hansch C, Steller EE. Inhibition of dihydrofolate reductase. Structure-activity correlations of quinazolines. J Med Chem. 1976;19(5):605–611. [DOI] [PubMed] [Google Scholar]
- 93.Dahl G, Akerud T. Pharmacokinetics and the drug-target residence time concept. Drug Discov Today. 2013;18(15–16):697–707. [DOI] [PubMed] [Google Scholar]
- 94.Lourens AC, Gravestock D, van Zyl RL, et al. Design, synthesis and biological evaluation of 6-aryl-1,6-dihydro-1,3,5-triazine-2,4-diamines as antiplasmodial antifolates. Org Biomol Chem. 2016;14(33):7899–7911. [DOI] [PubMed] [Google Scholar]
- 95.Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol. 1998;54(6):1140–1147. [DOI] [PubMed] [Google Scholar]
- 96.Sirawaraporn W Dihydrofolate reductase and antifolate resistance in malaria. Drug Resist Updat. 1998;1(6):397–406. [DOI] [PubMed] [Google Scholar]
- 97.Hopfinger AJ. A general QSAR for dihydrofolate reductase inhibition by 2,4-diaminotriazines based upon molecular shape analysis. Arch Biochem Biophys. 1981;206(1):153–163. [DOI] [PubMed] [Google Scholar]
- 98.Coats EA, Genther CS, Dietrich SW, Guo ZR, Hansch C. Comparison of the inhibition of methotrexate-sensitive and -resistant Lactobacillus casei cell cultures with purified Lactobacillus casei dihydrofolate reductase by 4,6-diamino-1,2-dihydro-2,2-dimethyl-1-(3-substituted-phenyl)-s-triazines. Use of quantitative structure-activity relationships in making inferences about the mechanism of resistance and the structure of the enzyme is situ compared with the enzyme in vitro. J Med Chem. 1981;24(12):1422–1429. [DOI] [PubMed] [Google Scholar]
- 99.Hansch C, Dietrich SW, Fukunaga JY. Inhibition of bovine and rat liver dihydrofolate reductase by 4,6-diamino-1,2-dihydro-2,2-dimethyl-1-(4-substituted-phenyl)-s-triazines. J Med Chem. 1981;24(5):544–549. [DOI] [PubMed] [Google Scholar]
- 100.Hansch C, Hathaway BA, Guo ZR, et al. Crystallography, quantitative structure-activity relationships, and molecular graphics in a comparative analysis of the inhibition of dihydrofolate reductase from chicken liver and Lactobacillus casei by 4,6-diamino-1,2-dihydro-2,2-dimethyl-1-(substituted-phenyl)-s-triazine s. J Med Chem. 1984;27(2):129–143. [DOI] [PubMed] [Google Scholar]
- 101.Kim KH, Dietrich SW, Hansch C, Dolnick BJ, Bertino JR. Inhibition of dihydrofolate reductase. 3. 4.6-Diamino-1,2-dihydro-2,2-dimethyl-1-(2-substituted-phenyl)-s-triazine inhibition of bovine liver and mouse tumor enzymes. J Med Chem. 1980;23(11):1248–1251. [DOI] [PubMed] [Google Scholar]
- 102.Marlowe CK, Selassie CD, Santi DV. Quantitative structure-activity relationships of the inhibition of Pneumocystis carinii dihydrofolate reductase by 4,6-diamino-1,2-dihydro-2,2-dimethyl-1-(X-phenyl)-s-triazines. J Med Chem. 1995;38(6):967–972. [DOI] [PubMed] [Google Scholar]
- 103.Hathaway BA, Guo ZR, Hansch C, Delcamp TJ, Susten SS, Freisheim JH. Inhibition of human dihydrofolate reductase by 4,6-diamino-1,2-dihydro-2,2-dimethyl-1-(substituted-phenyl)-s-triazine s. A quantitative structure-activity relationship analysis. J Med Chem. 1984;27(2):144–149. [DOI] [PubMed] [Google Scholar]
- 104.Singla P, Luxami V, Paul K. Triazine-benzimidazole hybrids: anticancer activity, DNA interaction and dihydrofolate reductase inhibitors. Bioorg Med Chem. 2015;23(8):1691–1700. [DOI] [PubMed] [Google Scholar]
- 105.Vajda S, Beglov D, Wakefield AE, Egbert M, Whitty A. Cryptic binding sites on proteins: definition, detection, and druggability. Curr Opin Chem Biol. 2018;44:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Surade S, Blundell TL. Structural biology and drug discovery of difficult targets: the limits of ligandability. Chem Biol. 2012;19(1):42–50. [DOI] [PubMed] [Google Scholar]
- 107.Oyen D, Wechselberger R, Srinivasan V, Steyaert J, Barlow JN. Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihydrofolate reductase of Escherichia coli. Biochim Biophys Acta. 2013;1834(10):2147–2157. [DOI] [PubMed] [Google Scholar]
- 108.Mauldin RV, Lee AL. Nuclear magnetic resonance study of the role of M42 in the solution dynamics of Escherichia coli dihydrofolate reductase. Biochemistry. 2010;49(8):1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen J, Dima RI, Thirumalai D. Allosteric communication in dihydrofolate reductase: signaling network and pathways for closed to occluded transition and back. J Mol Biol. 2007;374(1):250–266. [DOI] [PubMed] [Google Scholar]
- 110.Reynolds EH, Shorvon SD. Monotherapy or polytherapy for epilepsy? Epilepsia. 1981;22(1):1–10. [DOI] [PubMed] [Google Scholar]
- 111.French JA, Faught E. Rational polytherapy. Epilepsia. 2009;50 Suppl 8:63–68. [DOI] [PubMed] [Google Scholar]
- 112.Morphy R, Kay C, Rankovic Z. From magic bullets to designed multiple ligands. Drug Discov Today. 2004;9(15):641–651. [DOI] [PubMed] [Google Scholar]
- 113.Vauquelin G, Charlton SJ. Exploring avidity: understanding the potential gains in functional affinity and target residence time of bivalent and heterobivalent ligands. Br J Pharmacol. 2013;168(8):1771–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell-surface interactions. Curr Opin Chem Biol. 2000;4(6):696–703. [DOI] [PubMed] [Google Scholar]
- 115.Long DD, Aggen JB, Chinn J, et al. Exploring the positional attachment of glycopeptide/beta-lactam heterodimers. J Antibiot (Tokyo). 2008;61(10):603–614. [DOI] [PubMed] [Google Scholar]
- 116.Long DD, Aggen JB, Christensen BG, et al. A multivalent approach to drug discovery for novel antibiotics. J Antibiot (Tokyo). 2008;61(10):595–602. [DOI] [PubMed] [Google Scholar]
- 117.Valant C, Robert Lane J, Sexton PM, Christopoulos A. The best of both worlds? Bitopic orthosteric/allosteric ligands of g protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2012;52:153–178. [DOI] [PubMed] [Google Scholar]
- 118.Arooj M, Sakkiah S, Cao G, Lee KW. An innovative strategy for dual inhibitor design and its application in dual inhibition of human thymidylate synthase and dihydrofolate reductase enzymes. PLoS One. 2013;8(4):e60470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rodrigues JV, Bershtein S, Li A, Lozovsky ER, Hartl DL, Shakhnovich EI. Biophysical principles predict fitness landscapes of drug resistance. Proc Natl Acad Sci U S A. 2016;113(11):E1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sela-Passwell N, Rosenblum G, Shoham T, Sagi I. Structural and functional bases for allosteric control of MMP activities: can it pave the path for selective inhibition? Biochim Biophys Acta. 2010;1803(1):29–38. [DOI] [PubMed] [Google Scholar]
- 121.Suplatov D, Svedas V. Study of Functional and Allosteric Sites in Protein Superfamilies. Acta Naturae. 2015;7(4):34–45. [PMC free article] [PubMed] [Google Scholar]
- 122.Veith N, Feldman-Salit A, Cojocaru V, Henrich S, Kummer U, Wade RC. Organism-adapted specificity of the allosteric regulation of pyruvate kinase in lactic acid bacteria. PLoS Comput Biol. 2013;9(7):e1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Usenik A, Legisa M. Evolution of allosteric citrate binding sites on 6-phosphofructo-1-kinase. PLoS One. 2010;5(11):e15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brown KM, Costanzo MS, Xu W, Roy S, Lozovsky ER, Hartl DL. Compensatory mutations restore fitness during the evolution of dihydrofolate reductase. Mol Biol Evol. 2010;27(12):2682–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abdel-Magid AF. Allosteric modulators: an emerging concept in drug discovery. ACS Med Chem Lett. 2015;6(2):104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wenthur CJ, Gentry PR, Mathews TP, Lindsley CW. Drugs for allosteric sites on receptors. Annu Rev Pharmacol Toxicol. 2014;54:165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grover AK. Use of allosteric targets in the discovery of safer drugs. Med Princ Pract. 2013;22(5):418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Swinney DC. Biochemical mechanisms of drug action: what does it take for success? Nat Rev Drug Discov. 2004;3(9):801–808. [DOI] [PubMed] [Google Scholar]
- 129.Holdgate GA, Meek TD, Grimley RL. Mechanistic enzymology in drug discovery: a fresh perspective. Nat Rev Drug Discov. 2018;17(1):78. [DOI] [PubMed] [Google Scholar]
- 130.Tonddast-Navaei S, Srinivasan B, Skolnick J. On the importance of composite protein multiple ligand interactions in protein pockets. J Comput Chem. 2017;38(15):1252–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Srinivasan B, Forouhar F, Shukla A, et al. Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila. FEBS J. 2014;281(6):1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153(2):293–305. [DOI] [PubMed] [Google Scholar]