Abstract
Metacaspases are distant relatives of animal caspases found in plants, protozoa and fungi. Some recent studies have demonstrated that metacaspases are involved in regulating the developmental and environmentally induced programmed cell death in plants. In this study, we identified metacaspase gene family in potato (Solanum tuberosum L.) and analyzed their expression pattern in various developmental tissues and stress responses of plants. There were eight metacaspase genes identified in the Peptidase (Cysteine protease) C14 family and based upon sequence alignment and phylogenetic analysis, a systematic nomenclature of potato metacaspases (SotubMCs) has been proposed. Three of the eight candidate genes showing homology with Arabidopsis thaliana type I metacaspase, AtMC1 were given name SotubMC1, SotubMC2 and SotubMC3 as per the degree of relatedness. Similarly, the next three being homologous to A. thaliana type I metacaspase, AtMC3 were named SotubMC4, SotubMC5, and SotubMC6. The remaining two were named SotubMC7 and SotubMC8, showing significant similarity with type II metacaspases of A. thaliana, AtMC4 and AtMC9, respectively. Evolutionary divergence analysis of SotubMCs from its orthologs in seven other members of Solanaceae family as well as with A. thaliana, Vitis vinifera and Oryza sativa was also carried out. The dN/dS ratios of the orthologous pairs suggested the SotubMCs were under purifying (negative) selection in course of plant evolution. Splicing patterns of potato metacaspases were also analyzed. Amongst all SotubMCs, SotubMC2, SotubMC4, SotubMC6 and SotubMC7 genes appeared to produce multiple alternative spliced variants of different lengths. Furthermore using protein modeling tools, we have predicted the protein structure of identified metacaspases. The cis-regulatory elements analysis was also performed exhibiting the presence of development, stress and hormones related cis-elements in the promoter regions of the SotubMCs. This indicates that potato metacaspases might be playing important roles in the development, stress and hormone responsive pathways. Moreover, relative expression analysis of identified genes was carried out using qRT-PCR in various developmental tissues that also include stolons and tubers. The eight metacaspases showed differential expression in different tissues. Some of the tissues such as leaf undergoing senescence among different leaf developmental stages (immature, mature and senescent) displayed higher relative expression of some of the metacaspases, implying their involvement in leaf senescence. The expression pattern of SotubMCs under various abiotic, biotic and hormonal stresses was also analysed. The results showed that many members of the potato metacaspase gene family displayed differential expression patterns under various stress conditions. Taken together, the study could provide crucial resources for further investigations to understand the functional roles of the identified metacaspases in potato.
Keywords: Molecular biology, Plant biology, Bioinformatics, Genetics
1. Introduction
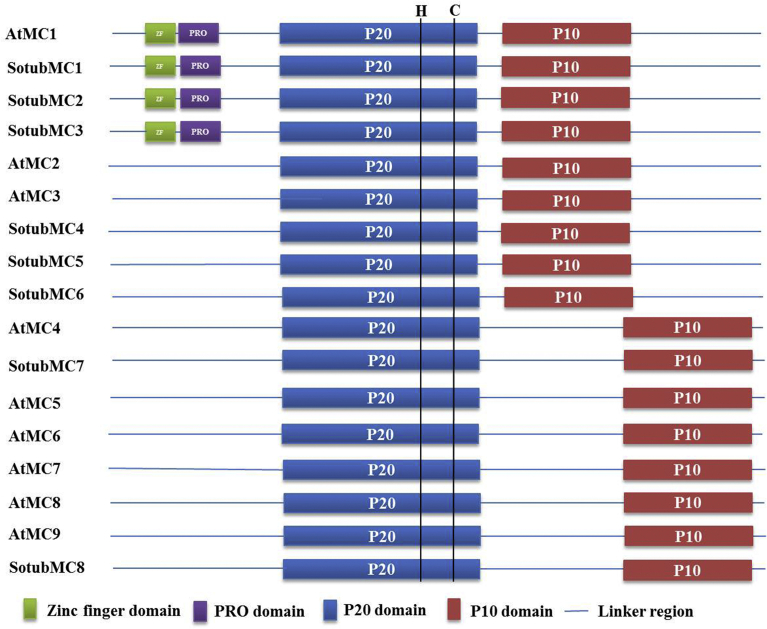
Programmed cell death (PCD) is an indispensable genetically controlled process for the growth and development of plants, and it also plays important roles in combating various biotic and abiotic stresses. As compared to animals, not much was known about the regulators of PCD until caspase-like proteins, metacaspases were discovered in plants (Uren et al., 2000). Metacaspases are structurally similar to caspases due to the presence of caspase-haemoglobinase fold, but functionally they differ on the basis of their different substrate specificity. Caspases are aspartate-specific, whereas metacaspases are specific to arginine or lysine. Metacaspases are divided into two types, type I and type II on the basis of structural and amino acid sequence similarities (Uren et al., 2000). Both type I and type II metacaspases consist of a large (p20) subunit having a His-Cys catalytic dyad and a small (p10) subunit. Type I metacaspases possess proline-rich prodomain in the N-terminal region. This proline-rich prodomain comprises zinc finger motif resembling Arabidopsis LSD1 which is induced by hypersensitive responses. Type II metacaspases, on the other hand, lack the prodomain at its N-terminal site, but harbor longer linker region between large (p20) and small (p10) subunits (Uren et al., 2000).
First evidence for the involvement of metacaspase in plant development comes from the study on Norway spruce (Picea abies). Suppression of mcII-Pa via RNA interference leads to termination of embryo pattern formation, thus suggesting the critical role it plays in embryogenesis (Suarez et al., 2004). In Arabidopsis thaliana genome, three type I (AtMC1-3) and six type II (AtMC4-9) metacaspases have been identified (Uren et al., 2000; Vercammen et al., 2004; Tsiatsiani et al., 2011). Arabidopsis type I AtMC1 and AtMC2 regulate the process of PCD antagonistically (Coll et al., 2010). AtMC1 acts as a positive regulator requiring conserved caspase-like putative catalytic residues for its function, whereas AtMC2, a negative regulator of PCD, acts independent of the putative catalytic residues. A predominant and constitutively expressing member of Arabidopsis metacaspase gene family, AtMC4 acts as a positive mediator of PCD under biotic as well as abiotic stress (Watanabe and Lam, 2011). Another type II metacaspase, AtMC8 positively regulates PCD induced by oxidative stress, UV and H2O2 (He et al., 2008). AtMC9, also a type II metacaspase, facilitates the post-mortem clearance of cell contents after vacuole rupture in xylem vessel elements (Bollhöner et al., 2013).
The roles of metacaspases have also been identified in many economically important vegetables and cereals. For instance, in wheat, a novel metacaspase TaMCA4 plays an important role in PCD induced by the fungal pathogen Puccinia striiformis f.sp. tritici (Wang et al., 2012) In pepper (Capsicum annuum), CaMC9 was suggested to act as a positive regulator of cell death upon infection by Xanthomonas campestris pv. Vesicatoria (Kim et al., 2013). In maize leaves, ozone treatment and aging resulted in significantly enhanced expression of type II metacaspases, thus suggesting the crucial role of the metacaspases in leaf response to ozone and age-mediated senescence (Ahmad et al., 2012). Upon infection of Nicotiana benthamiana with Colletotrichum destructivum, there are few possible roles suggested for NbMCA1, such as degradation of virulence factors of the pathogen, processing of pro-protein involved in stress response and eliminating damaged proteins created during stress (Hao et al., 2007). These reports clearly suggested the possible roles of metacaspases during growth, development and several biotic or abiotic stress-related responses in plants.
The genome-wide identification and expression analysis of metacaspase gene family have been reported in several plant species, such as Arabidopsis, grapes, rice, rubber, and tomato. Potato (Solanum tuberosum L.), one of the most important members of Solanaceae family, is the fourth most important food crop only after maize, rice and wheat; however, no study on potato metacaspases has been reported so far. Therefore, in this study, we have carried out a genome-wide survey of potato genome to identify metacaspases followed by its molecular, phylogenetic and evolutionary divergence analyses. Evolutionary divergences of S. tuberosum metacaspases in course of evolution was analyzed by comparing dN/dS ratios of its orthologs in seven other plant species of Solanaceae family and in A. thaliana, V. vinifera and O. sativa. The dN/dS ratios of SotubMCs and its orthologous pairs were calculated in order to detect evolutionary pressure acting on the genes. Furthermore, homology modeling of identified metacaspases was performed for detail 3-D structural analysis. Splice variants analysis along with expression profiling of metacaspases was also performed during stress responses and in various developmental stages of vegetative and reproductive tissues. Our study provides useful insights into the potato metacaspases for further investigations to be carried out to understand their functional roles in potato.
2. Materials and methods
2.1. Identification and nomenclature of metacaspases genes in Solanum tuberosum L.
Proteins and coding sequences of all nine metacaspases (AtMC1-AtMC9) of Arabidopsis thaliana were obtained from The Arabidopsis information resource (http://www.arabidopsis.org/). To identify the potato metacaspases (SotubMCs), protein and coding sequences of Arabidopsis metacaspases were subjected to ‘BLAST search’ against the potato genome databases (https://solgenomics.net/ and http://solanaceae.plantbiology.msu.edu/; The Potato Genome Sequencing Consortium, 2011) with default parameters.
In present work, we used a S. tubersosum specific five-letter prefix “Sotub” for nomenclature of identified genes. This was done to avoid overlapping gene names from sister taxa like Solanum tabascense, Solanum tenerum, Solanum tinctorium, Solanum triferum, Solanum trifoliatum, Solanum trilobum and Solanum torvum. If we use two letter prefix “St” for S. tuberosum, all other species will also have the same overlapping prefix. Therefore, to avoid overlapping gene names a more secure five-letter prefix was used by adding more letters from the genus and species names. Same has also been adopted by Sol genome and Spud genome databases in prefixing names of other Solanaceae plants.
The chromosomal location, length of gene and coding DNA sequence (CDS) of identified potato metacaspases were retrieved from Spud DB database (http://solanaceae.plantbiology.msu.edu/). Other physiochemical properties of potato metacaspases like protein length, molecular weight and Isoelectric point (pI) were calculated using ExPASy Prot-Param tool (http://web.expasy.org/protparam/). Pfam server (http://pfam.xfam.org/) was used to identify the conserved domains.
2.2. Multiple sequence alignment and phylogenetic analysis
Multiple sequence alignment of identified eight putative metacaspases of S. tuberosum along with nine Arabidopsis metacaspases was carried out using ClustalW with default parameters. Following alignment, a phylogenetic tree was constructed for potato metacaspases with Arabidopsis metacaspases using MEGA V6 (Tamura et al., 2013). In order to evaluate statistical reliability for each node, bootstrap analyses were performed with 1000 replications along with the use of Maximum-likelihood algorithm (Guindon and Gascuel, 2003).
2.3. Ortholog identification and evolutionary divergence analysis
Orthologs of SotubMCs were retrieved from genome databases of seven other members of Solanaceae family, viz., Solanum lycopersicum (Solyc; Tomato), Solanum pimpinellifolium (Sopim; wild tomato), Solanum pennellii (Sopen; wild relative of tomato), Solanum melongena (Sme, Eggplant), Capsicum annuum (Capana; Pepper), Nicotiana attenuata (Niatt; Wild tobacco), Nicotiana tabacum (Nitab; Tobacco) and Arabidopsis thaliana (At; Arabidopsis), Vitis vinifera (Vv; Grape) and Oryza sativa (Os; Rice) using best BLAST hits against the target species database in Solgenomics network (https://solgenomics.net/) and Phytozome V12.1 (https://phytozome.jgi.doe.gov/pz/portal.html). Confirmation of orthologous relationship was done through phylogenetic tree construction using UPGMA method in MEGA V6 (Tamura et al., 2013). All orthologous relationships data were visualized using the CIRCOS V 0.67 (Krzywinski et al., 2009). The dN (non-synonymous substitutions per non-synonymous site) and dS (synonymous substitutions per synonymous site) values of SotubMCs and its orthologous pairs were calculated using PAL2NAL (http://www.bork.embl.de/pal2nal/) which has codeml program of PAML package (Yang and Bielawski, 2000). The ds values of gene pairs so obtained were used to estimate their evolutionary divergence using formula T = dS/(2 × 6.5 × 10−9)×10−6 MYA. Divergence time of SotubMCs and its orthologs from Solanaceae family was compared with evolutionary time scale of speciation events in Solanaceae family (http://www.timetree.org/) (Kumar et al., 2017).
2.4. Structural prediction using protein modeling
Protein models of all the eight putative metacaspases of potato were built using i-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). The predicted models constructed by i-TASSER were investigated for the overall similarity in fold structure of these metacaspases. Putative protein sequences of eight SotubMCs were used as query sequences (Yang et al., 2014).
2.5. Cis-acting element analysis in potato metacaspases promoter regions
2 kb upstream region from 5′ genomic DNA of eight SotubMCs was obtained from the potato genome database, Spud DB (http://potato.plantbiology.msu.edu/). These sequences represent promoter regions in the metacaspase genes sequences. The sequences so obtained were then submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database for cis-element analysis (Lescot et al., 2002).
2.6. Plant material and growth conditions
S. tuberosum L. (Potato var. Kufri Badshah) was cultivated in potted soil in plant growth room under long day growth condition (16 hours light and 8 hours dark) at 22 ± 2 °C. Eight weeks old potato plants were used for sampling. Vegetative tissues, such as root, immature leaf, mature leaf, senescing leaf, stem, stolon and tuber were sampled followed by a quick freeze in liquid nitrogen. Similarly, reproductive tissues like flower bud, open flower, sepal, petal, stamen and carpel were sampled and immediately frozen in liquid nitrogen. Subsequently, total RNA extraction was performed.
2.7. RNA isolation and cDNA synthesis
RNA isolation was carried out using Ambion Pure link™ RNA mini kit (Life technologies, USA) according to the instructions provided. The genomic DNA contamination was removed by using DNAse treatment kit Ambion TURBO DNAse free™ (Life technologies, USA). The RNA samples were quantified and 0.5 μg of RNA was taken for cDNA synthesis using PrimeScript™ 1st strand cDNA synthesis kit (Takara, Clontech, Japan) as per manufacturer's instructions.
2.8. Quantitative real-time PCR analyses
Quantitative RT-PCR was carried out using real-time primer pair specific to each metacaspases using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus; Takara, Clontech, Japan) (Supplementary Table 1). The conditions for qRT-PCR were as follows: 1 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 55–58 °C (varying with different primers), 20 s at 72 °C. Melting curve analysis was also performed to verify that primer dimers were not being quantified. The relative expressions of metacaspases were calculated by normalizing the PCR threshold cycle (Ct) values to the expression of reference genes SotubUBQ5 (Sotub12g030900.1.1), SotubEF1A (Sotub06g010770.1.1) and SotubAPT1 (Sotub08g024880.1.1). Each sample was used in three biological and technical triplicates for expression analysis using comparative 2ˆ-ΔΔCt method (Schmittgen and Livak, 2008).
2.9. Expression analysis of SotubMC genes under biotic, abiotic stress and hormonal treatments
The SotubMCs genes expression under biotic and abiotic stresses and in different hormonal treatments was analyzed using Illumina RNA-Sequencing datasets generated by the PGSC (The Potato Genome Sequencing Consortium, 2011; Massa et al., 2011). For biotic stress, detached potato leaves were inoculated with Phytophthora infestans inoculum (Pi isolate US8: Pi02-007), 100 mg/ml acibenzolar-S-methyl (BTH) and 2 mg/ml DL-β-amino-n-butyric acid (BABA). To mimic herbivory response, primary and secondary leaves were wounded. For abiotic stress treatments 35 °C heat, 150 mM NaCl and 260 mM mannitol were used. To induce hormonal stress, 50 mM Abscisic acid (ABA), 10 mM indole-3-acetic acid (IAA), 50 mM gibberellic acid (GA3), and 10 mM 6- benzyl amino purine (BAP) was used. The treatments were given for 24 h on in vitro grown plants.
2.10. Subcellular localization prediction
Subcellular localization prediction of putative SotubMCs proteins was carried out using in silico prediction tools such as: AtSubP (Kaundal et al., 2010), CELLO (Yu et al., 2006), MultiLoc2 (Blum et al., 2009), Plant-mPloc (Chou and Shen, 2010), Wolf-pSORT (Horton et al., 2007) and SherLoc2 (Briesemeister et al., 2009).
3. Results
3.1. Identification, nomenclature and gene structure of S. tuberosum metacaspases
In total eight metacaspase genes were identified in potato genome from ITAG 3.1 based annotated database using Arabidopsis thaliana metacaspase genes as query sequences. The locus ID of eight SotubMCs along with their corresponding designated names has been shown in Table 1. We observed that two of the eight SotubMCs with ITAG locus ID Sotub01g042370.1.1 and Sotub01g042380.1.1 were represented in PGSC genomic database with a single gene ID PGSC0003DMG400012554. InterProScan data in Spud DB exhibited the presence of caspase-like domains in all the eight putative metacaspases of potato. The length of gene coding DNA sequences of this gene family ranged from 984 bp (SotubMC1) to 1251 bp (SotubMC8). The encoded polypeptides length ranged from 327 amino acids to 416 amino acids with their molecular weight in the range of 36.65–45.67 kDa. Isoelectric points of the gene family were found to be varying according to the classification of members, i.e., type I metacaspases has its pI in the basic pH range (8.26–9.10) for nearly all the metacaspases except SotubMC1 (pI-6.49) and SotubMC2 (pI-6.78), while type II metacaspases, SotubMC7 and SotubMC8, have its pI on the acidic pH scale with values 4.84 and 5.77, respectively (Table 1).
Table 1.
Characteristics of metacaspase gene family members identified in S. tuberosum.
| Gene name | Gene ID | Chromosomal location | A. thaliana orthologs locus | Protein |
||
|---|---|---|---|---|---|---|
| Protein length (amino acids) | Molecular weight (kD) | Isoelectric point (pI) | ||||
| SotubMC1 | Sotub01g028840.1.1 | 1:68565979-68559863 | At1g02170.1 | 370 | 39.86 | 6.49 |
| SotubMC2 | Sotub03g018580.1.1 | 3:36672404-36669360 | At1g02170.1 | 364 | 39.76 | 6.78 |
| SotubMC3 | Sotub05g024740.1.1 | 5:47383253-47386176 | At1g02170.1 | 362 | 40.27 | 8.79 |
| SotubMC4 | Sotub01g042370.1.1 | 1:81977513-81975918 | At5g64240.2 | 365 | 40.84 | 8.82 |
| SotubMC5 | Sotub01g042380.1.1 | 1:81982794-81981164 | At5g64240.2 | 365 | 40.66 | 8.26 |
| SotubMC6 | Sotub01g042390.1.1 | 1:81990877-81989297 | At5g64240.2 | 311 | 39.16 | 9.10 |
| SotubMC7 | Sotub09g030720.1.1 | 9:60913732-60911099 | At1g79340.1 | 416 | 45.67 | 4.84 |
| SotubMC8 | Sotub10g026810.1.1 | 10:56644039-56646068 | At5g04200.1 | 327 | 36.65 | 5.77 |
3.2. Chromosomal location of potato metacaspases
Potato metacaspase genes were found on five chromosomes out of twelve. Out of eight metacaspases, four were anchored on chromosome number 3, 5, 9 and 10, while the remaining four were detected on the same chromosome, chromosome number 1. The chromosome locations of the eight metacaspases have been mentioned in Table 1 and the distribution of all the eight SotubMCs on their corresponding chromosomes have been represented in Fig. 1. Out of the eight metacaspases, the shortest intron was found to be in SotubMC6, while the longest ones were seen in SotubMC1 and SotubMC2 (Fig. 2). Interestingly, variation in the number of introns and exons among the metacaspases was found according to the type of class these genes belonged to. The first six metacaspases (SotubMC1, SotubMC2, SotubMC3, SotubMC4, SotubMC5, and SotubMC6) of type I class showed the presence of five exons interrupted by four introns. The type II metacaspases, SotubMC7 and SotubMC8 exhibited the least number of introns and exons in the gene family. SotubMC7 consists of two exons separated by one intron, while SotubMC8 had three exons interrupted by two introns.
Fig. 1.
Chromosomal distribution of potato metacaspases. Vertical bars indicate locus of SotubMCs on potato chromosomes. Black dots at the center of the bars represent centromere. Red horizontal lines on the bars indicate positions of each gene (gene name written at the bottom of each bar) in the chromosome. Chromosome number is mentioned at the top of each chromosome.
Fig. 2.
Gene structure of potato metacaspases. Diagram represents the gene models of all SotubMC genes identified. Box indicates exon and black line represent intron. Exon-intron structure of all eight metacaspase genes was drawn schematically.
3.3. Multiple sequence alignment and phylogenetic analysis
Sequence alignment and protein structure analysis showed the presence of conserved sequences and domains between already known Arabidopsis metacaspases and the eight identified SotubMCs (Supplementary Fig 1; Fig 3). Similar to Arabidopsis metacaspases (AtMC1-AtMC9) caspase-like domains (p20 and p10) were found in all the eight metacaspases of potato with catalytic dyad His-Cys being present in the p20 subunit. In addition to this, type I Arabidopsis metacaspases (AtMC1-AtMC3), consisting of N-terminal prodomain showed close resemblance to six SotubMCs (SotubMC1, SotubMC2, SotubMC3, SotubMC4, SotubMC5, and SotubMC6). Out of the six SotubMCs having prodomain at their N-terminal region, three of them (SotubMC1, SotubMC2 and SotubMC3) had zinc finger domain-LSD1 (Lesion simulating disease-1). Furthermore, SotubMC7 and SotubMC8 in potato were highly homologous to type II Arabidopsis metacaspases AtMC4 and AtMC9, respectively, and showed comparatively long linker region between their p20 and p10 subunits than type I metacaspases. Moreover, protein sequence similarity matrix exhibited the sequence similarity between any two SotubMCs range from 42% to 92%, where the highest similarity was found between SotubMC4 and SotubMC5, 92% and the least identity between SotubMC4 and SotubMC8, 42%. A phylogenetic tree was constructed to determine evolutionary relationship between Arabidopsis and potato metacaspases (Fig. 4). Phylogenetic distribution in the resulting tree showed the presence of two types of metacaspases in potato. Type I metacaspases in Arabidopsis, AtMC1 and AtMC3 were found to be clustered with six SotubMCs altogether, with AtMC1 being clustered together with SotubMC1, SotubMC2 and SotubMC3, and AtMC3 was found to be in close association with SotubMC4, SotubMC5 and SotubMC6 in the phylogenetic distribution. Likewise, SotubMC7 and SotubMC8 were grouped with AtMC4 and AtMC9, respectively. Thus, a systematic nomenclature was proposed for metacaspase genes in potato based on phylogenetic analysis and amino acid sequence similarity between subfamily members (Table 1).
Fig. 3.
Schematic representation of the conserved motif compositions of metacaspase proteins in S. tuberosum. Blue line represent the non-conserved sequences, and each conserved motif is represented by a box.
Fig. 4.
Phylogenetic tree of metacaspase gene family of S. tuberosum and A. thaliana. Multiple sequence alignment of full-length proteins was performed using ClustalW, and the phylogenetic tree was constructed using MEGA6 by the Neighbor-joining method with 1000 bootstrap replicates.
3.4. Evolutionary divergence analysis of SotubMCs and its orthologs
Altogether 75 orthologs of eight SotubMCs were found from seven selected Solanaceae family members, and Arabidopsis, grape and rice genome database (Fig. 5; Supplementary Figs 2 & 3). Out of 75 orthologs, 52 were from within the Solanaceae family members. S. lycopersicum and S. pimpinellifolium both contain eight orthologs of them, while S. pennellii showed nine orthologs in its genome. The other four members S. melongena, C. annuum, N. attenuata and N. tabacum showed five, eight, six and eight orthologs, respectively. Besides Solanaceae family, in three other species, A. thaliana, V. vinifera and O. sativa there were nine, six and eight orthologs found, respectively.
Fig. 5.
Comparative orthologous relationships of SotubMCs with metacaspase genes from A. thaliana and S. lycopersicum. Metacaspase genes connecting S. tuberosum genome and A. thaliana and S. lycopersicum genomes are shown in colored links.
For evolutionary divergence quantification of the gene pairs under study, we estimated values of dN/dS ratios (Supplementary Tables 2 & 3). The dN/dS ratios of the SotubMCs with orthologs from seven other Solanaceae family members were found to be in the range 0.0740–0.5451, whereas the ratio values ranged 0.0034–0.1458, 0.0088–0.2532 and 0.0081–0.0486 when compared with A. thaliana, V. vinifera and O. sativa, respectively. The values of dN/dS ratio is less than 1 which strongly suggest that gene pairs are under purifying (negative) selection, i.e. natural selection here suppresses protein changes. The divergence time of the SotubMCs and its orthologs from the Solanaceae members ranged from ∼5.3 to 45.8 MYA (Table 2). The evolutionary divergence time of the SotubMCs and its orthologs was found in consistence with the species divergence time of the Solanaceae family members calculated using time tree software (Kumar et al., 2017 (Table 2)).
Table 2.
Estimated evolutionary divergence time of S. tuberosum metacaspases from its orthologs in Solanaceae family.
| Solanum Species pair | Metacaspase genes divergence time (MYA) | Molecular speciation time (MYA) |
|---|---|---|
| S. tuberosum-S. pennellii | 5.2–13.2 | 5.09–10.25 |
| S. tuberosum-S. lycopersicum | 5.3–15.9 | 5.09–10.25 |
| S. tuberosum-S. pimpinellifolium | 5.3–15.9 | 5.09–10.25 |
| S. tuberosum-S. melongena | 14.4–20.1 | 13.70–15.50 |
| S. tuberosum-C. annuum | 16.1–39.8 | 14.4–37.0 |
| S. tuberosum-N. attenuata | 13.1–41.9 | 17.5–39.1 |
| S. tuberosum-N. tabacum | 25.3–45.8 | 17.5–45.1 |
3.5. Analysis of transcript splicing patterns of potato metacaspases
To gain a better understanding of the gene structures of potato metacaspases different splicing patterns exhibited by these genes were studied (Fig. 6). Every member of the potato metacaspase gene family showed the presence of canonical dinucleotides GT and AG for donor and acceptor sites, respectively, in their introns. Among the gene family, SotubMC2, SotubMC4, SotubMC6 and SotubMC7 appeared to have spliced variants of different lengths. In SotubMC2 transcripts (SotubMC2.1, SotubMC2.2, SotubMC2.3, SotubMC2.4, SotubMC2.5, SotubMC2.6), SotubMC2.1, SotubMC2.2 and SotubMC2.4 showed intron retention in their 3′ UTR region with SotubMC2.1 and SotubMC2.2 retaining 37 bp second intron sequences in their second exon, while SotubMC2.4 the smallest transcript synthesized from SotubMC2 pre-mRNA retains first intron sequence of 265 bp in its 3′ UTR. One of the SotubMC6 transcripts, SotubMC6.2 exhibited 5′ alternative splicing in its third exon and during this splicing event 61 bp of exon sequence towards the donor site of the third intron is spliced along with the flanking third intron. Furthermore, loss of the first exon in transcripts of SotubMC2.4, SotubMC2.5 and SotubMC2.6 appear to have not involved any of the alternative splicing mechanisms, instead the presence of alternative promoters in these transcripts seems more plausible here.
Fig. 6.
Gene structure and different spliced variants of potato metacaspases. Dark blue and grey colored boxes indicate exon and intron, respectively. Light blue colored box represent UTRs. Dashed lines above or below exons and introns indicate splicing event and symbol represent start and codons, respectively.
3.6. Cis-acting elements analysis in the promoter regions of potato metacaspases
In order to understand the transcriptional regulation of potato metacaspase genes during development and stress responses in plants, cis-elements in their promoter regions were analyzed (Table 3). Around 200 cis-elements were identified in the promoter regions of eight SotubMCs using PlantCare database. Almost all of them had Skn-1-motif which is involved in endosperm expression. Another cis-element involved in endosperm expression, GCN4-motif, was found to be present in only four metacaspases- SotubMC2, SotubMC4, SotubMC5 and SotubMC8. Light responsive cis-element ACE, showed its presence in nearly all the metacaspases. Besides ACE, there found to be present several other cis-elements which were light responsive, such as Box-4, G-box, GT1-motif etc. Another developmental related cis-element as-2-box involved in shoot and meristem expression was identified in only SotubMC2.
Table 3.
Putative cis-acting elements identified in promoter region of potato metacaspases.
| Gene | Plant development related cis-elements | Stress and hormonal related cis-elements |
|---|---|---|
| SotubMC1 | Skn-1-motif | ABRE2; ARE5; Box-W1; CGTCA-motif3; HSE4; MBSI; TGACG-motif3 |
| SotubMC2 | Skn-1-motif2; GCN4-motif2; as-2-box | ERE; TC-rich repeats2; TCA-element |
| SotubMC3 | Skn-1-motif; O2-site; circadian; ACE | ABRE; ARE; CGTCA-motif2; ERE; HSE; TC-rich repeats; TGACG-motif2 |
| SotubMC4 | Skn-1-motif; GCN4-motif; O2-site; CAT-box; ACE | ABRE2; ARE2; CGTCA-motif; ERE; GARE-motif; GC-motif; HSE; LTR; MBS; TC-rich repeats; TGACG-motif |
| SotubMC5 | Skn-1-motif2; GCN4-motif; O2-site; CAT-box; ACE | ARE; CGTCA-motif2; ERE; GARE-motif; GC-motif; HSE; MBS; TC-rich repeats; TGACG-motif |
| SotubMC6 | Skn-1-motif3; CAT-box2; ACE | ARE; Box-W1; CGTCA-motif; TGACG-motif |
| SotubMC7 | ACE | ARE2; HSE2; MBS; P-box |
| SotubMC8 | Skn-1-motif2; GCN4-motif; ACE | ABRE2; ARE2; HSE; MBS; TC-rich repeats2; EIRE |
Note: Number shown in superscript is total number of each cis-element present, whereas cis-elements without any superscript indicates only one copy of corresponding cis-element. Skn-1-motif and GCN-4-motif, involved in endosperm expression; as-2-box, involved in shoot specific expression and responsiveness; O2-site, involved in zein metabolism regulation; circadian, responsible for circadian control; CAT-box, related to meristem expression; ACE-light responsive element. ABRE, Abscisic acid responsive element; ARE, essential for anaerobic induction; Box-W1, fungal elicitor responsive element; CGTCA-motif, involved in Methyl-jasmonic acid (MeJA) response; HSE, heat stress responsive element; MBS, involved in drought inducibility; MBSI, MYB binding site involved in flavonoid biosynthetic gene regulation; TGACG-motif, involved in MeJA responsiveness; ERE, Ethylene responsive element; TCA-element, Salicylic acid responsive element; TC-rich repeats, involved in defense and stress responsiveness; GARE-motif, Gibberellin responsive element; LTR, low temperature responsive element; GC-motif, involved in anoxic specific inducibility; P-box, Gibberellin responsive element; EIRE, elicitor responsive element.
Furthermore, along with the development-related cis-elements, metacaspases also contained several hormonal related cis-elements, such as ABRE, GARE-motif, ERE and TGCAG-motif. ABRE, Abscisic acid-responsive cis-element, was identified in SotubMC1, SotubMC3, SotubMC4 and SotubMC8; GARE, involved in gibberellin responsiveness, showed its presence in SotubMC4 and SotubMC5; Ethylene-responsive element (ERE) was identified in SotubMC1, SotubMC2, SotubMC3, SotubMC4 and SotubMC5; Methyl jasmonic acid responsive element (MeJA) was prevalent in SotubMC1, SotubMC3, SotubMC4, SotubMC6 and SotubMC6. Additionally, some stress related elements, such as ARE, HSE and LTR were also discovered in the 5′ upstream region of the metacaspases. Anaerobic induction element (ARE) was present in all the metacaspases except SotubMC2. Heat stress element (HSE) was identified in SotubMC1, SotubMC3, SotubMC4, SotubMC5, SotubMC7 and SotubMC8, while Low-temperature responsive element (LTR) showed its presence only in SotubMC4. From this result, it could be clearly indicated that the eight potato metacaspases might have significant roles in the growth, developmental and stress-related pathways in potato.
3.7. Expression analysis of potato metacaspases in developmental tissues
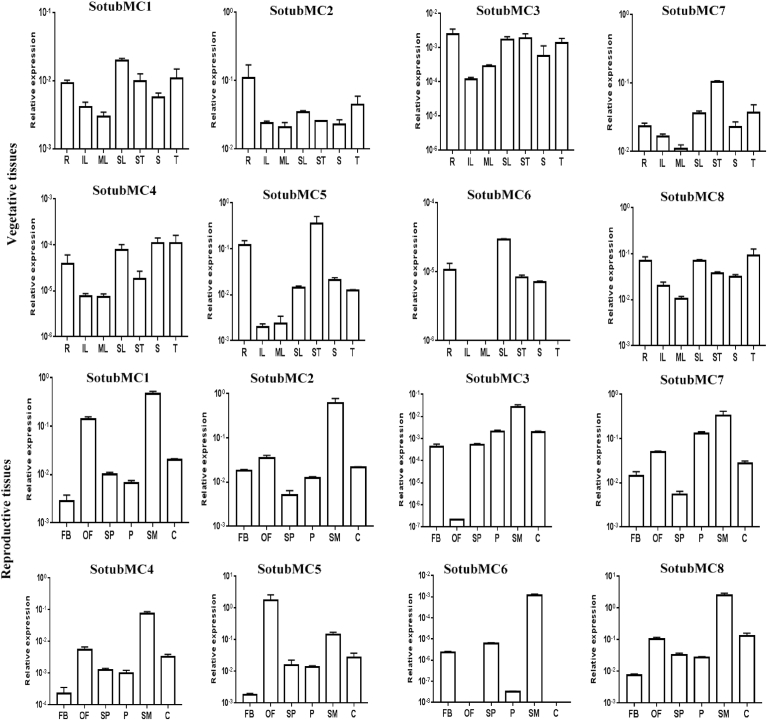
Expression patterns of potato metacaspases were analyzed in various vegetative and reproductive tissues using qRT-PCR (Fig. 7). Due to unavailability of some tissue samples, RNA-Seq expression data for those samples were obtained from transcriptome of the reference Potato Genome Solanum tuberosum Group “Phureja” Clone DM1-3 516R44 (PGSC, 2011; Massa et al., 2011). Out of the eight potato metacaspases, SotubMC1, SotubMC2 and SotubMC7 were found to be constitutively expressed in both vegetative as well as reproductive tissues under normal growth conditions. SotubMC1 showed higher relative expression in senescing leaf compared with immature and mature leaf. The expression of SotubMC1 in root and tuber was comparatively higher than stolon. Among reproductive tissues, SotubMC1 showed the highest relative expression in stamen. Compared with stamen, expression of the gene was significantly low in a carpel. In open flower, transcript level of the gene was found to be increased nearly two-fold vis-à-vis flower bud. Similar expression pattern was observed for SotubMC2 and SotubMC3 in these tissues. Nevertheless, according to the RNA-Seq data (Fig. 8), expression of SotubMC3 was not detected in fruit and tuber tissues. SotubMC4, one of the least expressed members of a potato metacaspase gene family, was found to show higher relative expression in stolon and tuber than root. SotubMC5, however, showed contradictory result, i.e. higher expression in root than stolon and tuber. Furthermore, SotubMC6 expression was observed only in the root, senescing leaf, stem and stolon with senescing leaf showing the highest expression. Expression of SotubMC6 was not detected in a few vegetative and reproductive tissues. Among reproductive tissues, SotubMC4, SotubMC5 and SotubMC6, all of them showed high transcript accumulation in stamen. Moreover, SotubMC4, SotubMC5 and SotubMC6 exhibited relatively high expression in open flower compare with flower bud.
Fig. 7.
The relative expression patterns of potato metacaspases in vegetative and reproductive tissues of potato determined by qRT-PCR. qRT-PCR data were normalized using potato UBQ5 gene. The name of the gene is written on the top of each bar diagram. FB-flower bud; OF-open flower; SP-sepal; P-petal; SM-stamen; C-carpel; R-root; IL-immature leaves; ML-mature leaves; SL-senescing leaves; ST-stem; S-stolon; T-tuber. Error bars represent the standard deviation of three biological and technical replicates.
Fig. 8.
Heat-map showing expression patterns of potato metacaspases in tuber and fruit tissues based on RNA seq data. The Illumina RNA-seq data were reanalyzed, and the relative expression was calculated with respect to control samples.
Type II metacaspase members, SotubMC7 and SotubMC8, highly expressed genes in the potato metacaspase gene family, also exhibited distinct expression pattern in different tissues. SotubMC7, for example, showed high transcript accumulation in stem and tuber, whereas in reproductive tissues stamen, petals and fruits are the ones with high transcript accumulation of the gene. Mature whole fruit exhibited a nearly two-fold increase in transcript level of the gene with respect to immature whole fruit. SotubMC8 was found to be expressed at a relatively high level in the root, senescing leaf and tuber, and in reproductive tissues stamen and mesocarp and endocarp of fruit, it showed high relative expression.
In different stages of leaf tissues, the expression of almost all the potato metacaspases increases in senescing leaf, thus suggesting their involvement in PCD of leaf cells as they age. Among flower tissue samples flower bud exhibited low transcript level of metacaspases vis-à-vis open flower. While among reproductive organs- stamen and carpel, stamen showed high transcript accumulation of all the eight metacaspases. Tuber, the most important organ of the potato plant in terms of economic perspective, showed transcript accumulation of selective members of the gene family. SotubMC4 and SotubMC5, for example, despite being the least expressing members displayed noticeable expression in sprouting tubers. Furthermore, among type II metacaspases, SotubMC7 showed significantly high expression in various tuber tissues, whereas SotubMC8 was found to be the least expressed in tuber tissues.
3.8. Expression of SotubMCs under biotic, abiotic stresses and hormonal treatments
Metacaspases have been reported to play significant roles during stress responses in various plants (Fagundes et al., 2015; Huang et al., 2015). Expression profiles of SotubMCs under various stress conditions could reveal about their possible involvement in stress responses. To investigate the expression profile of SotubMCs under biotic and abiotic stress, and hormonal treatment, RNA Sequencing data from reference Potato Genome S. tuberosum Group “Phureja” Clone DM1-3 516R44 was utilized (The Potato Genome Sequencing Consortium, 2011; Massa et al., 2011). From the expression results (Fig. 9) it could be clearly seen that SotubMC7 was most highly expressed amongst all member of the SotubMC gene family under the stress conditions (biotic as well as abiotic) and the hormonal treatments. In biotic stress, the potato leaves were infected with Phytopthora infestans and treated with BTH and BABA. In P. infestans infected leaves, SotubMC7 showed high upregulation, while in BTH treatment, expression of SotubMC7 was only slightly upregulated. In BTH and BABA treatments, SotubMC2 displayed a slight increase in their transcripts. In case of wounding, expression of SotubMC7 was found to be downregulated. Under both NaCl salt and mannitol stress (osmotic), expression of SotubMC7 was also downregulated. Furthermore, plants under heat stress were showing a slight increase in SotubMC4 and SotubMC5 transcripts.
Fig. 9.
Heat map showing the expression levels of potato metacaspases in various biotic and abiotic stresses and hormonal treatments based on the Illumina RNA-seq data. RNA-seq data were reanalyzed, and the relative expression of various stresses was calculated with respect to control (untreated samples).
To study the effects of hormonal stress on the expression of the SotubMC family, hormonal treatments of IAA, ABA, GA3 and BAP were given to the plants. In IAA, expression of all SotubMC genes was nearly unaffected, while in ABA treated plants only SotubMC2 expression was unregulated. However, SotubMC7, SotubMC4 and SotubMC5 showed increased expression in GA3 treated plants. In contrast, SotubMC7 showed slight downregulation in BAP treated plants. These results clearly indicated that expression of several of SotubMCs family members was modulated by the various stresses in plants.
3.9. Predicted subcellular localization of potato metacaspases
To predict the subcellular localization of putative potato metacaspase proteins, altogether six different prediction tools were employed. AtMC4 protein was used as a control as it has been experimentally shown to be localized in the cytoplasm (Watanabe and Lam, 2011) and all the tools used also exhibited the presence of Arabidopsis ortholog of potato SotubMC7 in the cytoplasm, thus validating the use of AtMC4 as a control. Nevertheless, the localization prediction result of SotubMCs proteins appears to be a bit ambiguous (Table 4; Supplementary Table 4). Despite the ambiguity in the result, it could be clearly seen that overall the prediction tools indicate that proteins of type I SotubMCs are present in the cytoplasm, nucleus, chloroplast, mitochondria, peroxisomes, plasma membrane and extracellular region. Whereas, type II SotubMCs proteins appear to be nuclear and cytoplasmic.
Table 4.
Subcellular location of potato metacaspases. AtMC4 was used here as a control for prediction accuracy. N- Nucleus, Ct- Cytoplasm, Cl- Chloroplast, PM-Plasma membrane, M-Mitochondria, P-Peroxisomes and ER- Endoplasmic reticulum. aChou and Shen (2010), bHorton et al. (2007).
| Metacaspase Protein | Plant-mPLoca | Wolf-pSORTb |
|---|---|---|
| SotubMC1 | Cl/N | Cl/Ct |
| SotubMC2.1 | N | Ct |
| SotubMC2.2 | N | Ct |
| SotubMC2.3 | Ct/N | Ct |
| SotubMC2.4 | N/PM | Ct |
| SotubMC2.5 | Ct | Ct/P |
| SotubMC2.6 | PM | Ct/P |
| SotubMC3 | Cl | Cl/N/P |
| SotubMC4.1 | N | Ct |
| SotubMC4.2 | N | Ct |
| SotubMC5.1 | N | Ct |
| SotubMC6.1 | Cl/N | Ct |
| SotubMC6.2 | Cl/N | Ct |
| SotubMC7.1 | Ct | Ct |
| SotubMC7.2 | Ct | Ct |
| SotubMC8 | Ct | ER |
| AtMC4 | Ct | Ct |
3.10. Structural modeling of potato metacaspases
Full-length three-dimensional protein structures were predicted using iterative threading assembly method (i-TASSER) to determine the structural similarity between the members of the potato metacaspase gene family (Fig. 10). Templates of each member of SotubMCs were detected by LOMETS (Wu and Zhang, 2007) from PDB library. Threading template search generated templates with the average value of Z score = 4.27. Z score is a measure of the significance of templates and templates with the highest significance are selected by i-TASSER for threading alignments. RMSD (Root mean square deviation) of the protein models averaged 9.26. RMSD is the quantitative measure of similarity of three-dimensional structure of two or more protein by calculating the average distance between Cα atomic coordinates of superimposed proteins. Type I SotubMCs predicted structures average value of RMSD = 8.4, whereas average RMSD in type II SotubMCs protein models is 11.8. TM-score (the metric for global fold similarity of two protein models) of the predicted protein structures averaged 0.57. TM-score values range between 0 and 1 and TM-score >0.5 generally assumes the proteins are in the same fold in SCOP/CATH database (Xu and Zhang, 2010). The estimated value of TM-score of modeled proteins averaged 0.61 for type I SotubMCs and in type II SotubMCs TM-score average = 0.46. Furthermore, C-score (confidence score for estimating the quality of the predicted models by i-TASSER) averaged -1.14. C-score value ranges from -5 to 2, where higher C-score indicates a model with higher confidence and vise versa. C-score cutoff > -1.5 signifies correct model. The average confidence scores for potato metacaspases of type I and type II protein structure models were -0.8 and -2.17, respectively.
Fig. 10.
Potato metacaspases structural modeling. Proteins structures of eight metacacapses of S. tuberosum were predicted based on iterative threading assembly refinement method (i-TASSER).
4. Discussion
Metacaspases, caspase-like cysteine proteases are ubiquitously present in the plant kingdom. From the results so far, it has been found that the number of these metacaspase genes vary in different plant species. For instance, in Arabidopsis, there are nine members of this metacaspases gene family (Tsiatsiani et al., 2011), whereas in plants such as grapes (Zhang et al., 2013), rubber (Liu et al., 2016a) and tomato (Liu et al., 2016b) there are six, nine and eight metacaspase genes found, respectively. From our study in potato, there are eight members of metacaspase gene family identified. The first six on account of their N-terminal prodomain and close resemblance to type I Arabidopsis metacaspases were categorized as type I. Among these six type I potato metacaspases, the first three, SotubMC1, SotubMC2, and SotubMC3 exhibited the presence of LSD1 type zinc finger domain at their N-terminal site. In Arabidopsis and rice it was found that the corresponding genes AtMC1 and OsMC1, respectively, interact with LSD1, a negative regulator of cell death under hypersensitive response, via their LSD1 type zinc finger domain (Dietrich et al., 1997; Coll et al., 2010; Huang et al., 2015). Whether these three type I SotubMCs interact with LSD1 or LSD1 –like proteins with their zinc finger domain is yet to be investigated. Furthermore, SotubMC4 and SotubMC9, lacking the prodomain at their N-terminal site and showing longer linker region between p20 and p10 subunits, thus exhibiting all the features of type II class of metacaspases were grouped into the same. In this study, we also found that SotubMC4, SotubMC5 and SotubMC6 were showing significant sequence similarity and the genes were tightly linked within chromosome 1. This implicates the possibility of tandem duplication event of these metacaspases in potato genome.
To gain further insights into the structural diversity of the potato metacaspase genes and to determine genetic relatedness in terms of length and number of introns/exons with certain other plant metacaspases, SotubMCs gene structures were analyzed. The study revealed a striking similarity in the number and length of exon/intron between potato and tomato metacaspases (Liu et al., 2016b). Both the plant species exhibited the presence of six type I metacaspases with five exons separated by four introns, and also lengths of introns/exons of these metacaspases were highly similar except the length of SotubMC3 and SotubMC6, which were showing noticeable difference in first and last introns, respectively, with their corresponding counterparts SlMC3 and SlMC6 in tomato metacaspase gene family. Similarly, type II metacaspases, SotubMC7 and SotubMC8 showed significant similarity in their gene structure vis-à-vis their tomato counterparts, SlMC7 and SlMC8, respectively (Liu et al., 2016b). The gene structure analysis so performed supports the classification of potato metacaspases. Furthermore, the significant similarity between the predicted structures of SotubMCs belonging to the same class provides an additional supporting data for their classification.
To gain better insights into the structural aspects of potato metacaspases, predicted protein structures of these metacaspases were built based on threading approach (i-TASSER). Based on the predicted secondary structures and predicted protein models (Fig. 10; Supplementary Fig 4) with high confident scores (C-score), the significant similarity was found in the overall fold structure of type I SotubMCs. Structural models of type II metacaspases were also found in close agreement with one another. It should be noted here that the structural models exhibited stark differences in the N-terminal region between type I and type II SotubMCs. In type I metacaspases, predicted tertiary structures at the N-terminal region (indicated by dark blue colored coils) lacks alpha-helix, however, protein models of the members of type II metacaspases in potato conspicuously show the presence of alpha-helix at the N-terminal region (indicated by dark blue colored helices). Thus, the predicted structural analysis further supports the nomenclature of SotubMCs proposed on the basis of phylogenetic and amino acid sequence similarity analysis.
The dN/dS ratio is one of the most widely used method to measure evolutionary pressure on genes. If the ratio exceeds 1, selection pressure acting on the gene is considered positive, i.e. it favours change in gene product. On the other hand, if the ratio is less than 1, the selection pressure acting on the gene would be stabilizing (negative) in nature, i.e. it resists the change in gene product (Kimura, 1977; Yang and Bielawski, 2000). The results from our analysis strongly suggest that SotubMCs and its orthologs are under stabilizing selection as the dN/dS ratio of all the paired genes in our study was found to be less than 1.
Synonymous substitution rate (ds) provides a metric for estimation of evolutionary divergence of homologous genes (Kimura, 1977). In our study, the dS values were used in evolutionary divergence analysis of SotubMCs and its orthologs from other members of Solanaceae family. The result indicated that most of the SotubMCs and its orthologs might have evolved after the speciation events of the Solanaceae plants.
Potato is found as tetraploid in nature, and the finding of metacaspases in potato with their spliced variants add on to the already complex nature of these genes. Alternative splicing events such as intron retention and 5′ alternative splicing, and introns in certain metacaspases possibly acting as alternate promoters are some of the phenomena appear to be involved in the synthesis of spliced variants of certain SotubMCs. Intron retention in SotubMC2.1 and SotubMC2.2 may lead to the generation of transcripts with premature stop codons (PTC), which would consequently result in loss of p20 or p10 domains of these SotubMCs or the decay of proteins altogether. However, introns retained in SotubMC7.1, SotubMC2.5, and SotubMC2.6 transcripts as their 5′ UTR are likely to play the role of alternate promoters in these genes instead of being spliceable entities (Morello and Breviario, 2008). Furthermore, synthesis of SotubMC6.1 from its pre-mRNA employs 5′ alternative splicing event at the third exon sequence (Syed et al., 2012). Evidence suggests 3′ or 5′ alternative splicing event might possibly lead to the generation of unproductive mRNAs with PTCs, which could further result in degradation of these mRNAs via non-sense mediated mRNA decay pathway (NMD). Thus, the production of spliced variants of metacaspases via different modes of alternative splicing contribute to the diversity of transcriptome/ proteome, which could be playing essential role in stress adaptive mechanism of plants (Syed et al., 2012; Reddy, 2007; Ali and Reddy, 2008; Reddy et al., 2013); however the signaling pathways per se relaying information of stress conditions to the splicing machinery are poorly understood and thereby provides an opportunity to further delineate the molecular players involved in this signal transduction.
Cis-regulatory elements acting as molecular switches play crucial roles in transcription initiation events. Upon systematic analysis of the promoter regions, elements related to various stress and growth or developmental processes have been identified in the eight SotubMCs. ACE, light responsive element, was prevalent in the SotubMCs. Along with ACE, there found to be present several other cis-elements involved in light responsiveness such as Box-4, G-box, GT1-motif, etc. Another cis-element called Skn-1-motif, involved in endosperm responsiveness, exhibited its presence in nearly all the SotubMCs similar to a previous study in grapes, wherein all the six metacaspases of grapes were found to consist of Skn-1-motif, and its involvement in endosperm development was supported by qRT-PCR results of ovule developmental stages of seed and seedless varieties of grapes (Zhang et al., 2013). These results indicate the possible involvement of SotubMCs in endosperm development.
SotubMCs also consist of cis-elements that indicate its involvement in various stress and hormonal responsiveness. Also, there have been reports suggesting that metacaspase gene family in rice, rubber and tomato exhibits differential expression induced by abiotic stresses such as cold, drought and salt (Huang et al., 2015; Liu et al., 2016a, Liu et al., 2016b). Abiotic stress responsive cis-elements such as HSE (heat stress-responsive element), LTR (low temperature-responsive element) and ARE (anaerobic responsive element), along with several essential hormonal responsive cis-elements- ABRE (abscisic acid responsive element), ERE (ethylene-responsive element), TGCAG-motif (Methyl jasmonic acid responsive element), and GARE-motif (Gibberellin responsive element) were found conspicuously in the 5′ upstream of not only SotubMCs but also in Arabidopsis, grapes, rubber and tomato metacaspases. From this result, it seems highly plausible that the stress and hormonal responsive cis-elements would be evolutionarily conserved in the promoter regions of metacaspases of all the above-mentioned plant species. Furthermore, plant hormones such as ABA, ethylene, jasmonic acid and gibberellin are known to be involved in plant defense and many diverse developmental pathways like seed germination, root growth, fruit ripening and dehiscence, etc. (Bari and Jones, 2008; Verhage et al., 2010). This implies that SotubMCs could be involved in cross-talk with these hormones and could also be responsible for induction of various stress-related responses directly or indirectly.
Expression profiles of metacaspases in different tissues and developmental stages of a plant could provide us with important cues that can direct us towards the essential roles that certain metacaspases may have in specific tissues, which could further be elucidated by their in-depth analysis using state-of-the-art molecular tools. There have been reports of the expression analysis of metacaspases in plants such as Arabidopsis, grapes, rice, rubber and tomato, indicating tissue specific differential expression pattern (Kwon and Hwang, 2013; Zhang et al., 2013; Wang and Zhang, 2014; Liu et al., 2016a, Liu et al., 2016b). Similarly, in the present study, the expression profile of eight SotubMCs in different vegetative and reproductive tissues of potato exhibited tissue specific differential expression pattern, thus showing consistency with the aforementioned reports. In our results, several of SotubMCs showed comparatively increased expression in senescing leaf tissues, suggesting a developmental role of SotubMCs in leaves especially in leaf senescence. In tomato, however, only SlMC2 of the tomato metacaspase gene family showed increased expression during leaf senescing stage indicating the only metacaspase in tomato to be involved in leaf development (Liu et al., 2016b). From the RNA seq expression results, SotubMC7 was found to exhibit significantly high expression in different tuber tissues, whereas SotubMC8 merely showed any expression in these tissues, thus implicating the involvement of SotubMC7 in physiological development of different tuber tissues. Results from grapes and tomato metacaspases study show tissue-specific expression pattern of type I metacaspases in these plants (Zhang et al., 2013; Liu et al., 2016b). For instance, all type I metacaspases of grapes showed relatively high expression in the stem, whereas in tomato, all its type I metacaspases were highly expressed in the root. Our result, however, indicates high relative expression of most of the SotubMCs in both roots as well as stem tissues. Among reproductive tissues, stamen showed significantly high relative expression of all metacaspases, thus indicating the possible essential role of SotubMCs in this male reproductive organ.
Metacaspases have been identified to be involved in biotic and abiotic stress responses in a number of plant species. For instance, AtMC4 is found to play a positive mediator in both biotic and abiotic stress in A. thaliana (Watanabe and Lam, 2011). Similarly, metacaspases in several other plants, such as tomato, rice and rubber were reported to show differential expression during different stress conditions (Huang et al., 2015; Liu et al., 2016a, Liu et al., 2016b). In order to investigate the possible involvement of SotubMCs in stress responses in potato, RNA seq data was utilized. Amongst all SotubMCs, SotubMC7 was found to be significantly upregulated in biotic stress (P. infestans), while in wounded potato leaves its expression was downregulated. In abiotic stress (salt and mannitol) treatments downregulated expression of SotubMC7 was observed. This differential expression in biotic and abiotic stress could be attributed to the significance of the role played by the SotubMC7 in these two different stress responses. In hormonal stress, SotubMC7 was highly expressed by GA3, whereas its expression was slightly downregulated by BAP. In addition to SotubMC7, a couple of other SotubMCs (SotubMC4 and SotubMC5) also showed increased expression in GA3. Our cis-element analysis further supported this increased expression of SotubMC7, SotubMC4 and SotubMC5 under GA3 treatment as their promoter sequences also possess gibberellic acid responsive cis-regulatory element.
In silico subcellular localization tools predicted that type II SotubMC proteins are strictly restricted to cytoplasm and nucleus, whereas type I SotubMCs are prevalent in the cytoplasm, nucleus, chloroplast, mitochondria, peroxisomes, plasma membrane and extracellular region. However, the delineating of their physiological roles could only be achieved after the experimental demonstration of their localization.
In summary, we identified a total of eight metcaspases in the genome of potato (S. tuberosum L.) and on the basis of the phylogenetic analysis and sequence similarity, a suitable nomenclature has also been given to them. Some of the identified metacaspase genes were appeared to evolve from tandem duplication events. Splicing pattern analysis of potato metacaspase gene family showed that some members appeared to produce multiple alternative spliced variants of various lengths. Evolutionary divergence analysis suggested the SotubMCs were under purifying (negative) selection in course of plant evolution. The expression analysis showed that transcript of each gene was differentially regulated in various developmental tissues and by various stresses and plant hormones, suggesting their distinct and important role in plants. Given the importance of potato as a major food crop, this study provided a systematic and comprehensive analysis of the metacaspase gene family in potato, and could be utilized as valuable resources for the further functional characterization of each member of this important family in plants.
Declarations
Author contribution statement
Nehal Dubey, Maitri Trivedi: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Suresh Varsani: Performed the experiments; Analyzed and interpreted the data.
Vishal Vyas, Manisha Farsodia: Analyzed and interpreted the data.
Sunil Kumar Singh: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was partially supported by grants from Science and Engineering Research Board, Govt. of India (EMR/2017/000043 and SERB/SR/SO/PS/53/2012) and Department of Biotechnology, Govt. of India (BT/PR1357/BRB/10/882/2010) (to SKS). Authors also acknowledged the UGC-DRS and DST-FIST programs for financial support.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Ahmad R., Zuily-Fodil Y., Passaquet C., Bethenod O., Roche R., Repellin A. Ozone and aging up-regulate type II metacaspase gene expression and global metacaspase activity in the leaves of field-grown maize (Zea mays L.) plants. Chemosphere. 2012;87:789–795. doi: 10.1016/j.chemosphere.2011.12.081. [DOI] [PubMed] [Google Scholar]
- Ali G.S., Reddy A.S. Regulation of alternative splicing of pre-mRNAs by stresses. Curr. Top. Microbiol. Immunol. 2008;326:257–275. doi: 10.1007/978-3-540-76776-3_14. [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J. Role of plant hormones in plant defense responses. Plant Mol. Biol. 2008;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- Blum T., Briesemeister S., Kohlbacher O. MultiLoc2: integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinf. 2009;10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollhöner B., Zhang B., Stael S., Denancé N., Overmyer K., Goffner D., Van Breusegem F., Tuominen H. Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 2013;200:498–510. doi: 10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- Briesemeister S., Blum T., Brady S., Lam Y., Kohlbacher O., Shatkay H. SherLoc2: a high-accuracy hybrid method for predicting subcellular localization of proteins. J. Proteome Res. 2009;8:5363–5366. doi: 10.1021/pr900665y. [DOI] [PubMed] [Google Scholar]
- Chou K.-C., Shen H.-B. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J., Epple P. Arabidopsis type I metacaspases control cell death. Science. 2010;330:1393–1397. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- Dietrich R., Richberg M., Schmidt R., Dean C., Dangl J. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- Fagundes D., Bohn B., Cabreira C., Leipelt F., Dias N., Bodanese-Zanettini M.H., Cagliari A. Caspases in plants: metacaspase gene family in plant stress responses. Funct. Integr. Genom. 2015;15:639e649. doi: 10.1007/s10142-015-0459-7. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hao L., Goodwin P.H., Hsiang T. Expression of a metacaspase gene of Nicotiana benthamiana after inoculation with Colletotrichum destructivum or Pseudomonas syringae pv. tomato, and the effect of silencing the gene on the host response. Plant Cell Rep. 2007;26:1879–1888. doi: 10.1007/s00299-007-0387-7. [DOI] [PubMed] [Google Scholar]
- He R., Drury G., Rotari V., Gordon A., Willer M., Farzaneh T., Woltering E., Gallois P. Metacaspase-8 modulates programmed cell death induced by ultra-violet light and H2O2 in Arabidopsis. J. Biol. Chem. 2008;283:774–783. doi: 10.1074/jbc.M704185200. [DOI] [PubMed] [Google Scholar]
- Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Zhang H., Hong Y., Liu S., Li D., Song F. Stress-responsive expression, subcellular localization and protein-protein interactions of the rice metacaspase family. Int. J. Mol. Sci. 2015;16:16216–16241. doi: 10.3390/ijms160716216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal R., Saini R., Zhao P.X. Combining machine learning and homology-based approaches to accurately predict subcellular localization in Arabidopsis. Plant Physiol. 2010;154:36–54. doi: 10.1104/pp.110.156851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Bae C., Oh S., Choi D. A pepper (Capsicum annuum L.) metacaspase 9 (Camc9) plays a role in pathogen-induced cell death in plants. Mol. Plant Pathol. 2013;14:557–566. doi: 10.1111/mpp.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Kwon S., Hwang D. Expression analysis of the metacaspase gene family in Arabidopsis. J. Plant Biol. 2013;56:391–398. [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Deng Z., Chen J., Wang S., Hao L., Li D. Genome-wide identification and expression analysis of the metacaspase gene family in Hevea brasiliensis. Plant Physiol. Biochem. 2016;105:90–101. doi: 10.1016/j.plaphy.2016.04.011. [DOI] [PubMed] [Google Scholar]
- Liu H., Liu J., Wei Y. Identification and analysis of the metacaspase gene family in tomato. Biochem. Biophys. Res. Commun. 2016;479:523–529. doi: 10.1016/j.bbrc.2016.09.103. [DOI] [PubMed] [Google Scholar]
- Massa A., Childs K., Lin H., Bryan G., Giuliano G., Buell R. The Transcriptome of the reference potato genome Solanum tuberosum group Phureja clone DM1-3 516R44. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello L., Breviario D. Plant spliceosomal introns: not only cut and paste. Curr. Genom. 2008;9:227–238. doi: 10.2174/138920208784533629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- Reddy A., Marquez Y., Kalyna M., Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25:3657–3683. doi: 10.1105/tpc.113.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Suarez M., Filonova L., Smertenko A., Savenkov E., Clapham D., Arnold S., Zhivotovsky B., Bozhkov P. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr. Biol. 2004;14:R339–R340. doi: 10.1016/j.cub.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Syed N., Kalyna M., Marquez Y., Barta A., Brown J. Alternative splicing in plants – coming of age. Trends Plant Sci. 2012;17:616–623. doi: 10.1016/j.tplants.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Potato Genome Sequencing Consortium Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P. Metacaspases. Cell Death Differ. 2011;18:1279–1288. doi: 10.1038/cdd.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A., O’Rourke K., Aravind L., Pisabarro M.T., Seshagiri S., Koonin E., Dixit V. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- Vercammen D., Cotte B., Jaeger G., Eeckhout D., Casteels P., Vandepoele K., Vandenberghe I., Beeumen J., Inzé D., Van Breusegem F. Type II metacaspases AtMC4 and AtMC9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 2004;279:45329–45336. doi: 10.1074/jbc.M406329200. [DOI] [PubMed] [Google Scholar]
- Verhage A., Van Wees S., Pieterse C. Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol. 2010;154:536–540. doi: 10.1104/pp.110.161570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang X., Feng H., Tang C., Bai P., Wei G., Huang L., Kang Z. TaMCA4, a novel wheat metacaspase gene functions in programmed cell death induced by the fungal pathogen Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2012;25:755–764. doi: 10.1094/MPMI-11-11-0283-R. [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang H. Genome-wide survey and characterization of metacaspase gene family in rice (Oryza sativa) J. Genet. 2014;93:93–102. doi: 10.1007/s12041-014-0343-6. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 2011;66:969–982. doi: 10.1111/j.1365-313X.2011.04554.x. [DOI] [PubMed] [Google Scholar]
- Wu S., Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang Y. How significant is a protein structure similarity with TM-score = 0.5. Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Bielawski J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2014;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.S., Chen Y.C., Lu C.H., Hwang J.K. Prediction of protein subcellular localization. Proteins: Struct. Funct. Bioinf. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- Zhang C., Gong P., Wei R., Li S., Zhang X., Yu Y., Wang Y. The metacaspase gene family of Vitis vinifera L.: characterization and differential expression during ovule abortion in stenospermocarpic seedless grapes. Gene. 2013;528:267–276. doi: 10.1016/j.gene.2013.06.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.