Abstract
There is compelling evidence that synergistic drug combinations have become promising strategies for combating complex diseases, and they have evident predominance comparing to traditional one drug - one disease approaches. In this paper, we develop a computational method, namely SyFFM, that takes pharmacological data into consideration and applies field-aware factorization machines to analyze and predict potential synergistic drug combinations. Firstly, features of drug pairs are constructed based on associations between drugs and target, and enzymes, and indication areas. Then, the synergistic scores of drug combinations are obtained by implementing field-aware factorization machines on latent vector space of these features. Finally, synergistic combinations can be predicted by introducing a threshold. We applied SyFFM to predict pairwise synergistic combinations and three-drug synergistic combinations, and the performance is good in terms of cross-validation. Besides, more than 90% combinations of the top ranked predictions are proved by literature and the analysis of parameters in model shows that our method can help to investigate and explain synergistic mechanisms underlying combinatorial therapy.
Keywords: Synergistic drug combinations, Factorization machines, Computational methods
1. Introduction
Synergistic drug combinations consist of two or more active pharmaceutical ingredients (APIs), which are developed to target multiple diseases or conditions, such as Exforge (amlodipine/valsartan). Accumulated studies have shown that synergistic drug combinations are widely used in the treatment of cancer, AIDs and other complex diseases [12,16]. Christie M. Ballantyne et al. found that the therapeutic effect of the combination of ezetimibe and simvastatin (Vytorin) was significant for treatment of hypercholesterolemic patients [3]. Zhong et al. have found that Homoharringtonine combining with arsenic trioxide could induce apoptosis of RPMI 8226 cell line [63]. Drug combinations could improve medication compliance by reducing the pill burden of patients. Moreover, drug combinatorial therapy has better efficacy and smaller side effect than single drugs because of the fact that the development and progression of systemic diseases often involve in complex biological processes. Investments in synergistic drug combinations have emerged and become an increasingly important strategy because of the high failure rate in new drug development pipeline and high costs in marketing new drugs [15]. It also has been argued that synergistic drug combinations played vital roles in drug development as a new paradigm of drug discovery [24].
Despite of more attentions have been paid to research on synergistic drug combinations, there are still a lack of effective prediction methods. Most synergistic combinations were found using exhaustive methods, such as high-throughput screening or pure experimental tests, which are time-consuming and costly. Therefore, a number of computational approaches for synergistic combinations prediction have been emerged with the growth of biological datasets [36]. For example, Zhao et al. proposed a computational method to predict novel combinations by integrating molecular and pharmacological data, but its performance is limited by the feature constructed in this model [62]. Ligeti et al. presented a prediction model by applying a propagation algorithm on a protein-protein association network [38], but the construction of network is affected by incompleteness of biological data. Sun et al. constructed support vector machines and naïve Bayesian classifiers on Hadoop for drug combination prediction, but it cannot explain synergistic mechanisms [51]. Chen et al. collected synergistic antifungal drug combinations and construct the first Antifungal Synergistic Drug Combination Databases (ASDCD) [10] and developed a semi-supervised algorithm implemented on drug-target interactions and drug chemical structures data [11], but its predictive performance is evaluated only on antifungal drugs. Liu constructed features of drug pairs using some feature selection techniques and predicted potential synergistic drug combinations by applying random forest algorithm [41]. Based on drug target network and drug induced gene expression profiles [26], Li built a prediction model named by SyDRa using random forest algorithm [37], which is a simple and effective computational model. However, it is a cancer-specific model and it was not tested on other diseases. Besides, most of these quantitative and qualitative prediction methods are proposed to pairwise synergistic combinations and cannot be applied to predict multi-drug synergistic combinations.
In this paper, by considering synergistic mechanisms as combinations of their pharmacological characteristics [8], we proposed a new model, namely SyFFM, to infer potential pairwise synergistic combinations and three-drug synergistic combinations. Drug targets, metabolic enzymes and ATC (Anatomical Therapeutic Chemical Classification System) were collected for constructing features of drug combinations and FFM (Field-aware Factorization Machines) [30] were applied to calculate the synergistic scores. FFM models pairwise interactions via inner products of respective feature latent vectors, and it has good performance under huge sparsity. Using factorized parameters, FFM provides a new perspective to investigate interactions between drugs. SyFFM was evaluated on various datasets, and the good predictive performance suggests that it would help to identify novel drug combinations effectively. Analysis on the parameters of SyFFM demonstrates that the new model can provide insights to uncover the synergistic mechanisms.
2. Methods
Let denotes the drug set, be the target set, be the enzyme set and be the ATC set. For each drug pair , is defined to indicate their synergistic relationship, if is an approved synergistic combination, and if not.
2.1. Features construction
For each drug pair , we constructed its feature vector as follows,
-
1.
Target pairs of , : (i) Denote the targets of as and the target pairs of as . (ii) Collect the target pairs of all synergistic drug combinations, written . In other words, each pair in is a target pair of at least one synergistic drug combination and the target pairs in were sorted order by target ID. (iii) Construct a 0–1 row vector and , where the k-th components of is 1 if the k-th target pair is one target pair of . Otherwise, the k-th components of is 0.
-
2.
Enzyme pairs of , : The enzyme pairs of all synergistic drug combinations could be obtained according to drug-enzyme associations, recorded as . Sort the enzyme pairs in by enzyme ID and construct a 0–1 row vector . The components of is set in a similar way as described above for the construction of .
-
3.
ATC pairs of , : The ATC pairs of all synergistic drug combinations could be obtained according to drug-ATC associations, recorded as . Sort the ATC pairs in by ATC ID and construct a 0–1 row vector . The components of is set in a similar way as described above for the construction of .
-
4.
Feature vector of , : Concatenate three features together, and the feature vector of was denoted as . Here, is referred to as a filed of feature and each components is referred to as a feature. It is worth noting that the features constructed by our method are very sparse due to incomplete pharmacological data and the fact that most drugs do not have the same targets, enzymes or ATC.
2.2. SyFFM on synergistic drug combinations prediction
Degree-2-polynomial mappings (Poly2) has been recorded as an effective approach to learn information implicit in feature conjunctions [7,34]. Based on this, we can simplify synergistic mechanisms as combinations of pharmacological characteristics, and for each drug pair , the combinations of features can be represented by a linear function
| (1) |
and denoted the contribution values of feature interaction and a single feature to the synergistic relationship between and , respectively. is modeled to be an indicator to measure the strength of synergy between and , so should be learned approximating to the real label .
Unfortunately, the above function could not reflect the real interactions between different features accurately. Take feature interactions and for example, may affect the interaction between and . However, is independent from in the above function and they could not capture this mutually dependent relationship.
So, we improved ϕ by implementing FFM, which replaces all interactions by mapping similar features to be embedded near each other into several latent spaces according to fields it belongs to. Thus, dot product between latent vectors would capture interactions related to similar features and interactions not recorded in the available dataset are also allowed to caught. Although higher order of interactions could be estimated, we only considered first and second order due to high computational complexity. In our dataset, each feature can be represented by three latent vectors and the contribution value of feature interaction can be represented by product of their corresponding latent vectors, writren by . The latent vectors would help to extract the potential interactions between two drugs and can be rewritten as
| (2) |
For example, the feature vector of a drug combination is , and , , , then the interaction of features can be represented as follows,
| (3) |
Indicator should be learned to approximate the real synergistic relationship . Logistic loss function is chosen to learn feature interactions, which is a widely used loss function to penalize incorrect predictions in classification problem. In addiction, -regularization terms are introduced to prevent overfitting of SyFFM and generalize SyFFM to other drugs not included in the available dataset. Thus, the problem of synergistic drug combinations prediction would be converted to learn v and w by solving the following optimization problem,
| (4) |
We implemented stochastic gradient methods to derive the optimal solution of this optimization problem. Let be the optimum solution, and the synergistic score between and was defined as . Given a threshold θ, was predicted as a synergistic combination if its synergistic score was larger than θ, and θ was selected by cross validations. The code of field-aware factorization machines can be got at https://www.csie.ntu.edu.tw/∼cjlin/libffm/ contributed by the Machine Learning Group at National Taiwan University.
3. Results
3.1. Prediction on pairwise synergistic drug combinations
SyFFM was implemented on three datasets respectively, DCDB data (including 946 approved pairwise synergistic drug combinations, 759 drugs from DCDB [40]), NData (68 approved pairwise synergistic combinations and 92 drugs from paper [28]) and FDA data (184 approved pairwise synergistic combinations and 238 drugs from FDA until November 2010). In DCDB, there are 287661 possible pairs of 759 drugs and 3‰ were approved to be synergistic combinations. Similarly, there are 1% pairs in NData and 6‰ pairs in FDA data were approved to be synergistic combinations. In each dataset, we regarded these approved synergistic combinations as ’positive samples', and the ’negative samples’ were obtained by sampling from the random pairs of all drugs. We set different ratios of positive-to-negative samples (1:1, 1:2, 1:3, 1:4, 1:5) and analysis the impact of ratios on performance of SyFFM.
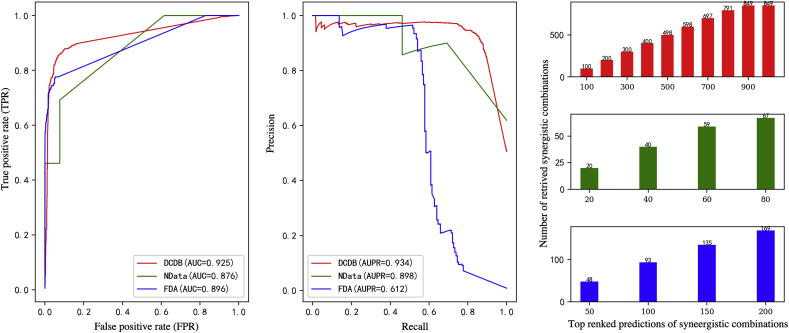
Firstly, 5-fold cross validation was implemented to evaluate our method. To guarantee the nonrandom of the prediction results, we implemented our method on 50 different negative samples and the optimal parameters were adjusted by cross validation. We got average AUC (DCDB) = 0.925 (0.87 ∼ 0.93), AUC (NData) = 0.876 (0.84 ∼ 0.92), AUC (FDA) = 0.896(0.86 ∼ 0.92) (Fig. 1). Secondly, the synergistic scores obtained by our method were sorted in descending order, for DCDB data, SyFFM correctly retrieved all associations in the top-400 records, 697 in the top-700 records and 791 in the top-800 records. For NData, SyFFM correctly retrieved 40, 59, 67 in the top-40, 60, 80 records. For FDA data, SyFFM correctly retrieved 48 93, 135 in the top-50, 100, 150 records. (Fig. 1).
Fig. 1.
The results of SyFFM on pairwise synergistic drug combinations.
Synergistic combinations in FDA were updated after November 2010 and the predictive ability of SyFFM for potential synergistic drug combinations is evaluated by implemented on FDA data (until November 2010). 11 potential synergistic combinations are predicted by SyFFM and 9 are confirmed by literature, which is shown in Table 1.
Table 1.
The 11 potential synergistic drug combinations predicted by SyFFM and 9 are confirmed by literature.
| Drug 1 | Drug 2 | Evidence of synergy combinations |
|---|---|---|
| ampicillin sodium | tazobactam | PMID:24041466 |
| esomeprazole magnesium | sodium bicarbonate | None |
| trandolapril | hydrochlorothiazide | PMID: 7527107 |
| dienogest | ethinyl estradiol | PMID: 20394455 |
| eprosartan mesylate | amlodipine besylate | None |
| tenofovir disoproxil fumarate | lamivudine | PMID:13130407 |
| niacin | atorvastatin calcium | PMID:10095800 |
| trandolapril | amlodipine besylate | None |
| norelgestromin | Ethinyl Estradiol | PMID: 11849630 |
| Ethinyl estradiol | ethynodiol diacetate | PMID: 2046084 |
| desogestrel | Ethinyl estradiol | PMID: 9673846 |
3.2. The impact of ratios of positive-to-negative samples on performance of SyFFM
To test the impact of different proportions of positive and negative samples on the performance of SyFFM, we randomly selected different numbers of negative samples from all non-approved drug pairs. AUC, AUPR and F1 score are used and the result is shown in Table 2, which means that the performance of SyFFM is getting worse as the number of negative samples increased. According to the above analysis, we set the ratio of positive-to-negative to be 1:1.
Table 2.
The performance of SyFFM of different ratios of positive-to-negative samples.
| Ratio of positive-to-negative | AUC | AUPR | F1 score |
|---|---|---|---|
| 1:1 | 0.925 | 0.934 | 0.7611 |
| 1:2 | 0.8773 | 0.8287 | 0.7405 |
| 1:3 | 0.8318 | 0.7629 | 0.7162 |
| 1:4 | 0.7435 | 0.6959 | 0.6963 |
| 1:5 | 0.7377 | 0.6976 | 0.6759 |
3.3. Prediction on three-drug synergistic combinations
We also evaluated the performance of SyFFM on 295 approved three-drug synergistic combinations collected from DCDB, including 368 drugs. Each three-drug combination was regarded as three pairwise drug combination, for example, three-drug combination was regarded as , and . If is a synergistic combination, then three pairwise combinations are treated as synergistic combinations (we record this hypothesis as ). In this way, we converted three-drug synergistic combinations prediction to pairwise synergistic drug combinations prediction, and the three-drug combination is predicted as synergistic if and only if all the corresponding three pairwise drug combinations are predicted as synergistic combinations.
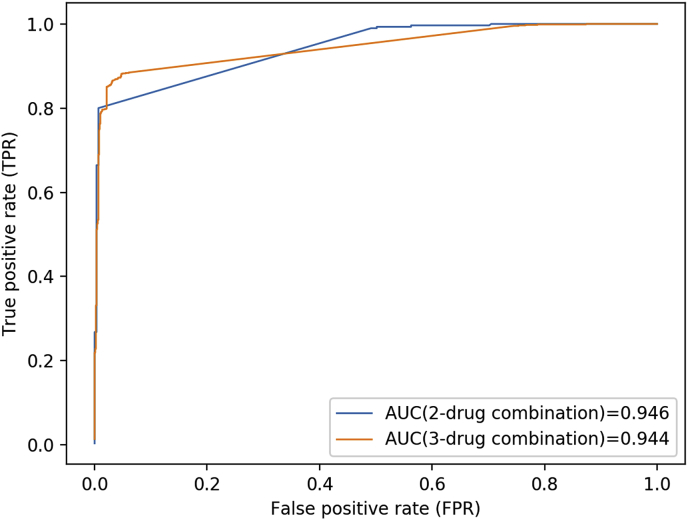
By decomposing entire three-drug combinations of 368 drugs into pairwise combinations, we implemented SyFFM and the pairwise combinations were predicted. Then three-drug combinations could be predicted by screening the corresponding pairwise combinations. We got AUC of 0.946 in terms of 5-fold cross validation on the entire pairwise combinations (Fig. 2(blue curve)) and AUC of 0.944 on the entire three-drug combinations (Fig. 2(yellow curve)).
Fig. 2.
The results of SyFFM on three-drug synergistic combinations.
3.4. Analysis of feature vectors of drug pairs
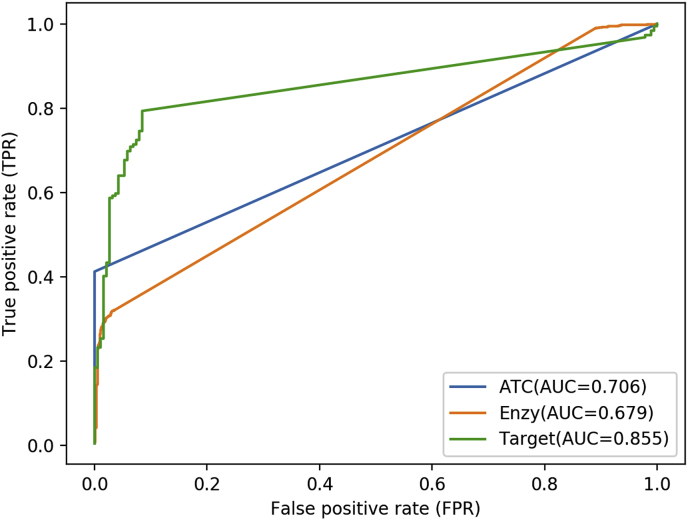
In order to verify the influences of features defined in our method, we used target pairs , enzyme pairs or ATC pairs to be the feature vector of drug pair . Then, field-aware factorization machines were implemented on pairwise drug combinations using the new feature vectors respectively. The average AUCs achieved using new feature vectors were 0.706, 0.679 and 0.855, respectively, which was shown in Fig. 3. The performance of our method was not satisfying in terms of AUCs under 5-fold cross validation, which indicates that interactions between features from different fileds play important roles in synergistic mechanisms.
Fig. 3.
The results of SyFFM using three new feature vectors.
3.5. Analysis of parameters of SyFFM
The coefficients in function ϕ indicate the contribution values of the corresponding features or feature interactions. Incorporating biological knowledge, we analyzed coefficients obtained on the prediction of pairwise drug combinations.
We sorted coefficients of single features (i.e. the first order coefficients w) in descending way and found that the features corresponding to top-10 coefficients are all target pairs. The result is shown in Table 3.
Table 3.
Features and approved synergistic drug combinations corresponding to top-10 coefficients.
| Target 1 | Target 2 | Approved synergistic combination | Disease |
|---|---|---|---|
| Progesterone receptor | Estrogen receptor | Estrogens, conjugated; Medroxyprogesterone acetate | Symptoms of menopause [18] |
| Ethinyl Estradiol; Norethindrone | Contraception [1] | ||
| Angiotensin-converting enzyme (ACE) | Type-1 angiotensin II receptor (AR) | Lisinopril; Losartan | Kidney Diseases [17] |
| DDP-4 | PPAR-γ | Rosiglitazone; Saxagliptin | Diabetes Mellitus, Type 2 [23,32,33] |
| Alogliptin; Pioglitazone | Type 2 Diabetes Mellitus [14,48] | ||
| Glucocorticoid receptor | Beta-2 adrenergic receptor | Fluticasone Propionate; Formoterol | Asthma, Bronchial [5,13,42] |
| Budesonide; Formoterol | Chronic Obstructive Pulmonary Disease; Asthma; Bronchiectasis [46,59] | ||
| 5′-AMPK | DDP-4 | Alogliptin benzoate; Metformin Hydrochloride | Type 2 Diabetes Mellitus [2] |
| Metformin; Sitagliptin | Type 2 diabetes who do not use daily insulin injections; Diabetes Mellitus, Type 2 [57] | ||
| DNA | Ribonucleoside-diphosphate reductase subunit M2 | Fluorouracil; SC144 | Colorectal cancer [40] |
| Renin | AR | Aliskiren; Irbesartan | Hypertension [40] |
| Progesterone receptor | GnRH | Leuprorelin acetate; Acetynone acetonidone | For initial management of the painful symptoms of endometriosis and for management of recurrence of symptoms [40] |
| Follicle-stimulating hormone receptor | GnRH-R | Buserelin; Pregnyl | Ovarian Hyperstimulation [25] |
| Angiotensin-converting enzyme | Voltage-dependent calcium channel gamma-1 subunit | None approved in DCDB |
It is shown from the table that these target pairs are keys to the mechanism of synergy. Since a target is involved in different biological pathways [9,19], the multiple interactions between different targets may be applied in treatment of other diseases. For example, the activation of progesterone receptor can also inhibit the growth of breast tumors by changing the synthesis of estrogen receptor chromatin [49,53]; both ACE and AR play an important roles in the regulation of plasma, and the American Diabetes Association also suggested using ACE inhibitors or AR inhibitors to treat diabetes [22,47]; sitagliptin, inhibitors of DDP-4, can regulate the expression of PPAR and may have a protective effect on myocardial cells [35]; both 5′-AMPK and DDP-4 are targets of polycystic ovary syndrome [55]. These findings show that our model finds important features of synergism and may help to explain the synergistic mechanism between two drugs.
3.6. Comparison with previously methods for predicting synergistic drug combination
Using DCDB data, we compared the prediction performance between SyFFM and Bai's method proposed in Ref. [4]. Bai's method is a synergistic drug combination prediction model by applying a novel improved naïve Bayesian algorithm on various types of features (targets, side effects, pathways, enzymes, transporters). The comparison result of leave-one-out cross validation is shown in Table 4.
Table 4.
Comparison result of SyFFM and Bai's method using leave-one-out cross validation.
| Method | Precision | Recall | F1 score | Accuracy |
|---|---|---|---|---|
| SyFFM | 0.9983 | 0.4573 | 0.9213 | 0.9255 |
| Bai's method with targets | 0.9008 | 0.5000 | 0.6431 | 0.7034 |
| Bai's method with side effects | 0.6957 | 0.6400 | 0.6667 | 0.6800 |
| Bai's method with pathways | 0.6133 | 0.6216 | 0.6174 | 0.6238 |
| Bai's method with enzymes | 0.6018 | 0.8095 | 0.6904 | 0.6115 |
| Bai's method with transporters | 0.4815 | 0.7500 | 0.5865 | 0.5339 |
4. Conclusions
By representing drug combinations as combinations of their pharmacological characteristics, we proposed a computational synergistic drug combination prediction approach, namely SyFFM. The features of drug combinations are constructed by integrating pharmacologic data such as drug targets and enzymes, and the synergistic scores between drugs are obtained by implementing field-aware factorization machines. SyFFM achieves good performances by predicting pairwise synergistic drug combinations and three-drug synergistic combinations, and plenty of prediction results are validated by various databases and literature. We demonstrated that SyFFM provides biological explanations for the synergistic principles underlying combinatorial therapy and offers a new method for studies on synergistic mechanisms.
However, several potential limitations exist in SyFFM. Firstly, the performance of model is limited due to the incompletion data about synergistic drug combinations. Secondly, SyFFM relies on the feature patterns enriched in synergistic combinations and other feature construction approaches may be more preferable to be adopted. It is worth noting that the strict hypothesis would introduce unreasonable bias for n-drug combinations with n increases and the computation complexity would present to be an exponential growth. So we only test SyFFM on pairwise and three-drug combinations, and the application of SyFFM to multi-drug synergistic combinations still has to be improved further. How to build a better synergistic drug combinations prediction model by utilizing diverse knowledge on drugs remains be an challenge for future research.
Acknowledgements
We thank anonymous reviewers for valuable suggestions and funding support by National Natural Science Foundation of China under Grant No.11631014.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2018.10.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Archer D.F., Nakajima S.T., Sawyer A.T., Wentworth J., Trupin S., Koltun W.D., Gilbert R.D., Ellman H. Norethindrone acetate 1.0 milligram and ethinyl estradiol 10 micrograms as an ultra low-dose oral contraceptive. Obstet Gynecol. 2013;122:601–607. doi: 10.1097/AOG.0b013e3182a1741c. [DOI] [PubMed] [Google Scholar]
- 2.Aschner P., Chan J., Owens D., Picard S., Wang E., Dain M., Pilorget V., Echtay A. Insulin glargine versus sitagliptin in insulin-naive patients with type 2 diabetes mellitus uncontrolled on metformin (easie): a multicentre, randomised open-label trial. Lancet. 2012;379:2262–2269. doi: 10.1016/S0140-6736(12)60439-5. [DOI] [PubMed] [Google Scholar]
- 3.Ballantyne C.M., Abate N., Yuan Z., King T.R., Palmisano J. Dose-comparison study of the combination of ezetimibe and simvastatin (Vytorin) versus atorvastatin in patients with hypercholesterolemia: the Vytorin versus Atorvastatin (VYVA) study. Am Heart J. 2005;149(3):464–473. doi: 10.1016/j.ahj.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Bai L.Y., Dai H., Xu Q. Prediction of effective drug combinations by an improved Na'´ie Bayesian algorithm. Int J Mol Sci. 2018;19(2):467. doi: 10.3390/ijms19020467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodzenta-Lukaszyk A., van Noord J., McAulay K., McIver T. Efficacy and safety profile of fluticasone/formoterol combination therapy compared to its individual components administered concurrently in asthma: a randomised controlled trial. Curr Med Res Opin. 2013;29:579–588. doi: 10.1185/03007995.2013.772506. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y.W., Hsieh C.J., Chang K.W. Training and testing low-degree polynomial data mappings via linear SVM. J Mach Learn Res. 2010;11(11):1471–1490. [Google Scholar]
- 8.Chen D., Liu X., Yang Y., Yang H., Lu P. Systematic synergy modeling: understanding drug synergy from a systems biology perspective. BMC Syst Biol. 2015;9(1):56. doi: 10.1186/s12918-015-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Li B.Q., Zheng M.Y., Zhang J., Feng K.Y., Cai Y.D. Prediction of effective drug combinations by chemical interaction, protein interaction and target enrichment of KEGG pathways. BioMed Res Int. 2013:1–10. doi: 10.1155/2013/723780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X., Ren B., Chen M., Liu M.X., Ren W., Wang Q.X. ASDCD: antifungal synergistic drug combination database. PloS One. 2014;9(1) doi: 10.1371/journal.pone.0086499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Ren B., Chen M., Wang Q., Zhang L., Yan G. NLLSS: predicting Synergistic Drug Combinations Based on Semi-supervised Learning. PLoS Comput Biol. 2016;12(7) doi: 10.1371/journal.pcbi.1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 13.Corren J., Mansfield L., Pertseva T., Blahzko V., Kaiser K. E—fficacy and safety of fluticasone/formoterol combination therapy in patients with moderate-to-severe asthma. Respir Med. 2013;107:180–195. doi: 10.1016/j.rmed.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 14.DeFronzo R., Burant C., Fleck P., Wilson C., Mekki Q., Pratley R. Efficacy and tolerability of the dpp-4 inhibitor alogliptin combined with pioglitazone, in metformin-treated patients with type 2 diabetes. J Clin Endocrinol Metabol. 2011;97:1615–1622. doi: 10.1210/jc.2011-2243. [DOI] [PubMed] [Google Scholar]
- 15.Dickson M., Gagnon J.P. The cost of new drug discovery and development. Discov Med. 2009;4(22):172–179. [PubMed] [Google Scholar]
- 16.Foucquier J., Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3(3) doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fried L., Emanuele N., Zhang J., Brophy M., Conner T., Duckworth W., Leehey D., McCullough P. Combined angiotensin inhibition for the treatment of diabetic nephropathy. N Engl J Med. 2013;369:1892–1903. doi: 10.1056/NEJMoa1303154. [DOI] [PubMed] [Google Scholar]
- 18.Gambacciani M., Ciaponi M., Cappagli B., Genazzani A.R. Effects of low-dose continuous combined conjugated estrogens and medroxyprogesterone acetate on menopausal symptoms, body weight, bone density, and metabolism in postmenopausal women. Am J Obstet Gynecol. 2001;185:1180–1185. doi: 10.1067/mob.2001.117669. [DOI] [PubMed] [Google Scholar]
- 19.Goto S., Okuno Y., Hattori M., Nishioka T., Kanehisa M. LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res. 2002;30(1):402–404. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng J., Zhang W., Zhang X., Han F., Li X., He X., Li Q., Chen J. Effect of angiotensin-converting enzyme inhibitors and angiotensin II receptor Blockers on all-cause mortality, cardiovascular deaths, and cardiovascular events in patients with diabetes mellitus. JAMA Internal Med. 2014;174:773–785. doi: 10.1001/jamainternmed.2014.348. [DOI] [PubMed] [Google Scholar]
- 23.Hollander P., Li J., Frederich R., Allen E., Chen R. Safety and efficacy of saxagliptin added to thiazolidinedione over 76 weeks in patients with type 2 diabetes mellitus. Diabetes Vasc Dis Res. 2011;8:125–135. doi: 10.1177/1479164111404575. [DOI] [PubMed] [Google Scholar]
- 24.Hopkins A.L. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 25.Humaidan P., Polyzos N., Alsbjerg B., Erb K., Mikkelsen A., Elbaek H., Papanikolaou E., Andersen C. Gnrha trigger and individualized luteal phase hcg support according to ovarian response to stimulation: two prospective randomized controlled multi-centre studies in ivf patients. Hum Reprod. 2013;28:2511–2521. doi: 10.1093/humrep/det249. [DOI] [PubMed] [Google Scholar]
- 26.Iwata H., Sawada R., Mizutani S., Kotera M., Yamanishi Y. Large-scale prediction of beneficial drug combinations using drug efficacy and target profiles. J Chem Inf Model. 2015;55(12):2705–2716. doi: 10.1021/acs.jcim.5b00444. [DOI] [PubMed] [Google Scholar]
- 28.Jia J., Feng Zh, Xiaohua Ma. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev. 2009;2(8):111. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 30.Juan Y.C., Zhuang Y., Chin W.S. Field-aware factorization machines for CTR prediction. ACM Conf Recommend Syst. 2016:43–50. [Google Scholar]
- 32.Karyekar C., Donovan M., Allen E., Fleming D. Efficacy and safety of saxagliptin combination therapy in us patients with type 2 diabetes. PGM (Postgrad Med) 2011;123:63–70. doi: 10.3810/pgm.2011.07.2305. [DOI] [PubMed] [Google Scholar]
- 33.Karyekar C., Frederich R., Ravichandran S. Clinically relevant reductions in HbA1c without hypoglycaemia: results across four studies of saxagliptin. Int J Clin Pract. 2013;67:759–767. doi: 10.1111/ijcp.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo K., Mastumoto Y. 2003. Fast methods for kenel-based text analysis in proceedings of the 41st annual meeting of the association of computational linguistics. [Google Scholar]
- 35.Lee T.I., Kao Y.H., Chen Y.C., Huang J.H., Hsu M.I., Chen Y.J. The dipeptidyl peptidase-4 inhibitor-sitagliptin modulates calcium dysregulation, inflammation, and ppars in hypertensive cardiomyocytes. Int J Cardiol. 2013;168:5390–5395. doi: 10.1016/j.ijcard.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 36.Li J., Zheng S., Chen B., Butte A.J., Swamidass S.J., Lu Z. A survey of current trends in computational drug repositioning. Briefings Bioinf. 2016;17(1):2–12. doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X., Xu Y., Hui C. Prediction of synergistic anti-cancer drug combinations based on drug target network and drug induced gene expression profiles. Artif Intell Med. 2017 Nov;83:35–43. doi: 10.1016/j.artmed.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Ligeti B., Vera R., Lukács G., Gyorffy B., Pongor S. Predicting effective drug combinations via network propagation. IEEE Biomed Circ Syst Conf (BioCAS) 2013:78–381. [Google Scholar]
- 40.Liu Y., Hu B., Fu C., Chen X. DCDB: drug combination database. Bioinformatics. 2010;26(4):587–588. doi: 10.1093/bioinformatics/btp697. [DOI] [PubMed] [Google Scholar]
- 41.Liu L., Chen L., Zhang Y.H. Analysis and prediction of drug-drug interaction by minimum redundancy maximum relevance and incremental feature selection. J Biomol Struct Dyn. 2017;35(2):312–329. doi: 10.1080/07391102.2016.1138142. [DOI] [PubMed] [Google Scholar]
- 42.Bodzenta-Lukaszyk A., Pulka G., Dymek A., Bumbacea D., McIver T. Efficacy and safety of fluticasone and formoterol in a single pressurized metered dose inhaler. Respir Med. 2011;105:674–682. doi: 10.1016/j.rmed.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Noonan M., Rosenwasser L., Martin P. Efficacy and safety of budesonide and formoterol in one pressurised metered-dose inhaler in adults and adolescents with moderate to severe asthma: a randomised clinical trial. Drugs. 2006;66:2235–2254. doi: 10.2165/00003495-200666170-00006. [DOI] [PubMed] [Google Scholar]
- 47.Hamon M., Amant C., Bauters C., Richard F., Helbecque N., Fadden E. Association of angiotensin converting enzyme and angiotensin II type 1 receptor genotypes with left ventricular function and mass in patients with angiographically normal coronary arteries. Heart. 1997;77:502–505. doi: 10.1136/hrt.77.6.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratley R., Reusch J., Fleck P., Wilson C., Mekki Q. Efficacy and safety of the dipeptidyl peptidase-4 inhibitor alogliptin added to pioglitazone in patients with type 2 diabetes: a randomized, double-blind, placebocontrolled study. Curr Med Res Opin. 2009;25:2361–2371. doi: 10.1185/03007990903156111. [DOI] [PubMed] [Google Scholar]
- 49.Sofi G.N., Sofi J.N., Nadeem R., Shiekh R.Y., Khan F.A., Sofi A.A., Bhat H.A., Bhat R.A. Estrogen receptor and progesterone receptor status in breast cancer in relation to age, histological grade, size of lesion and lymph node involvement. Asian Pac J Cancer Prev APJCP. 2012;13:5047–5052. doi: 10.7314/apjcp.2012.13.10.5047. [DOI] [PubMed] [Google Scholar]
- 51.Sun Y., Xiong Y., Xu Q., Wei D. A hadoop-based method to predict potential effective drug combination. BioMed Res Int. 2014:1–15. doi: 10.1155/2014/196858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas C., Gustafsson J.A. Progesterone receptor-estrogen receptor crosstalk: a novel insight. Trends Endocrinol Metabol: TEM. 2015;26:453–454. doi: 10.1016/j.tem.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Fu X., Xu J., Wang Q., Kuang H. Systems pharmacology to investigate the interaction of berberine and other drugs in treating polycystic ovary syndrome. Sci Rep. 2016;6:28089. doi: 10.1038/srep28089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams-Herman D., Johnson J., Teng R., Luo E., Davies M., Kaufman K., Goldstein B. Efficacy and safety of initial combination therapy with sitagliptin and metformin in patients with type 2 diabetes: a 54week study. Curr Med Res Opin. 2009;25:569–583. doi: 10.1185/03007990802705679. [DOI] [PubMed] [Google Scholar]
- 59.Worth H., Fórster K., Eriksson G., Nihlén U., Peterson S. Budesonide added to formoterol contributes to improved exercise tolerance in patients with copd. Respir Med. 2010;104:1450–1459. doi: 10.1016/j.rmed.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 62.Zhao X.M., Iskar M., Zeller G., Kuhn M., Van Noort V., Bork P. Prediction of drug combinations by integrating molecular and pharmacological data. PLoS Comput Biol. 2011;7(12) doi: 10.1371/journal.pcbi.1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou X.J., Zhou Y.H., Chen X.H., Qian W.B. Homoharringtonine combined arsenic trioxide induced apoptosis in human multiple myeloma cell line RPMI 8226: an experimental research. Chin J Integrated Tradit West Med. 2013;33(6):834–839. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.