Abstract
Extracellular vesicles (EVs) carry various molecules involved in intercellular communication and have raised great interest as drug delivery systems. Several engineering methods have been investigated for vesicle loading. Here, we studied the electroporation of EVs isolated from plasma to load antitumor microRNAs (miRNAs). First, we optimized the transfection protocol using miRNA cel-39 by evaluating different parameters (voltage and pulse) for their effect on vesicle morphology, loading capacity, and miRNA transfer to target cells. When compared with direct incubation of EVs with miRNA, mild electroporation allowed more efficient loading and better protection of miRNA from RNase degradation. Moreover, electroporation preserved the naive vesicle cargo, including RNAs and proteins, and their ability to be taken up by target cells, supporting the absence of vesicle damage. EVs engineered with antitumor miRNAs (miR-31 and miR-451a) successfully promoted apoptosis of the HepG2 hepatocellular carcinoma cell line, silencing target genes involved in anti-apoptotic pathways. Our findings indicate an efficient and functional miRNA encapsulation in plasma-derived EVs following an electroporation protocol that preserves EV integrity.
Keywords: exosome, electroporation, vesicle engineering, drug delivery, miRNA loading, miRNA enrichment, miR-31, miR-451a, hepatocarcinoma, cancer therapy
Graphical Abstract
Introduction
Extracellular vesicles (EVs) are a heterogeneous population of membrane-surrounded particles released by all virtually living cells. They primarily include microvesicles, released through budding of plasma membrane, and exosomes derived from the endosomal compartment.1 EVs are fundamental players in cell-to-cell communication, being natural carriers of a complex cargo that includes proteins, lipids, and nucleic acids. The discovery of EVs as natural vehicles of functional nucleic acids has raised great interest in their use as drug delivery carriers for gene therapy.2 In particular, increasing efforts have been made to exploit EVs as carriers of microRNAs (miRNAs). These molecules are well-known crucial regulators of biological processes and represent promising powerful tools for therapeutic interventions against several diseases. Negatively regulating the expression of multiple target genes, miRNAs modulate several processes, including cell cycle, apoptosis, migration, inflammation, and angiogenesis.3 miRNA dysregulation is actively involved in cancer pathogenesis and progression, representing a promising targeting for therapeutic strategies. In this setting, miRNAs function either as oncomiRs, promoting cancer growth, or as tumor suppressors, inhibiting cancer development.4 It has been shown that miRNA alterations robustly contribute to the progression of hepatocellular carcinoma (HCC), one of the most flagrantly aggressive and invasive human cancers.5 Several tumor suppressor miRNAs were demonstrated to inhibit HCC progression, such as miR-337, miR-214-5p, miR-31, miR-223, miR-451a, and miR-199a-3p.6, 7, 8, 9, 10, 11 However, miRNAs are susceptible to environmental degradation in the human body, with the consequent loss of their biological activity. This remains a major obstacle to their clinical applicability.12 Thus, EVs represent a promising vehicle for therapeutic miRNA delivery because they are easily packaged in EVs because of their small size and are protected from enzymatic degradation.13
In recent years, evidence has demonstrated the feasibility of engineering EVs to deliver therapeutic molecules.14 Different methods of nucleic acid loading in EVs have been investigated, including passive loading such as incubation or active loading such as electroporation, sonication, transfection, extrusion, saponin permeabilization, and hypotonic dialysis.15, 16, 17, 18 Recently, Zhang et al.18 showed successful miRNA loading following a calcium chloride-mediated transfection or electroporation in EVs for in vitro and in vivo delivery. However, transfection has been reported to be associated with the presence of contaminating transfection reagents in samples.19 Electroporation was shown to be superior in EV loading in comparison with sonication, incubation, extrusion, saponin permeabilization, and hypotonic dialysis.16, 20 Electroporation of plasma-derived EVs has been successfully investigated to transfer small interfering RNA (siRNA) in monocytes and lymphocytes.21 On the other hand, incubation was reported to maintain EV size and morphology better than sonication and freeze-thaw cycles.22 However, recent evidence suggests that EV electroporation for loading of nucleic acids may be inefficient because of protocol variability and molecular aggregation.23, 24 In fact, the formation of aggregates of insoluble nucleic acids could reduce their uptake and activity, leading to inefficacy of loaded EVs on target cells.23, 25 Moreover, electroporation may trigger the aggregation of EVs and change their morphological features, highlighting the importance of electroporation protocol optimization to allow efficient particle engineering.24, 26, 27
In this study, we compared miRNA loading of EVs from human plasma of healthy donors using electroporation and incubation protocols. We optimized the electroporation protocol by evaluating different parameters (voltage and pulse) on vesicle morphology, loading capacity, protection from enzymatic degradation, loss of EV endogenous content, and miRNA transfer to target cells. Moreover, we evaluated the functional delivery of selected miRNAs by engineering EVs with antitumor miRNAs to promote apoptosis of HepG2 HCC cell line.
Results
Optimization of Electroporation Protocol to Load miRNA in EVs
We used, as a potential vehicle for miRNA delivery, EVs isolated from human plasma of healthy donors because they were per se ineffective on target cells used in this study. To define the most efficient electroporation protocol, we electroporated EVs (EVe) with different electroporation parameters, using different voltages (500, 750, and 1,000 V) and different numbers of pulses (1 or 10 pulses) of 20 ms. We engineered EVs with a synthetic miRNA (cel-39) derived from Caenorhabditis elegans, which is easily detectable in human EVs and cells. Electroporation was compared with simple incubation of EVs and miRNAs (EVi+miRNA).
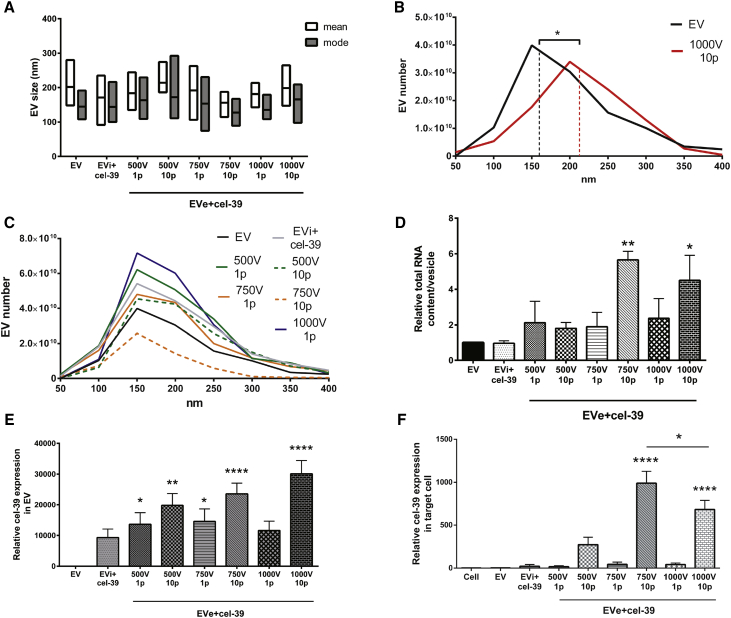
Because electrical fields can induce EV aggregation, EVs were examined using NanoSight (NTA) (Figures 1A–1C) after electroporation.24 The analysis of EV size, as mean and mode parameters, among all electroporation protocols did not show differences between electroporated and control EVs (Figure 1A). However, deeper investigation of EV size distribution revealed a shift of EV profile after electroporation with the protocol using 1,000 V and 10 pulses, with a significant increase in median size compared with control EVs (Figure 1B). This shift was not observed with other electroporation protocols (Figure 1C).
Figure 1.
Comparison of Electroporation Parameters to Load miRNA cel-39 in EVs
EVs derived from plasma were electroporated (EVe) with different parameters, voltages and pulses, or simply incubated (EVi) with miRNA cel-39. Size analysis of engineered EVs by NanoSight measured the mean and mode size (A) and the particle size distribution in EV population (B and C) in all samples. The median size of control EVs (EV) and EVs electroporated using 1,000 V and 10 pulses (1,000 V 10 p) is shown in EV size distribution (B). The size profile of control EVs and all other engineering protocols is also shown (C). (D) After engineering, total RNA was extracted from all samples and quantified. The total RNA content in EVs was evaluated in comparison with naive EVs. ANOVA with Dunnett’s multiple-comparisons test (n = 3). (E) qRT-PCR analysis of engineered miRNA cel-39 relative expression in EVs following different loading protocols. Kruskal-Wallis ANOVA with Dunn’s multiple-comparisons test versus control EVs (n = 3). (F) miRNA cel-39 expression in target TEC cells after 24 h of treatment with engineered EVs (2.5 × 109 EVs/mL). miRNA cel-39 RQ value is shown in recipient cells comparing samples with untreated cells (Cell). ANOVA with Tukey’s multiple comparisons test (n = 3). EVs were loaded with a miRNA dose of 10 pmol. Data are expressed as mean ± SEM. *p < 0.05, **p < 0.01, and ****p < 0.001.
To assess the efficiency of loading protocols, we evaluated the accumulation of the miRNA cel-39 in EVs. First, the total RNA content analysis revealed a significantly increase when EVs were electroporated with higher voltages (750 and 1,000 V) and 10 pulses compared with control EVs. In particular, the setting with 750 V and 10 pulses allowed major RNA enrichment (Figure 1D). The measurement of miRNA cel-39 expression by qRT-PCR showed high miRNA loading in EVs using all protocols (Figure 1E). Unlike incubation, almost all electroporation protocols induced significant miRNA cel-39 accumulation, with the best efficiency using higher voltages (750 and 1,000 V) and 10 pulses. Next, we compared all protocols evaluating the capacity of engineered EVs to transfer the exogenous miRNA in target cells. miRNA expression was measured in tumor endothelial cells (TECs) after 24 h of EV treatment using qRT-PCR assay. As shown in Figure 1F, electroporation protocols with higher voltages (750 or 1,000 V) and more pulses allowed significant accumulation of miRNA cel-39 in recipient cells. Interestingly, the middle electroporation setting (750 V and 10 pulses) induced major miRNA transfer rather than the highest protocol (1,000 V and 10 pulses), indicating its superiority in EV functional loading.
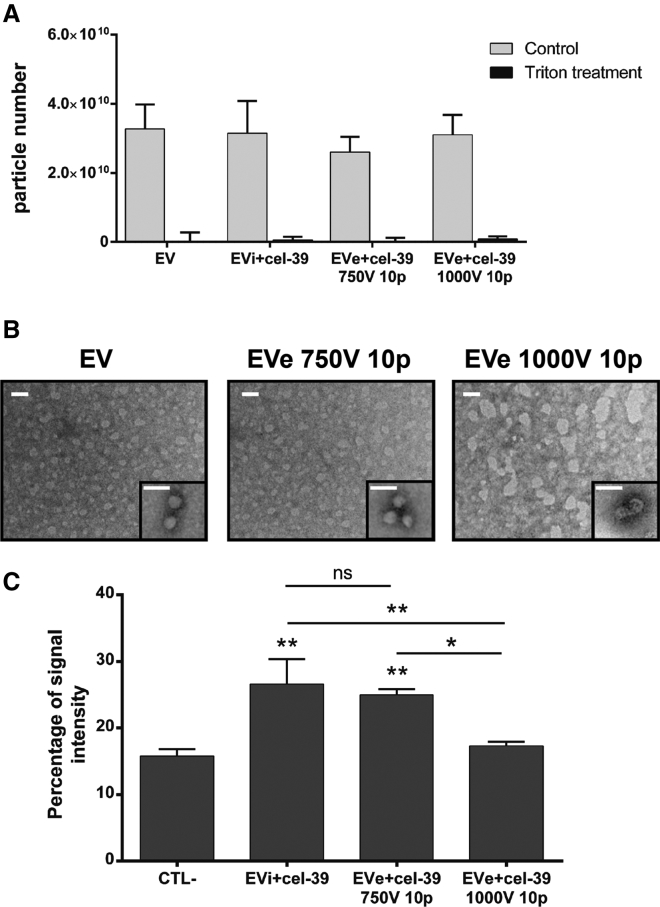
Because reduced miRNA transfer may be due to miRNA aggregation or EV damage, we more closely investigated the 750 or 1,000 V and 10 pulse settings. NanoSight analysis demonstrated that both electroporation protocols did not increase particle number in comparison with control EVs and that there was clear EV degradation after treatment with 0.1% Triton, indicating the absence of miRNA aggregates (Figure 2A). Transmission electron microscopy analysis showed a spheroid morphology of purified EVs and EV damage after the electroporation with 1,000 V and 10 pulses but not with the mild protocol (750 V and 10 pulses) (Figure 2B). EV alteration following the 1,000 V and 10 pulse protocol was also correlated to reduced uptake by target cells in comparison with incubation and the mild electroporation protocols (Figure 2C). On the basis of these results, the mild electroporation protocol using 750 V and 10 pulses was considered the most suitable and efficient method and was selected for subsequent experiments.
Figure 2.
Evaluation of miRNA Aggregates and EV Damage following Different Electroporation Settings
(A) EVs and miRNA cel-39 were incubated (EVi+cel-39) or electroporated (EVe+cel-39) using 750 or 1,000 V and 10 pulse settings. NanoSight analysis of control EVs or engineered EVs treated or not with 0.1% Triton X-100 (n = 3). (B) Representative transmission electron microscopy of control EVs, EVs electroporated with 750 V and 10 pulse setting (EVe 750V 10p) or 1,000 V and 10 pulse setting (EVe 1,000V 10p). Three experiments were performed with similar results (scale bar, 100 nm). (C) EVs labeled with a PKH-26 dye for membrane staining were engineered with miRNA cel-39 and used to stimulate target TEC cells for 24 h (4.2 × 109 EVs/mL). EV uptake was evaluated using FACS as percentage of EV membrane-staining signal compared with untreated cells (CTL−) (n = 3). ANOVA with Turkey’s multiple-comparisons test. *p < 0.05 and **p < 0.01.
Finally, we determined loading efficiency using a calibration curve, comparing the miRNA amount used to engineer EVs and that detected in EVs after engineering. The optimized electroporation protocol (750 V and 10 pulses) allowed efficiency of 31.63 ± 5.94% in comparison with the simple incubation of about 7.97 ± 4.27%, corresponding to a mean of 21.09 ± 3.96 or 5.31 ± 2.85 molecules/EV, respectively.
Engineered EV Incorporation into Target Cells and miRNA Protection from Enzymatic Digestion
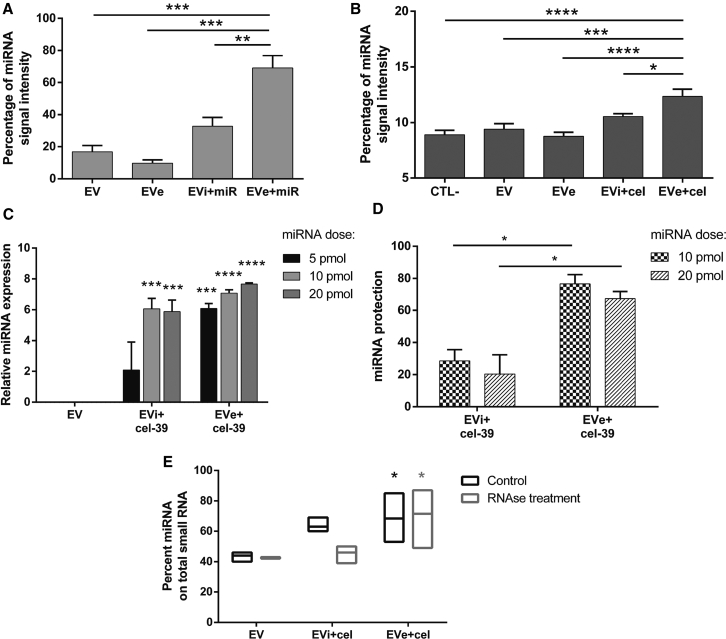
Successful miRNA loading was confirmed through fluorescence-activated cell sorting (FACS) analysis. To this purpose, EVs were engineered with an Alexa Fluor 555-labeled control miRNA. The measurement of signal intensity revealed significant miRNA enrichment following electroporation in EVs (EVe+miR), in comparison with naive EV, EVs electroporated alone (EVe), and EVs incubated with miRNA (EVi+miR) (Figure 3A). Next, we also evaluated engineered miRNA uptake in target TECs after 24 h of treatment using FACS analysis (Figure 3B). miRNA transfer to cells reflected direct loading in EVs, with more significant miRNA transfer after the electroporation protocol in comparison with incubation.
Figure 3.
miRNA Loading in EVs and Protection from Enzymatic Degradation
EVs were engineered with an Alexa Fluor 555-labeled control miRNA (miR) to perform FACS analysis. (A) miRNA enrichment in EVs was measured as percentage of signal intensity in EVs electroporated or incubated with miRNA (EVe+miR and EVi+miR, respectively) in comparison with controls, naive EVs, and vesicles electroporated alone (EVe) (n = 3). ANOVA with Turkey’s multiple-comparisons test. (B) EVs electroporated or incubated with the miRNA were used to treat target TECs for 24 h (4.2 × 109 EVs/mL). Engineered miRNA transfer was measured as miRNA percentage signal intensity in untreated cells (CTL−), cells treated with control EVs, vesicles electroporated alone (EVe), and EVs electroporated or incubated with miRNA (EVe+miR and EVi+miR, respectively) (n = 5). ANOVA with Dunnett’s multiple-comparisons test versus EVe+miR. (C) EVs were engineered with different doses of miRNA cel-39 (5, 10, and 20 pmol), and miRNA loading was analyzed in qRT-PCR experiments. Data are represented as ln(RQ) of miRNA cel-39 in comparison with control EVs (n = 3). (D) Engineered EVs with miRNA cel-39 were treated with RNase A for 30 min at 37°C. miRNA protection was evaluated using qRT-PCR analysis as percentage of miRNA expression (ΔCt values) after RNase degradation in comparison with untreated samples. EVs engineered with 10 or 20 pmol miRNA dose were tested, and the different effect of electroporation and incubation was analyzed (n = 3). ANOVA with Turkey’s multiple-comparisons test. (E) EVs incubated or electroporated with miRNA cel-39 were treated or not with RNase and analyzed for their small RNA profile using Bioanalyzer in comparison with each control EV sample. miRNA enrichment was expressed as percentage of miRNA on total small RNA content. ANOVA with Dunnett’s multiple-comparisons test (n = 3). Data are expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.001.
To further evaluate miRNA loading, we measured the accumulation of the miRNA cel-39 in EVs using qRT-PCR with increasing miRNA doses (Figure 3C). We engineered the same number of EVs with different doses of miRNA cel-39: 5, 10, or 20 pmol. NanoSight analysis did not show an increase in particle number, suggesting an absence of miRNA aggregation with all miRNA doses (data not shown). Measurement of miRNA relative expression demonstrated more efficient loading in EVe+cel-39 than EVi+cel-39 for all tested doses. As expected, using the electroporation protocol, increasing miRNA dose clearly induced a correlated increase in loading. For the incubation protocol, in contrast, the increase in miRNA amount did not correspond to a loading increase, suggesting saturation of miRNA bound to EV surface.
EVs are extensively reported to protect their cargo from the microenvironmental degradation mediated by RNase enzymes. We tested loaded miRNA resistance to RNase A by comparing EVi+cel-39 and EVe+cel-39 (Figure 3D). Analysis of miRNA expression using qRT-PCR allowed the measurement of miRNA protection, as the amount of miRNA detected after RNase treatment in comparison with untreated samples. As shown in Figure 3D, miRNA electroporated in EVs was significantly more protected by enzymatic digestion compared with incubation, with a rate of about 70% in comparison with 25%, respectively. These data were also confirmed on bioanalyzer analysis (Figure 3E).
Taken together, these results confirmed the superiority of the electroporation protocol in EV loading and the protection of engineered miRNA in comparison with the incubation protocol.
Electroporation Does Not Affect EV Cargo
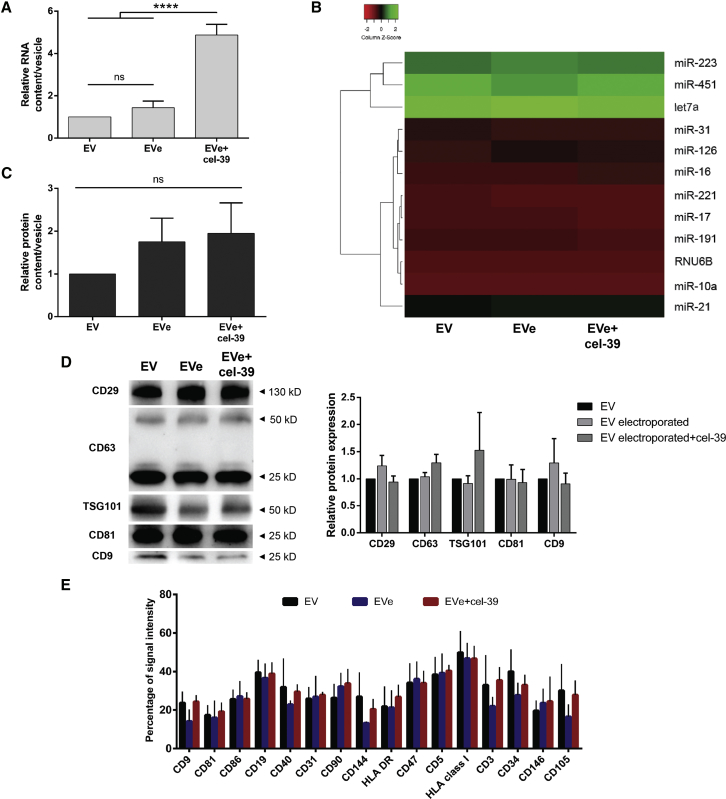
Permeabilization of EV membrane induced by electroporation may induce the leakage of molecules contained in EVs, altering their cargo and biological function. To determine whether electroporation could modify endogenous EV content, we analyzed RNA, miRNA, and protein cargo in control EVs and EVs electroporated in the presence or absence of miRNA cel-39 (EVe+cel-39 and EVe, respectively) (Figure 4).28 First, total RNA content was similar in EVs and EVe, suggesting the absence of RNA loss after engineering (Figure 4A). As expected, we detected a significant increase in the total RNA cargo in EVe+cel-39 compared with EVs and EVe (Figure 4A). Because the RNA cargo of EVs is various, we decided to evaluate the miRNA content because these small molecules are suitable to cross membrane pores during electroporation, both entering EVs, as miRNA cel-39, or exiting them. Thus, we performed qRT-PCR analysis of a panel of miRNAs expressed in plasma-derived EVs.29, 30, 31 Electroporation in the presence or absence of miRNA cel-39 did not significantly alter miRNA endogenous expression compared with EVs (Figure 4B), confirming the absence of RNA molecule loss after electroporation. Next, we evaluated EV protein content without observing a decrease of the total amount of protein associated with EVe and EVe+cel-39 (Figure 4C). To more closely investigate the EV protein cargo, evaluation of classical vesicular protein markers was performed using western blot (Figure 4D). Analysis of tetraspanins CD63, CD81, and CD9, the integrin β1 (CD29), and tumor susceptibility gene 101 (TSG101) among all samples indicated the absence of protein content alteration in EVe and EVe+cel-39 compared with EV. Finally, to evaluate whether electroporation could qualitatively alter protein composition of EV membrane markers, FACS analysis was performed. Our results did not show significant variation in the expression of surface markers between EVs, EVe, and EVe+cel-39 (Figure 4E), suggesting that electroporation preserves EV membrane protein composition.
Figure 4.
EV Content Analysis after Electroporation
EVs were electroporated alone or with the miRNA cel-39 (EVe and EVe+cel-39, respectively), and their endogenous cargo was compared with control EVs. (A) Total RNA contained in EVs was quantified, and the relative data are shown as nanograms in each EV compared with control EVs (n = 6). ANOVA with Turkey’s multiple-comparisons test. (B) Heatmap representation of the expression of a panel of miRNAs in all samples, using average linkage as clustering method and Euclidean distance measurement. miRNA relative expression was evaluated as 2−ΔCt values normalized to global miRNA expression of each sample (n = 3). (C) Total protein content in samples refers to micrograms of total protein in EVs normalized to control EVs (n = 4). ANOVA with Dunnett’s multiple-comparisons test. (D) Western blot analysis of integrin β1 (CD29), CD63, TSG101, CD81, and CD9 in all samples. Representative image (left) and quantitative analysis (right) (n = 3). (E) Surface markers were evaluated in FACS experiments. The percentage of signal intensity for each protein is shown in all samples (n = 3). Data are expressed as mean ± SEM. ****p < 0.001.
Taken together, these data demonstrated that electroporation did not modify endogenous EV cargo, including total RNAs, miRNAs, total proteins, and vesicular markers. Moreover, also the expression of surface markers was preserved after electroporation, supporting the safety of electroporation protocol on EV composition.
Biological Activity of EV Engineered with Antitumor miRNAs on HepG2
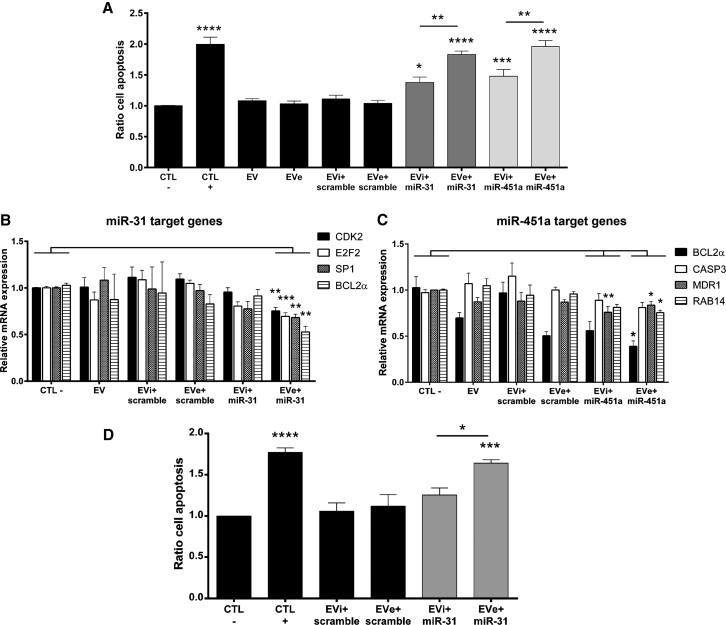
Next, we tested the functionality of engineered miRNAs to provide an antitumor effect of EVs. We engineered EVs with antitumor miRNAs and evaluated their capacity to induce apoptosis in a HCC cell line, HepG2 cells. For this purpose, we electroporated plasma-derived EVs, biologically inactive in our model, with two synthetic miRNAs (miR-451a and miR-31-5p) which we previously demonstrated to promote apoptotic signals in HepG2.32 We evaluated cell apoptosis after 24 h of treatment with EVs (2.5 × 109 EVs/mL), comparing the activity of EVs electroporated with an antitumor miRNA with controls: untreated cells (CTL−), naive EVs, EVs electroporated alone (EVe), EVs incubated with antitumor miRNA (EVi+miR-31/miR-451a), and EVs incubated and electroporated with a miRNA scramble (EVi+scramble and EVe+scramble) (Figure 5A). The apoptosis assay demonstrated a statistically significant increase of cancer cell apoptosis with EVs engineered with miR-31 and miR-451, with a higher effect following electroporation compared with the incubation protocol (Figure 5A). Moreover, the same amount of miRNA, both miR-31 and miR-451a, was ineffective to promote cancer cell apoptosis without EV engineering (Figure S1).
Figure 5.
Anti-cancer Effect of EVs Engineered with miRNAs
EVs loaded with two antitumor miRNAs (10 pmol), miR-31 and miR-451a, were used to stimulate HepG2 target cells for 24 h (2.5 × 109 EVs/mL). EVs were engineered using electroporation (EVe) and incubation (EVi) protocols. (A) Cell apoptosis was detected by Muse Annexin V kit, and total apoptosis is shown as a ratio in comparison with untreated cells (CTL−) (n = 4). ANOVA with Turkey’s multiple-comparisons test. (B and C) The effect on miRNA target genes in recipient cells was analyzed using qRT-PCR and is expressed as RQ value. For miR-31, the following genes were evaluated: CDK2, E2F2, SP1, and BCL2α (B). For miR-451a, the expression of BCL2α, CASP3, MDR1, and RAB14 genes was measured (C). Kruskal-Wallis ANOVA with Dunn’s multiple-comparisons test (n = 4). (D) The effect on cell apoptosis was also analyzed after RNase treatment of EVs engineered with miR-31. Total cell apoptosis was evaluated using Muse Annexin V kit and is expressed as a ratio in comparison with untreated cells (CTL−) (n = 4). ANOVA with Turkey’s multiple-comparisons test. In all experiments, the activity of electroporated EVs was compared with untreated cells (CTL−) and incubated EVs with the same miRNA. Cells were also treated with other control samples: control EVs, EVs electroporated alone (EVe), EVs incubated or electroporated with control miRNAs (scramble), and doxorubicin as positive apoptosis control (150 ng/mL) (CTL+). Data are expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.001.
To confirm the biological effect, we evaluated the ability of engineered EVs to silence target genes of miR-31 and miR-451. To this aim, we measured gene expression in recipient cells by qRT-PCR after 24 h of treatment with loaded EVs (Figures 5B and 5C). The treatment with EVe+miR-31 induced a significant downregulation of its target genes, including the cyclin-dependent kinase 2 (CDK2), the transcription factor E2F2, the Sp1 transcription factor (SP1), and the anti-apoptotic protein BCL2α (Figure 5B). In contrast, EVi+miR-31 did not induce significant gene inhibition (Figure 5B). For EVs engineered with miR-451, we analyzed the expression of BCL2α, caspase-3 (CASP3), the multidrug resistance protein 1 (MDR1), and the ras-related protein 14 (RAB14) genes (Figure 5C). The incubation protocol induced the significant reduction only of MDR1, whereas the electroporation protocol significantly downregulated BCL2α, MDR1, and RAB14. Although the change was not significant, EVe+miR-451a also reduced the expression of CASP3 (Figure 5C). Overall, molecular analysis of the effect of engineered EVs on target cells confirmed that the electroporation protocol was more efficient than incubation. Finally, we investigated the capacity of EVs engineered with a selected antitumor miRNA, miR-31, to maintain their anticancer effect after RNase treatment. Figure 5D shows cancer cell apoptosis after 24 h of EV stimuli treated with RNase A, demonstrating that the electroporation protocol maintained a statistically significant effect after enzymatic digestion that was superior to the incubation protocol.
Discussion
EVs are crucial mediators of cell-cell communication, and growing evidence suggests their promising use as drug delivery systems. This is particularly important for molecules with a difficult in vivo administration due to naive susceptibility to degradation and/or significant side effects. For instance, EVs have been successfully used to vehicle siRNA molecules directed against key genes for cancer therapy.16, 19 Numerous methods to load isolated EVs have been recently investigated, and several of them were proved effective. However, obstacles limit the application of EVs as a delivery system, such as engineering efficiency and EV damage. Electroporation is a common technique used to engineer cells that leads to higher EV loading capacity compared with other protocols. In a study by Fuhrmann et al.,20 EVs of different cellular origins (cancer, endothelial, and stem cells) were loaded with porphyrins using various engineering methods (electroporation, dialysis, saponin, and extrusion), and electroporation was proved to improve the encapsulation efficiency. In addition, EV electroporation was reported to efficiently vehicle therapeutic siRNA to treat lung injury and bladder cancer and miR-26a to reduce HepG2 proliferation.33, 34, 35
In the present study, we used electroporation to enrich EVs from plasma with miRNAs, comparing its efficiency with a simple incubation protocol. During electroporation, applied electric fields increase membrane permeability and induce the formation of temporary pores on the membrane lipid bilayer. The opening of membrane allows the entrance of exogenous molecules into EVs, but it may also account for the loss of endogenous molecules.36 However, EV electroporation is characterized by high variability, and numerous groups have recently investigated factors that can affect protocol efficacy. Transfection efficiency could be reduced by nucleic acid aggregation, EV aggregation, nucleic acid, and EV sizes.23, 24, 25 Interestingly, a recent study by Yang et al.27 highlighted how electroporation parameters such as intensity of voltage and number of pulses can affect the loading of synthetic particles. Thus, in this study, we tested different electroporation settings (500, 750, and 1,000 V and 1 or 10 pulses) to find the more efficient protocol to load plasma-derived EVs with miRNAs. We demonstrated that a higher voltage and a greater number of pulses induced increased miRNA loading in EVs, clearly superior to simple incubation. However, electroporation with the highest voltage (1,000 V and 10 pulses) damaged EVs. Thus, we selected the electroporation protocol with the medium voltage level (750 V) and the larger pulse number (10) as the most efficient and suitable. This protocol allowed efficient EV loading, with more than 30% of transfection yield, superior to other studies on EV electroporation (0.09%, 2%, >15%, and 20%).23, 25, 27, 37 Because different membrane composition can affect increased permeability during electroporation, we hypothesize that EVs isolated from different sources could be differentially sensible to electroporation, and the protocol should be optimized for each type of EV.
Next, to more closely investigate the potential miRNA loading in EVs, we electroporated the same amount of EVs with different miRNA doses. As expected, we detected higher miRNA loading with increasing miRNA doses (5, 10, and 20 pmol) whereas increasing doses in the incubation protocol did not increase miRNA associated with EVs, suggesting a binding saturation. An important feature of EVs as therapeutic carriers is their capacity to protect loaded miRNAs from enzymatic digestion.13 We found that electroporation was significantly superior to incubation in protecting miRNA, suggesting that most of it was encapsulated into EVs. The incubation, instead, provided only about 30% of miRNA protection, possibly because of the miRNA bound to RNA-binding protein on EV surface.38, 39
Membrane pores generated during electroporation could favor the leakage of endogenous EV content or modify the membrane composition, affecting the EV biological activity and target cell uptake. Previous studies analyzed EVs before and after electroporation, and Liang et al.35 showed the maintenance of CD63 expression after electroporation. With the electroporation protocol used in the present study, no significant loss of native molecules was observed. The absence of alteration in surface markers supported the maintenance of EV membrane composition, as confirmed also by their unaltered capacity to be taken up by target cells. In fact, tetraspanins (CD81 and CD9) and integrins (CD29) are reported to have a role in EV cell binding and uptake.40, 41, 42 The absence of EV content alteration could help characterize the quality of EV electroporation protocols.
miRNA administration has been demonstrated to restore the altered expression typical of cancers.43 Recent studies investigated the use of synthetic particles to vehicle miRNA as therapeutic drug to suppress HCC.44, 45, 46, 47, 48 Our group previously demonstrated that miRNAs contained in hepatic stem cell-derived EVs contribute to the suppression of HCC, including miR-31 and miR-451.29 Here, we engineered plasma-derived EVs with miR-31 and miR-451a, demonstrating their effectiveness in promoting HepG2 apoptosis. The effect of loaded miRNAs was also verified as transcription inhibition of their target genes. In particular, EVs electroporated with miR-31 significantly downregulated CDK2, typically overexpressed in HCC and implicated in the regulation of cell cycle and SP1, an important gene involved in the regulation of HepG2 apoptosis, proliferation, and invasion.49, 50 Moreover, EV-mediated delivery of miR-31 significantly downregulated the expression of E2F2, involved in cell proliferation, apoptosis, and migration in gastric cancer.51 Significant EV-mediated silencing was also observed for BCL2α, identified as miR-31 target by bioinformatic analysis and involved in the intrinsic apoptotic pathway.52 Similarly, EV electroporated with miR-451a directly targeted BCL2α, reducing also CASP3 expression and thereby playing an effective role in HepG2 apoptosis.53 In addition, these vesicles significantly inhibited miR-451a target RAB14, which mediates the effect of miRNA on the increased radio-sensitivity of nasopharyngeal carcinoma and the suppression of human non-small cell lung cancer, and it has been also involved in the suppression of breast cancer.54, 55, 56 Finally, EVs loaded with miR-451a efficiently suppressed the expression of MDR1, a key target gene that has been demonstrated to regulate chemosensitivity in several cancers, including HCC.57, 58, 59
In conclusion, we defined a protocol of electroporation suitable for engineering EVs derived from plasma. Plasma-derived EVs are a promising drug delivery system because they can be easily isolated from patients and used in an autologous manner. Moreover, we provided evidence for the effectiveness of engineered plasma-derived EVs in inducing HCC in vitro apoptosis by shuttling antitumor miRNAs.
Materials and Methods
Cell Culture
A human TEC was established and cultured in EndoGRO basal complete medium (Merck Millipore, Burlington, Massachusetts, USA) plus 10% fetal bovine serum (FBS). Briefly, TECs were isolated from renal clear-cell carcinomas and were characterized as endothelial cells by morphology, positive staining for von Willebrand factor (vWF) antigen, CD146, CD105, and vascular endothelial-cadherin and negative staining for desmin and cytokeratin, as previously described.60 Human hepatocellular carcinoma cell line HepG2 was purchased from ATCC (American Type Culture Collection, Manassas, Virginia, USA) and maintained in culture in low-glucose DMEM (EUROCLONE, Milan, Italy) containing 10% FBS.
EV Isolation
Plasma-derived EVs were isolated from frozen human plasma of healthy blood donors provided by the Blood Bank of Città della Salute e della Scienza di Torino. All samples were obtained after informed consent and approval by the internal review board of the Blood Bank. EVs from each donor were obtained from about 250 mL plasma bags and isolated by differential ultracentrifugation as previously described, with a recovery rate of about 5.33 × 109 ± 2.40 × 109 EVs/mL plasma.58 In brief, plasma samples were centrifuged at 1,500 × g for 20 min to remove debris, apoptotic bodies, and platelets. The supernatant was subsequently ultracentrifuged at 10,000 × g, followed by ultracentrifugation at 100,000 × g for 2 h at 4°C using a 70 mL polycarbonate tube (SW 45 Ti rotor, Beckman Coulter Optima L-90 K ultracentrifuge, Brea, California, USA). Samples were then washed with saline buffer solution and ultracentrifuged at 100,000 × g for 2 h at 4°C. EV pellets were then resuspended in saline buffer solution with 1% DMSO and stored at −80°C. EVs were then thawed and used for biological assays or molecular analysis. EVs were characterized as previously described by NanoSight analysis, expression by western blot of CD63, CD81, CD9 tetraspanins and TSG101 as positive markers and ribosomal protein S29 as negative marker, and by electron microscopy (data not shown).61 Each plasma bag was sufficient to prepare one set of control EVs and engineered EVs, and different plasma bags were used to verify donor variability for functional and loading experiments.
EV Analysis by NanoSight
EVs were analyzed by nanoparticle tracking analysis (NTA), using the NanoSight LM10 system(NanoSight, Salisbury, UK). To define EV concentration and size profile, the NanoSight system was equipped with a 405 nm laser and NTA 3.1 analytic software. The Brownian movements of EVs existing in each sample were subjected to a laser light source and were recorded by a camera. The analytic software converted this information into size and concentration parameters using the Stokes-Einstein equation. For each sample, three videos of 30 s duration were recorded, and camera levels were set for all the acquisition at 16. Briefly, EVs were diluted (1:1,000 for purified plasma-derived EVs and 1:200 for engineered EVs) in 1 mL vesicle-free saline solution (Fresenius Kabi, Bad Homburg vor der Höhe, Germany). NTA post-acquisition settings were optimized and maintained constant among all samples, and each video was then analyzed to measure EV mean, mode, median (D50), and concentration. For the analysis of the presence of miRNA aggregates, control or engineered EVs were treated with 0.1% Triton X-100 (Bio-Rad, Hercules, CA, USA) for 1 h at 37°C, and the number of particles was measured using NTA as described above.
FACS Characterization of EVs
Plasma-derived EVs were characterized by cytofluorimetric analysis using the CytoFLEX flow cytometer (Beckman Coulter) with CytExpert software. The following fluorescein isothiocyanate (FITC) or allophycocyanin (APC) conjugated antibodies were used: CD9, CD19, CD81, CD86, CD90, HLA DR, CD47, CD34 (BD Biosciences, Franklin Lakes, New Jersey, USA), CD40, CD31, CD144, CD3, CD146, CD105 (Miltenyi Biotec, Bergisch Gladbach, Germany), CD5 (Thermo Fisher Scientific, Waltham, Massachusetts, USA), and HLA class I (BioLegend, San Diego, California, USA). Conjugated mouse non-immune isotypic immunoglobulin G (IgG) (Miltenyi Biotec) was used as control. In brief, EVs (5 × 108 particles) were labeled for 15 min at 4°C with antibodies and immediately diluted 1:3 and acquired.62
EV Loading Protocols
EVs were engineered using electroporation performed on a Neon Transfection System (Thermo Fisher Scientific) following the manufacturer’s protocol as previously described.18 Briefly, EVs and miRNA were mixed, and the final volume was adjusted to 10 μL using the electroporation buffer. Ratios of 3 × 109 EVs and different miRNA doses were used: 5, 10, or 20 pmol. EVs were engineered with different miRNAs in selected experiments: cel-39-3p, miR-31-5p, and miR-451a (QIAGEN, Hilden, Germany). The EV-miRNA mixture was electroporated using a pulse width of 20 ms and different voltages (500, 750, 1,000 V) and numbers of pulses (1–10), according to the manufacturer’s protocol. Then, the mixture was incubated for 30 min at 37°C and overnight at 4°C. Simple incubation of EVs with miRNAs or electroporation of EVs in the absence of miRNAs was used as controls in selected experiments. To remove unbounded miRNAs, samples were washed by ultracentrifugation at 100,000 × g for 2 h at 4°C using a 10 mL polycarbonate tube (SW 90 Ti rotor, Beckman Coulter Optima L-90 K ultracentrifuge). Finally, EV pellets were resuspended in saline buffer solution with 1% DMSO and stored at −80°C for downstream analysis.
Transmission Electron Microscopy
For transmission electron microscopy analysis, control EVs or EVs electroporated with different settings were placed on 200 mesh nickel formvar carbon-coated grids (Electron Microscopy Science, Hatfield, Pennsylvania, USA) and left to adhere for 20 min. Next, grids were incubated with 2.5% glutaraldehyde containing 2% sucrose. After washing in distilled water, samples were negatively stained with Nano-W and NanoVan (Nanoprobes, Yaphank, New York, USA) and analyzed using a Jeol JEM 1010 electron microscope (Jeol, Tokyo, Japan).61
RNase Treatment
In selected experiments, EVs were treated with RNase A (Thermo Fisher Scientific), using a concentration of 0.2 μg/mL, for 30 min at 37°C. The RNase inhibitor (Thermo Fisher Scientific) was used to stop the reaction as described by the manufacturer’s protocol, and EVs were washed by ultracentrifugation at 100,000 × g for 2 h at 4°C using a 10 mL polycarbonate tube (SW 90 Ti rotor, Beckman Coulter Optima L-90 K ultracentrifuge). Eventually, EV pellets were resuspended in saline buffer solution with 1% DMSO and stored at −80°C for molecular analysis and in vitro experiments.
RNA Isolation and qRT-PCR
Total RNA was isolated from EVs using the miRNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol. RNA concentration of samples was quantified using a spectrophotometer (mySPEC, VWR, Radnor, Pennsylvania, USA), and the small RNA composition of samples was measured using capillary electrophoresis on an Agilent 2100 Bioanalyzer using the small RNAs kit (Agilent Technologies, Santa Clara, California, USA). qRT-PCR for gene and miRNA expression analysis was performed in triplicate using a 96-well QuantStudio 12K Flex Real-Time PCR system (Thermo Fisher Scientific). For mRNA analysis, cDNA was obtained using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, California, USA). Five nanograms of cDNA was combined with SYBR GREEN PCR Master Mix (Applied Biosystems) as described by the manufacturer’s protocol, and GAPDH was used as housekeeping control. For miRNA analysis, miScript SYBR Green PCR Kit (QIAGEN) was used. Briefly, RNA samples were reverse transcribed using the miScript Reverse Transcription Kit (QIAGEN), and cDNA samples were used to quantify miRNAs of interest. Experiments were run using 3 ng of cDNA for each reaction as described by the manufacturer’s protocol (QIAGEN). The RNU6B small nucleolar RNA was used as control. miRNA and mRNA comparison between samples was calculated on relative expression data normalized using appropriate endogenous controls. Fold change expression (Rq = 2−ΔΔCt) with respect to controls was calculated for all samples. For RNase-treated EVs, percentage of miRNA protection was calculated on the basis of cycle threshold (Ct) differences between treated and untreated EVs. ΔCt of miRNA in the RNase-treated sample was compared with the untreated sample (untreated controls were considered 100%). For the generation of calibration curve for absolute quantification of miRNA, synthetic cel-miR-39 (QIAGEN) was quantified spectrophotometrically (mySPEC, VWR), and 200 ng were reverse transcribed using the miScript Reverse Transcription Kit (QIAGEN). cDNA was serially diluted 1:5 from a starting point of 2.4 ng to have ten dilutions. Serial dilutions were run in five replicates using Relative Standard Curve on 96-well QuantStudio 12K Flex Real-Time PCR system as described by the manufacturer’s protocol (Thermo Fisher Scientific). The calibration curve was used to convert the Ct values of each sample into the corresponding amount of miRNA molecules or mole. The Pearson correlation coefficient for the standard curve was determined as R2 > 0.99. The percentage of engineering yield was calculated as follows: (cel-39 mol/EV in sample)/(cel-39 mol/EV used to engineer EVs). To analyze miRNA transfer from EVs to target cells, TECs, or HepG2 were preplated in a 24-well plate (25,000 cells/well) and stimulated with 2.5 × 109 EVs/mL for 24 h in low-glucose DMEM in the absence of serum (300 μL/well). Then, samples were submitted to RNA extraction and qRT-PCR analysis as described above.
Protein Extraction and Western Blot Analysis
Proteins were extracted from EVs by RIPA buffer (150 nM NaCl, 20 nM Tris-HCl, 0.1% SDS, 1% deoxycholate, 1% Triton X-100, pH 7.8) supplemented with a cocktail of protease and phosphatase inhibitors (Sigma-Aldrich, St. Louis, Missouri, USA). The protein content was quantified using the BCA Protein Assay Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Briefly, 5 μL of sample were dispensed into wells of a 96-well plate, and total protein concentrations were determined using a linear standard curve established with BSA.
Thirty micrograms of proteins were separated by electrophoresis using a 7.5% gradient SDS-polyacrylamide gel. The proteins were transferred to a polyvinylidene fluoride (PVDF) membrane by the Trans-Blot Turbo Transfer System (Bio-Rad) and then immunoblotted with the following antibodies: CD63 and TSG101 (Santa Cruz Biotechnology, Dallas, Texas, USA), CD81 and CD9 (Abcam, Cambridge, UK), and CD29 (Thermo Fisher Scientific). The protein bands were visualized using a ChemiDoc (Bio-Rad) with an enhanced chemiluminescence (ECL) detection kit (GE Healthcare, Chicago, Illinois, USA). Protein quantification was performed normalizing the sample amount with total protein loaded detected by ponceau using Image Lab Software (Bio-Rad).
FACS Analysis of EV Loaded with miRNA and Their Incorporation into Target Cells
To trace EVs by FACS, EVs were labeled with PKH-26 dye (Sigma-Aldrich) for 30 min at 37°C and washed by ultracentrifugation at 100,000 × g for 2 h at 4°C using a 10 mL polycarbonate tube (SW 90 Ti rotor, Beckman Coulter Optima L-90 K ultracentrifuge). To trace miRNA loaded into EVs by FACS, EVs were electroporated with AllStar Negative siRNA Alexa Fluor 555-labeled (QIAGEN) and washed as described above. For EV incorporation, 25,000 cells/well TECs were plated in 24-well plates and treated with 4.2 × 109 EVs/mL for 24 h in low-glucose DMEM in the absence of serum. Quantitative analysis of engineered miRNA and EV uptake was performed by FACS using the CytoFLEX flow cytometer (Beckman Coulter) with CytExpert software.
Apoptosis Assay
HepG2 were seeded at 25,000 cells/well into 24-well plates and maintained in culture in serum-free low-glucose DMEM in the absence (CTR−) or presence of EVs (2.5 × 109 EVs/mL) for 24 h (300 μL/well). For experiments with free-miRNAs and transfected miRNAs, cells were plated 25,000 cells/well in a 24 multi-well plate in 600 μL of low-glucose DMEM plus 10% FBS. Transfection was performed following the manufacturer’s protocol. Briefly, 3 μL of HiPerFect transfection reagent (QIAGEN) was mixed with each miRNA (2.5 nM for miRNA dose comparable with engineered EV treatment and 10 nM as effective treatment) and added to each well. Cells maintained in low-glucose DMEM plus 150 ng/mL doxorubicin were used as positive control (CTR+). Apoptosis was measured after 24 h by Muse Annexin V and Dead Cell Assay Kit (Merck Millipore) following the instructions. The percentage of total apoptotic cells was measured.
Statistical Analysis
Data were analyzed using GraphPad Prism 6.0 Demo. Statistical analyses were performed using ANOVA with Dunnett’s or Turkey’s multiple-comparisons test as appropriate. The relative expression of miRNAs and mRNAs (RQ values) in samples was compared with apposite controls using Kruskal-Wallis ANOVA with Dunn’s multiple-comparisons test. Values were expressed as their mean ± SEM. Statistical significance was established at p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.001).
Author Contributions
Conceptualization, M.A.C.P., M.F.B., and G.C.; Methodology, M.A.C.P. and M.F.B.; Investigation, M.A.C.P.; Resources, S.D. and S.G.; Writing – Original Draft, M.A.C.P. and G.C.; Writing – Review & Editing, M.A.C.P., B.B., M.F.B., S.D, S.G., C.T., and G.C.; Supervision, C.T., B.B., M.F.B., and G.C.; Funding Acquisition, B.B. and G.C.
Conflicts of Interest
G.C. (Department of Medical Sciences and 2i3T Scarl, University of Turin, Turin, Italy) is a component of scientific advisory board of Unicyte AG. M.A.C.P. (Department of Medical Sciences, University of Turin, Turin, Italy) and G.C. (Department of Medical Sciences and 2i3T Scarl, University of Turin, Turin, Italy) are named inventors on a related European patent application (number 18188493.3). The authors declare no competing interests.
Acknowledgments
This study was supported by Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.), project IG2012; by a grant from Unicyte AG (Oberdorf, Nidwalden, Switzerland) REG2018, and by Regione Piemonte POR FESR 2014/2020 – Bando Piattaforma Tecnologica Salute e Benessere – Project “Terapie Avanzate per Processi Fibrotici Cronici (EVER).” We thank Chiara Pomatto for assistance with the graphical abstract creation.
Footnotes
Supplemental Information includes one figure and can be found with this article online at https://doi.org/10.1016/j.omtm.2019.01.001.
Supplemental Information
References
- 1.Yáñez-Mó M., Siljander P.R., Andreu Z., Zavec A.B., Borràs F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong J.P., Holme M.N., Stevens M.M. Re-engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano. 2017;11:69–83. doi: 10.1021/acsnano.6b07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero-Cordoba S.L., Salido-Guadarrama I., Rodriguez-Dorantes M., Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol. Ther. 2014;15:1444–1455. doi: 10.4161/15384047.2014.955442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen E., Xu X., Liu R., Liu T. Small but heavy role: microRNAs in hepatocellular carcinoma progression. BioMed Res. Int. 2018;2018:6784607. doi: 10.1155/2018/6784607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui H., Song R., Wu J., Wang W., Chen X., Yin J. MicroRNA-337 regulates the PI3K/AKT and Wnt/β-catenin signaling pathways to inhibit hepatocellular carcinoma progression by targeting high-mobility group AT-hook 2. Am. J. Cancer Res. 2018;8:405–421. [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Wang H., Ren Z. MicroRNA-214-5p inhibits the invasion and migration of hepatocellular carcinoma cells by targeting Wiskott-Aldrich syndrome like. Cell. Physiol. Biochem. 2018;46:757–764. doi: 10.1159/000488734. [DOI] [PubMed] [Google Scholar]
- 8.Du Z., Niu S., Xu X., Xu Q. MicroRNA31-NDRG3 regulation axes are essential for hepatocellular carcinoma survival and drug resistance. Cancer Biomark. 2017;19:221–230. doi: 10.3233/CBM-170568. [DOI] [PubMed] [Google Scholar]
- 9.Dong Z., Qi R., Guo X., Zhao X., Li Y., Zeng Z., Bai W., Chang X., Hao L., Chen Y. MiR-223 modulates hepatocellular carcinoma cell proliferation through promoting apoptosis via the Rab1-mediated mTOR activation. Biochem. Biophys. Res. Commun. 2017;483:630–637. doi: 10.1016/j.bbrc.2016.12.091. [DOI] [PubMed] [Google Scholar]
- 10.Huang J.Y., Zhang K., Chen D.Q., Chen J., Feng B., Song H., Chen Y., Zhu Z., Lu L., De W. MicroRNA-451: epithelial-mesenchymal transition inhibitor and prognostic biomarker of hepatocelluar carcinoma. Oncotarget. 2015;6:18613–18630. doi: 10.18632/oncotarget.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callegari E., D’Abundo L., Guerriero P., Simioni C., Elamin B.K., Russo M., Cani A., Bassi C., Zagatti B., Giacomelli L. miR-199a-3p modulates MTOR and PAK4 pathways and inhibits tumor growth in a hepatocellular carcinoma transgenic mouse model. Mol. Ther. Nucleic Acids. 2018;11:485–493. doi: 10.1016/j.omtn.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian H., Tay C.Y., Setyawati M.I., Chia S.L., Lee D.S., Leong D.T. Protecting microRNAs from RNase degradation with steric DNA nanostructures. Chem. Sci. (Camb.) 2017;8:1062–1067. doi: 10.1039/c6sc01829g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L., Sharples R.A., Scicluna B.J., Hill A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles. 2014;3:PMC3968297. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barile L., Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Didiot M.C., Hall L.M., Coles A.H., Haraszti R.A., Godinho B.M., Chase K., Sapp E., Ly S., Alterman J.F., Hassler M.R. Exosome-mediated delivery of hydrophobically modified siRNA for Huntingtin mRNA silencing. Mol. Ther. 2016;24:1836–1847. doi: 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamichhane T.N., Jeyaram A., Patel D.B., Parajuli B., Livingston N.K., Arumugasaamy N., Schardt J.S., Jay S.M. Oncogene knockdown via active loading of small RNAs into extracellular vesicles by sonication. Cell. Mol. Bioeng. 2016;9:315–324. doi: 10.1007/s12195-016-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan X., Sansanaphongpricha K., Myers I., Chen H., Yuan H., Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017;38:754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D., Lee H., Zhu Z., Minhas J.K., Jin Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017;312:L110–L121. doi: 10.1152/ajplung.00423.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Loughlin A.J., Mäger I., de Jong O.G., Varela M.A., Schiffelers R.M., El Andaloussi S., Wood M.J.A., Vader P. Functional delivery of lipid-conjugated siRNA by extracellular vesicles. Mol. Ther. 2017;25:1580–1587. doi: 10.1016/j.ymthe.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuhrmann G., Serio A., Mazo M., Nair R., Stevens M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control. Release. 2015;205:35–44. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 21.Wahlgren J., De L Karlson T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P., Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z., Patel T., Piroyan A., Sokolsky M., Kabanov A.V., Batrakova E.V. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kooijmans S.A.A., Stremersch S., Braeckmans K., de Smedt S.C., Hendrix A., Wood M.J.A., Schiffelers R.M., Raemdonck K., Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release. 2013;172:229–238. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Johnsen K.B., Gudbergsson J.M., Skov M.N., Christiansen G., Gurevich L., Moos T., Duroux M. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology. 2016;68:2125–2138. doi: 10.1007/s10616-016-9952-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamichhane T.N., Raiker R.S., Jay S.M. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol. Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood J.L., Scott M.J., Wickline S.A. Maximizing exosome colloidal stability following electroporation. Anal. Biochem. 2014;448:41–49. doi: 10.1016/j.ab.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z., Xie J., Zhu J., Kang C., Chiang C., Wang X., Wang X., Kuang T., Chen F., Chen Z. Functional exosome-mimic for delivery of siRNA to cancer: in vitro and in vivo evaluation. J. Control. Release. 2016;243:160–171. doi: 10.1016/j.jconrel.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter M.P., Ismail N., Zhang X., Aguda B.D., Lee E.J., Yu L., Xiao T., Schafer J., Lee M.L., Schmittgen T.D. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R.L., Liu Y., Liang M. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Contreras M., Shah S.H., Tamayo A., Robbins P.D., Golberg R.B., Mendez A.J., Ricordi C. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci. Rep. 2017;7:5998. doi: 10.1038/s41598-017-05787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fonsato V., Collino F., Herrera M.B., Cavallari C., Deregibus M.C., Cisterna B., Bruno S., Romagnoli R., Salizzoni M., Tetta C., Camussi G. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells. 2012;30:1985–1998. doi: 10.1002/stem.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ju Z., Ma J., Wang C., Yu J., Qiao Y., Hei F. Exosomes from iPSCs delivering siRNA attenuate intracellular adhesion molecule-1 expression and neutrophils adhesion in pulmonary microvascular endothelial cells. Inflammation. 2017;40:486–496. doi: 10.1007/s10753-016-0494-0. [DOI] [PubMed] [Google Scholar]
- 34.Greco K.A., Franzen C.A., Foreman K.E., Flanigan R.C., Kuo P.C., Gupta G.N. PLK-1 silencing in bladder cancer by siRNA delivered with exosomes. Urology. 2016;91:241.e1–241.e7. doi: 10.1016/j.urology.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 35.Liang G., Kan S., Zhu Y., Feng S., Feng W., Gao S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomedicine. 2018;13:585–599. doi: 10.2147/IJN.S154458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yarmush M.L., Golberg A., Serša G., Kotnik T., Miklavčič D. Electroporation-based technologies for medicine: principles, applications, and challenges. Annu. Rev. Biomed. Eng. 2014;16:295–320. doi: 10.1146/annurev-bioeng-071813-104622. [DOI] [PubMed] [Google Scholar]
- 37.Tian Y., Li S., Song J., Ji T., Zhu M., Anderson G.J., Wei J., Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 38.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Liang H., Zhang J., Zen K., Zhang C.Y. microRNAs are ligands of Toll-like receptors. RNA. 2013;19:737–739. doi: 10.1261/rna.036319.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hazawa M., Tomiyama K., Saotome-Nakamura A., Obara C., Yasuda T., Gotoh T., Tanaka I., Yakumaru H., Ishihara H., Tajima K. Radiation increases the cellular uptake of exosomes through CD29/CD81 complex formation. Biochem. Biophys. Res. Commun. 2014;446:1165–1171. doi: 10.1016/j.bbrc.2014.03.067. [DOI] [PubMed] [Google Scholar]
- 41.Mulcahy L.A., Pink R.C., Carter D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKelvey K.J., Powell K.L., Ashton A.W., Morris J.M., McCracken S.A. Exosomes: mechanisms of uptake. J. Circ. Biomark. 2015;4:PMC5572985. doi: 10.5772/61186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gambari R., Brognara E., Spandidos D.A., Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology (Review) Int. J. Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue H.Y., Liu Y., Liao J.Z., Lin J.S., Li B., Yuan W.G., Lee R.J., Li L., Xu C.R., He X.X. Gold nanoparticles delivered miR-375 for treatment of hepatocellular carcinoma. Oncotarget. 2016;7:86675–86686. doi: 10.18632/oncotarget.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X., Magnus J., Kaimal V., Karmali P., Li J., Walls M., Prudente R., Sung E., Sorourian M., Lee R. Lipid nanoparticle-mediated delivery of anti-miR-17 family oligonucleotide suppresses hepatocellular carcinoma growth. Mol. Cancer Ther. 2017;16:905–913. doi: 10.1158/1535-7163.MCT-16-0613. [DOI] [PubMed] [Google Scholar]
- 46.Dhanasekaran R., Gabay-Ryan M., Baylot V., Lai I., Mosley A., Huang X., Zabludoff S., Li J., Kaimal V., Karmali P., Felsher D.W. Anti-miR-17 therapy delays tumorigenesis in MYC-driven hepatocellular carcinoma (HCC) Oncotarget. 2017;9:5517–5528. doi: 10.18632/oncotarget.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F., Wang F., Zhu C., Wei Q., Zhang T., Zhou Y.L. miR-221 suppression through nanoparticle-based miRNA delivery system for hepatocellular carcinoma therapy and its diagnosis as a potential biomarker. Int. J. Nanomedicine. 2018;13:2295–2307. doi: 10.2147/IJN.S157805. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Varshney A., Panda J.J., Singh A.K., Yadav N., Bihari C., Biswas S., Sarin S.K., Chauhan V.S. Targeted delivery of microRNA-199a-3p using self-assembled dipeptide nanoparticles efficiently reduces hepatocellular carcinoma in mice. Hepatology. 2018;67:1392–1407. doi: 10.1002/hep.29643. [DOI] [PubMed] [Google Scholar]
- 49.Kim H.S., Lee K.S., Bae H.J., Eun J.W., Shen Q., Park S.J., Shin W.C., Yang H.D., Park M., Park W.S. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6:8089–8102. doi: 10.18632/oncotarget.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao G., Han C., Zhang Z., Wang L., Xu J. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochem. Biophys. Res. Commun. 2017;490:371–377. doi: 10.1016/j.bbrc.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 51.Wang H., Zhang X., Liu Y., Ni Z., Lin Y., Duan Z., Shi Y., Wang G., Li F. Downregulated miR-31 level associates with poor prognosis of gastric cancer and its restoration suppresses tumor cell malignant phenotypes by inhibiting E2F2. Oncotarget. 2016;7:36577–36589. doi: 10.18632/oncotarget.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nan Y., Han L., Zhang A., Wang G., Jia Z., Yang Y., Yue X., Pu P., Zhong Y., Kang C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 53.Gu X., Li J.Y., Guo J., Li P.S., Zhang W.H. Influence of MiR-451 on drug resistances of paclitaxel-resistant breast cancer cell line. Med. Sci. Monit. 2015;21:3291–3297. doi: 10.12659/MSM.894475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang T., Sun Q., Liu T., Chen J., Du S., Ren C., Liao G., Yuan Y. MiR-451 increases radiosensitivity of nasopharyngeal carcinoma cells by targeting ras-related protein 14 (RAB14) Tumour Biol. 2014;35:12593–12599. doi: 10.1007/s13277-014-2581-x. [DOI] [PubMed] [Google Scholar]
- 55.Wang R., Wang Z.X., Yang J.S., Pan X., De W., Chen L.B. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30:2644–2658. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 56.Yu J., Wang L., Yang H., Ding D., Zhang L., Wang J., Chen Q., Zou Q., Jin Y., Liu X. Rab14 suppression mediated by MiR-320a inhibits cell proliferation, migration and invasion in breast cancer. J. Cancer. 2016;7:2317–2326. doi: 10.7150/jca.15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu H., Wu H., Liu X., Evans B.R., Medina D.J., Liu C.G., Yang J.M. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem. Pharmacol. 2008;76:582–588. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou J.J., Deng X.G., He X.Y., Zhou Y., Yu M., Gao W.C., Zeng B., Zhou Q.B., Li Z.H., Chen R.F. Knockdown of NANOG enhances chemosensitivity of liver cancer cells to doxorubicin by reducing MDR1 expression. Int. J. Oncol. 2014;44:2034–2040. doi: 10.3892/ijo.2014.2347. [DOI] [PubMed] [Google Scholar]
- 59.Gao B., Yang F.M., Yu Z.T., Li R., Xie F., Chen J., Luo H.J., Zhang J.C. Relationship between the expression of MDR1 in hepatocellular cancer and its biological behaviors. Int. J. Clin. Exp. Pathol. 2015;8:6995–7001. [PMC free article] [PubMed] [Google Scholar]
- 60.Bussolati B., Deambrosis I., Russo S., Deregibus M.C., Camussi G. Altered angiogenesis and survival in endothelial cells derived from renal carcinoma. FASEB J. 2003;17:1159–1161. doi: 10.1096/fj.02-0557fje. [DOI] [PubMed] [Google Scholar]
- 61.Deregibus M.C., Figliolini F., D’Antico S., Manzini P.M., Pasquino C., De Lena M., Tetta C., Brizzi M.F., Camussi G. Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 2016;38:1359–1366. doi: 10.3892/ijmm.2016.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruno S., Tapparo M., Collino F., Chiabotto G., Deregibus M.C., Soares Lindoso R., Neri F., Kholia S., Giunti S., Wen S. Renal regenerative potential of different extracellular vesicle populations derived from bone marrow mesenchymal stromal cells. Tissue Eng. Part A. 2017;23:1262–1273. doi: 10.1089/ten.tea.2017.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.