Abstract
The influenza A virus poses serious public health challenges worldwide. Strikingly, small noncoding microRNAs (miRNAs) that modulate gene expression are closely involved in antiviral responses, although the underlying mechanisms are essentially unknown. We now report that microRNA-340 (miR340) is downregulated following influenza A and other RNA virus infections, implying that host cells deplete miR340 as an antiviral defense mechanism. Accordingly, the inhibition or knockdown of endogenous miR340 clearly prevents the infection of cultured cells, whereas the forced expression of miR340 significantly enhances virus replication. Using next-generation sequencing, we found that miR340 attenuates cellular antiviral immunity. Moreover, mechanistic studies defined miR340 as a repressor of RIG-I and OAS2, critical factors for the establishment of an antiviral response. Collectively, these data indicate that host cells may lower their viral loads by regulating miRNA pathways, which may, in turn, provide new opportunities for treatment.
Keywords: microRNA, miR340, RIG-I, antiviral response, influenza A virus
Introduction
The influenza A virus is a serious zoonotic pathogen that causes yearly epidemics and periodic pandemics with high death tolls.1 Although the pathogenesis of influenza has been investigated extensively,2 a more detailed understanding of virus-host interactions, especially of virus-induced changes in the host, may identify critical points in the virus life cycle that could be targeted to restrict replication.
MicroRNAs (miRNAs) are post-transcriptional regulators that mediate translational repression and/or mRNA degradation by complementary binding to the 3ʹ UTRs in target mRNAs.3 miRNA expression is frequently regulated in sophisticated ways in response to cellular changes, including during developmental transitions,4 cancer progression,5 interferon or poly(I:C) stimulation,6, 7 and viral infections.8 Accumulating evidence now suggests that miRNAs are aberrantly expressed during infection with influenza A virus.9, 10, 11 In some cases, viruses even induce miRNAs against immune factors to promote replication.12, 13, 14 Conversely, hosts also deploy miRNAs to restrict virus replication through interactions with the viral genome or by downregulating host factors required for viral proliferation.15, 16, 17, 18 Thus, the complex regulatory impact of miRNAs on influenza virus-host interactions requires further study.
The innate immune system senses pathogen-associated molecular patterns through an array of pattern recognition receptors.19 For example, retinoic acid-inducible gene I (RIG-I, also known as DDX58), the prototypical RIG-I-like receptor, detects viral RNA containing 5ʹ-triphosphates.20, 21 Subsequently, RIG-I is ubiquitinated by TRIM25 to propagate the signal via the mitochondrial protein MAVS (also known as VISA, IPS-1, or Cardif)22, 23, 24, 25 and to activate the nuclear factor κB (NF-κB), IRF-3, and IRF-7 transcription factors. This cascade boosts the production of type I interferons and downstream genes, thereby establishing a cellular antiviral state. Accordingly, RIG-I is frequently regulated or targeted by host and viral factors. Recently, miRNAs were reported to fine-tune the antiviral immune response by targeting various molecules in this cascade,13, 14, 26, 27 although the underlying mechanisms remain to be fully elucidated.
Previously, we reported that one such miRNA, miR340, is indeed involved in influenza A virus infection,28 although it is classically linked to tumor progression.29, 30, 31 We now provide new evidence that the depletion of miR340 stimulates the cellular immune response against viruses. Accordingly, the silencing or deletion of endogenous miR340 lowers the viral load in A549 cells, while ectopically expressed miR340 enhances viral replication. By using next-generation sequencing and luciferase reporter assays, we identified the antiviral factors RIG-I and OAS2 as bona fide miR340 targets. Our data indicate that miR340 mediates a regulatory feedback loop during host-virus interactions to control both antiviral responses and infection.
Results
Influenza A Virus and Poly(I:C) Downregulate miR340
In a previous study employing miRNA microarrays, we found that the influenza A virus (A/duck/Hubei/hangmei01/2006 [H5N1/HM]) suppresses miR340.28 We have again confirmed that miR340 is clearly diminished in A549 cells infected with H5N1/HM virus for 24 and 36 h at an MOI of 0.5 (Figure 1A). A viral load-dependent suppression of miR340 levels in H5N1/HM-infected cells was observed (Figure 1B). In addition, the expression of miR340 precursors in virus-infected cells followed a trend similar to that of the mature miRNA (Figure 1C).
Figure 1.
Viral Infection and Poly(I:C) Transfection Suppress miR340 Expression
(A) A549 cells were mock infected or infected with H5N1/HM for 24 and 36 h at an MOI of 0.5, and miR340 levels were quantified by qRT-PCR. (B) Relative expression of miR340 in A549 cells infected with different H5N1/HM concentrations. (C) Relative expression of miR340 precursors was measured in H5N1/HM-infected A549 cells. (D and E) A549 cells were mock infected or infected with H5N6 (D) or PR8 (E) viruses for 24 h, and miR340 was detected by qRT-PCR. (F) A549 cells were mock infected or infected with Sendai virus or vesicular stomatitis virus for 24 h, and miR340 was detected by qRT-PCR. (G) A549 cells seeded in 12-well plates were transfected with 0.01 μg poly(I:C), and the relative expression of miR340 was quantified after 24 h by qRT-PCR. (H and I) 293T cells were mock infected or infected with H5N1/HM for 24 h at an MOI of 0.5 or 1, and levels of mature miR340 (H) and its precursors (I) were quantified by qRT-PCR. Results represent the mean ± SD of three independent experiments. p value was calculated by using Student’s t test (A–E and G) or one-way ANOVA with Bonferroni multiple comparison test (F, H, and I) (*p < 0.05, **p < 0.01). SEV, Sendai virus; VSV, vesicular stomatitis virus.
To determine if downregulation of miR340 is specific to H5N1 influenza virus, we additionally detected miR340 levels in A549 cells infected with H5N6 or PR8 influenza viruses. As shown in Figures 1D and 1E, cellular miR340 expression was significantly suppressed by either virus. Similarly, the RNA abundance of mature miR340 was decreased by other RNA viruses, specifically the Sendai virus and vesicular stomatitis virus (Figure 1F). Poly(I:C), a synthetic mimetic of viral double-stranded RNA, also downregulated miR340 2-fold, as measured by qRT-PCR (Figure 1G). Finally, miR340 and its precursors were also diminished in 293T cells infected with H5N1/HM (Figures 1H and 1I). Collectively, these results strongly indicate that miR340 is highly sensitive to infection with influenza A virus and, thus, may be involved in the ensuing cellular response.
miR340 Overexpression Enhances the Replication of Influenza A Virus
In some cases, miRNAs that modulate antiviral pathways are strongly induced or suppressed by viral infection and, therefore, trigger feedback mechanisms that promote or restrict viral replication. Thus, we tested whether viral infection is sensitive to miR340 overexpressed from a pcDNA3.1+ vector encoding its precursor. Overexpression was verified by qRT-PCR after transfection into A549 cells for 24 h (Figure 2A). These cells were then infected with the influenza strain H5N1/HM at an MOI of 0.1. Culture medium was harvested at various time points to determine virus titers, whereas cellular proteins and RNA were extracted for western blot and qRT-PCR, respectively.
Figure 2.
miR340 Overexpression Promotes Replication of Influenza A Virus in A549 Cells
(A) Expression of miR340 by p340 was verified using qRT-PCR after transfection into A549 cells for 24 h. (B and C) A549 cells were transfected with 1 μg empty vector or vector encoding miR340. After 24 h, cells were infected with H5N1/HM at an MOI of 0.1, and the viral nucleoprotein, matrix protein M1, and polymerase acidic protein mRNAs (B) together with their proteins (C) were determined by qRT-PCR and western blot 24 h thereafter. (D) H5N1/HM virus titers in the media were also determined at the indicated time points using the TCID50 assay. (E) Nucleoprotein levels of PR8 and H5N1/HM strains were measured by western blotting in cells transfected with empty vector or vector encoding miR340. (F) H5N1/HM nucleoprotein increased levels in a concentration-dependent manner when using a vector encoding miR340. (G) A549 cells were transfected with 60 nM miR340 mimetics or control mimetics, infected with H5N1/HM for 24 h, and analyzed by western blotting to assess viral nucleoprotein levels. (H) A549 cells were also transfected with 60 nM miR340 inhibitors or control inhibitors, infected with H5N1/HM for 24 h, and analyzed by western blotting. Values represent the mean ± SD of three independent experiments. p value was calculated by using the Student’s t test (A and D) or two-way ANOVA with Bonferroni multiple comparison test (B) (*p < 0.05, **p < 0.01). hpi, hours post-infection; E-vec, empty vector; p340, pcDNA3.1-miR340; NP, influenza A virus nucleoprotein; A-NC, negative control inhibitor; A-miR340, miR340 inhibitor; RIOD, relative integrated optical density. In (E)–(H), each lane represents an independent experiment.
We found that the viral nucleoprotein (NP), matrix protein M1 (M1), and polymerase acidic protein (PA) mRNAs (Figure 2B) together with their proteins (Figure 2C) were markedly enriched in miR340-overexpressing cells. Similar results were obtained with the PR8 (Figure 2E, left) and H5N1/HM (Figure 2E, right) strains, the latter of which was used as a positive control. In addition, viral titers in the medium significantly increased 24 and 36 h post-infection in cells overexpressing miR340 when compared to their levels in cells transfected with an empty vector (Figure 2D). Furthermore, miR340 enhanced influenza A virus replication in a concentration-dependent manner (Figure 2F). Particularly, synthetic miR340 mimetics also enhanced the accumulation of viral NPs in infected A549 cells (Figure 2G). Conversely, transfection of miR340 inhibitors, which are single-stranded oligonucleotides that compete with endogenous miRNA for binding to target mRNA, restricted the replication of influenza A virus, as assessed by NP abundance (Figure 2H). Taken together, these results highlight miR340 as a potential regulator by which the host restricts virus production.
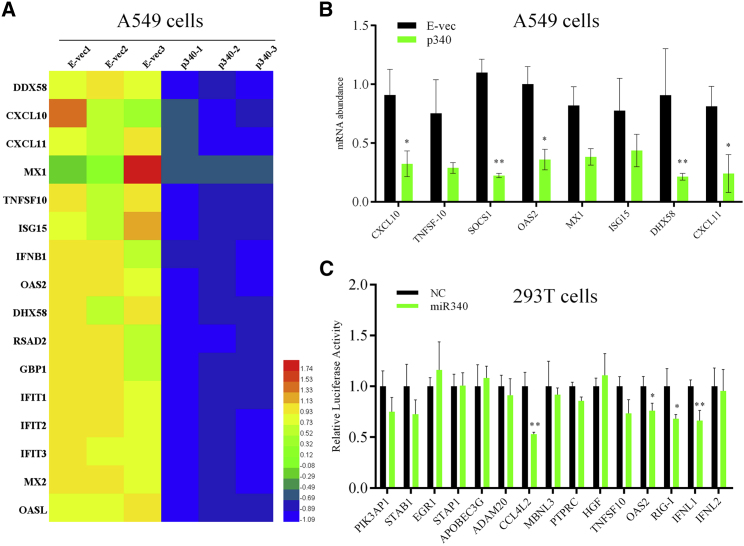
We next aimed to explore the target genes of miR340, whose expression could impact virus replication. In consequence, the expression of several interferon-stimulated genes, including CXCL10, interferon (IFN)β, RIG-I, and OAS2, was found to be downregulated in A549 cells overexpressing miR340, as assessed by comparing gene expression in these cells versus that of cells transfected with an empty vector (Figure 3A). Downregulation was confirmed by qRT-PCR, indicating that these genes are suppressed by miR340 (Figure 3B). In addition, potential miR340 targets were computationally identified using miRanda and TargetScan, and those associated with virus infection were tested using a reporter assay, in which full-length 3ʹ UTRs from target genes were subcloned downstream of firefly luciferase. Renilla luciferase was used as an internal control. These constructs were then cotransfected with miR340 mimetics or the negative control into 293T cells, and the relative luciferase activities were measured after 24 h. As shown in Figure 3C, the miR340 mimetics reduced luciferase expression in the constructs containing 3ʹ UTRs from OAS2, RIG-I, IFNL1, or CCL4L2, implying that these genes are physiological miR340 targets.
Figure 3.
RIG-I and OAS2 Are Putative miR340 Targets
(A) Heatmaps of partially downregulated genes in A549 cells following transfection with a vector encoding miR340. (B) Downregulation was validated by qRT-PCR. (C) 293T cells were cotransfected with miR340 or negative control and luciferase reporter constructs encoding 3′ UTRs from candidate genes. Luciferase was quantified 24 h post-transfection and normalized to the mean luciferase activity in cells cotransfected with the negative control. Results represent the mean ± SD of three independent experiments. p value was calculated by using two-way ANOVA with Bonferroni multiple comparison test (B) or the Student’s t test (C) (*p < 0.05, **p < 0.01). E-vec, empty vector; p340, pcDNA3.1-miR340.
miR340 Targets and Downregulates RIG-I and OAS2
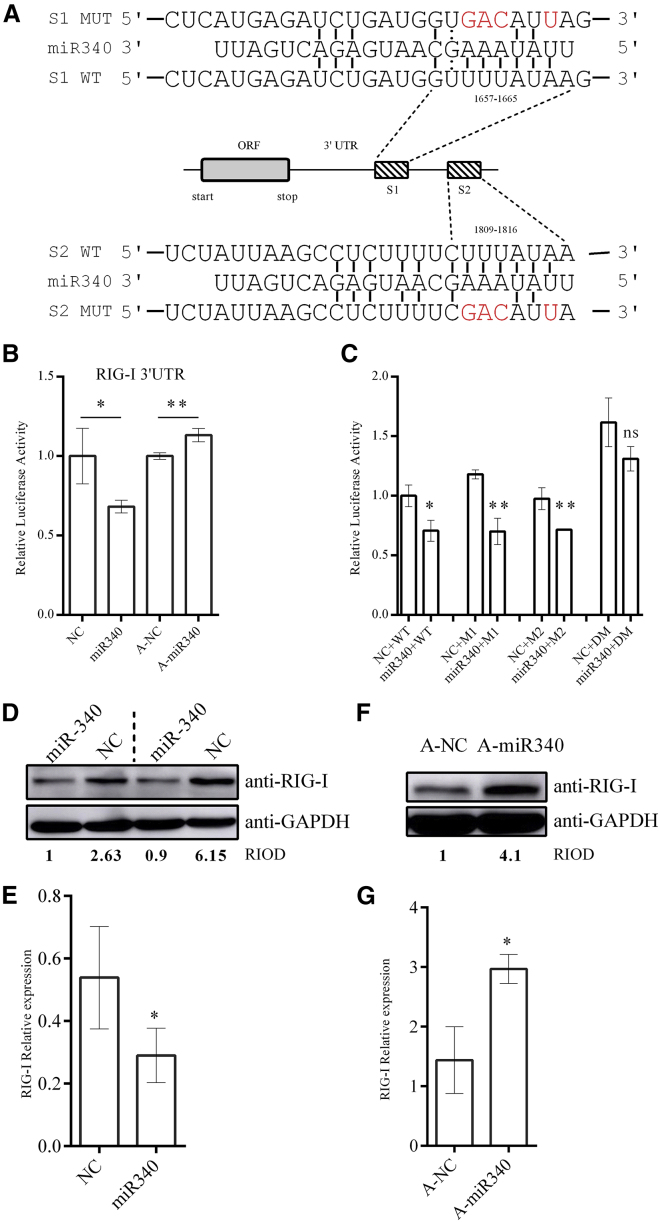
miR340 target sites were identified at positions 1,657 and 1,809 in the 3ʹ UTR of RIG-I (Figure 4A). Accordingly, luciferase expression from a reporter construct containing the wild-type 3ʹ UTR was significantly suppressed by miR340, but expression was stimulated following treatment with miR340 inhibitors (Figure 4B). Additionally, individual mutations in the miR340 target sites did not abrogate these effects, whereas mutations in both sites restored the suppressive effects of miR340 (Figure 4C), indicating that both sites contributed to miR340 function. Moreover, using western blots and RT-PCR, we demonstrated that the ectopic expression of miR340, but not that of the negative control miRNA, dramatically suppressed endogenous RIG-I (Figures 4D and 4E). Conversely, miR340 inhibitors, but not control inhibitors, clearly increased endogenous RIG-I expression at the protein and mRNA levels (Figures 4F and 4G).
Figure 4.
miR340 Inhibits RIG-I Expression in A549 Cells
(A) Predicted miR340 target sites in the 3′ UTR of RIG-I. Mutations are highlighted in red. (B) miR340, inhibitor, or the corresponding control was cotransfected into 293T cells along with a luciferase reporter vector encoding the 3′ UTRs from RIG-I. Relative luciferase activity was measured 24 h post-transfection. (C) Luciferase reporter vector encoding wild-type or mutated (M1, M2, and DM) 3′ UTRs from RIG-I were cotransfected into 293T cells with 60 nmol/L miR340 or negative control. After 24 h, cells were lysed and assayed for luciferase activity. The value of NC + WT was set to 1 for normalization. (D) Western blot and (E) RT-PCR for RIG-I in A549 cells transfected with 50 nM miR340 mimetics or negative control. GAPDH was used as a loading control, and each lane represents an independent experiment. (F) Western blot and (G) RT-PCR for RIG-I in A549 cells transfected with miR340 inhibitor or inhibitor control. GAPDH was used as a loading control. Results represent the mean ± SD of three independent experiments. p value was calculated by using Student’s t test (*p < 0.05, **p < 0.01). A-NC, negative control inhibitor; A-miR340, miR340 inhibitor; ns, not significant; RIOD, relative integrated optical density.
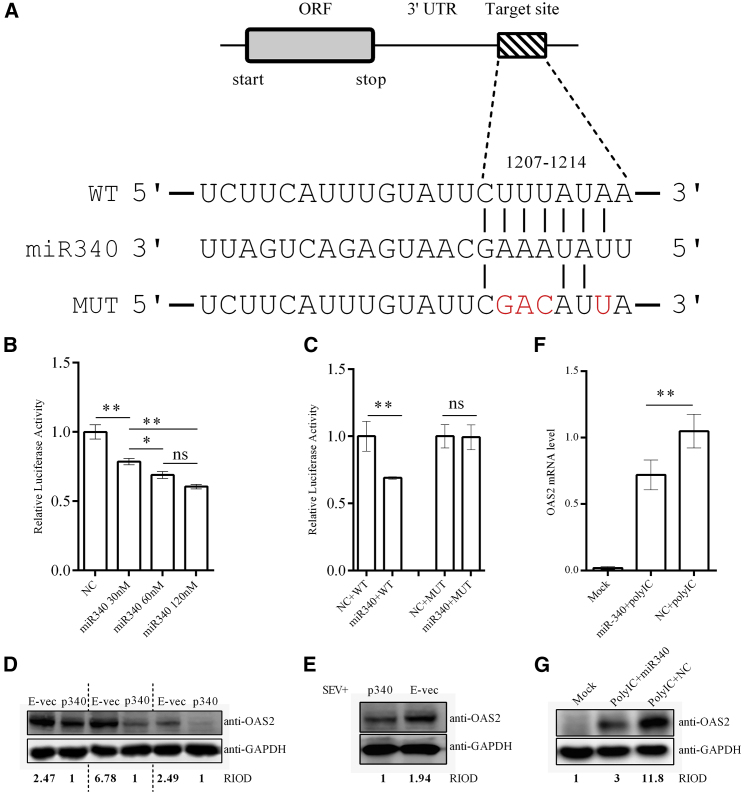
In OAS2, one target site of miR340 was predicted at position 1,207 in the 3ʹ UTR (Figure 5A). As shown in Figure 5B, luciferase expression from a reporter construct containing the wild-type sequence was significantly suppressed by transfected miR340 in a concentration-dependent fashion. As expected, mutation of several bases in the seed sequence (Figure 5A) abrogated this effect (Figure 5C). Similarly, transfected miR340 reduced OAS2 protein levels (Figure 5D), even in virus-infected cells or cells transfected with poly(I:C) (Figures 5E–5G). Collectively, these results strongly suggest that RIG-I and OAS2 represent physiological targets of miR340.
Figure 5.
OAS2 Is Targeted by miR340
(A) miR340 target site in the 3′ UTR of OAS2. Mutations targeting the seed complementarity to miR340 are highlighted in red. (B) Luciferase reporter vector encoding the OAS2 3′ UTR was cotransfected into 293T cells with the indicated concentrations of miR340 mimetics. Relative luciferase activity was quantified 24 h post-transfection. (C) Luciferase reporter vector encoding wild-type or mutated 3′ UTRs from OAS2 was cotransfected into 293T cells with miR340 mimetics or negative control, and luciferase activity was measured 24 h later. (D) Cellular OAS2 levels were quantified after transfection with empty vector or that encoding miR340 (three independent experiments are presented). (E) A549 cells were transfected for 24 h with empty vector or that encoding miR340 and then infected with Sendai virus. Endogenous OAS2 levels were quantified by western blotting 24 h thereafter. (F) qRT-PCR and (G) western blot for OAS2 in A549 cells cotransfected with 0.01 μg poly(I:C) and 60 nM miR340 mimetics or negative control. Data represent the mean ± SD from triplicate independent experiments. p value was calculated by using Student’s t test (C) or one-way ANOVA with Bonferroni multiple comparison test (B and F) (*p < 0.05, **p < 0.01). E-vec, empty vector; p340, pcDNA3.1-miR340; ns, not significant; RIOD, relative integrated optical density.
miR340 Suppresses RIG-I Signaling during Infection with Influenza A Virus
To test whether enhanced virus replication is associated with the miR340-mediated suppression of RIG-I and its downstream signaling, A549 cells were transfected with miR340 or control RNA for 24 h, followed by infection with H5N1/HM, and RIG-1 was quantified 12 h thereafter. As expected, RIG-I mRNA and protein levels were clearly decreased upon influenza A virus infection in the presence of miR340 (Figures 6A and 6B), whereas anti-miR340 enhanced the expression of RIG-I in A549 cells (Figures 6C and 6D). Similarly, cotransfection with miR340 attenuated the ability of poly(I:C) to induce RIG-I (Figures 6E and 6F). In contrast, miR340 inhibitors, but not inhibitor controls, markedly enhanced RIG-I expression upon cotransfection with poly(I:C) (Figures 6G and 6H). Finally, during H5N1/HM infection, miR340 depleted mRNAs downstream of RIG-I, including CCL5, IFNβ, ISG15, interleukin (IL)-6, IFNα, and CXCL10 (Figure 6I). Moreover, we found that miR340 inhibited the secretion of CCL5, CXCL10, IL-6, and IFNβ proteins by A549 cells infected with influenza virus (Figures 6J and 6K). Together, these results suggest the miR340 causes a severe decrease in the RIG-I-dependent expression of antiviral genes.
Figure 6.
miR340 Suppresses Virus-Induced RIG-I Expression and Downstream Signaling
(A) RT-PCR and (B) western blot for RIG-I in A549 cells transfected with 60 nM miR340 mimetics or negative control and then infected with H5N1/HM at an MOI of 0.1 for 12 h. (C and D) The same procedures as in (A) and (B) were performed to assess the effect of miR340 inhibitor on RIG-I mRNA (C) and protein (D) levels during H5N1/HM infection. (E) RT-PCR and (F) western blot for RIG-I in A549 cells cotransfected with 0.01 μg poly(I:C) and 60 nM miR340 mimetics or negative control. (G) RT-PCR and (H) western blot for RIG-I in A549 cells cotransfected with 0.01 μg poly(I:C) and 60 nM miR340 inhibitor or inhibitor control. (I) Quantification of expression of CCL5, ISG15, IL-6, IFNβ, CXCL10, and IFNα mRNA in A549 cells that were mock transfected or transfected with miR340 or negative control, and then infected with H5N1/HM at an MOI of 0.1 for 12 h. (J and K) Quantities of CCL5, CXCL10 (J), IL-6, and IFNβ (K) proteins secreted into cell culture medium were determined by multiplex sandwich immunoassays. Data represent the mean ± SD of three independent experiments. p value was calculated by ANOVA (*p < 0.05, **p < 0.01). A-NC, negative control inhibitor; A-miR340, miR340 inhibitor; ND, not determined; RIOD, relative integrated optical density.
miR340 Knockout Restricts Virus Replication by Inducing RIG-I
miR340 was knocked out in A549 cells using the lentiviral CRISPR-Cas9 system and a pair of suitable guide RNAs (Figure 7A) driven by two independent U6 promoters on the same plasmid. A control plasmid containing scrambled guide RNAs was also constructed. Clonal cell lines were then generated as described in the Materials and Methods, and the knockout of miR340 was confirmed by sequencing and RT-PCR (Figures 7A and 7B). The expression of RIG-I mRNA increased in the knockout cells in comparison with that in wild-type cells or cells transformed with control guide RNAs, further validating the regulatory relationship with miR340 (Figure 7C). Similarly, we found by western blotting that RIG-I protein was more abundant in the miR340-knockout cells (Figure 7D). In addition, transfection of miR340 into the knockout cells clearly downregulated RIG-I (Figure 7E). These results confirm that endogenous miR340 is a key suppressor of RIG-I.
Figure 7.
Knockout of miR340 Boosts RIG-I Expression and Restricts Virus Infection in A549 Cells
(A) Validation of miR340 deletion from a clonal cell line by PCR and sequencing. miR340 was deleted using the lentiviral CRISPR-Cas9 system. A pair of gRNAs is highlighted in blue and protospacer adjacent motifs (PAMs) are indicated in green. The dashes represent deleted regions of miR340 in the genome. (B) Knockout efficiency was tested by RT-PCR. (C and D) Wild-type A549 cells, miR340-knockout cells, and control cells edited with scrambled guide RNAs were counted and seeded in a 12-well plate at the same density. After reaching 90% confluence, cells were harvested and analyzed by RT-PCR and western blot for RIG-I mRNA (C) and protein (D) levels. (E) miR340-knockout cells were transfected with 60 nM miR340 or analyzed for RIG-I (each lane shows an independent experiment). (F–H) miR340-knockout cells and control knockout cells were infected for 24 h with or without H5N1/HM at an MOI of 0.1, and cellular RIG-I (G), viral nucleoprotein, and M1 mRNA (F) levels were quantified by RT-PCR. The protein levels of RIG-I and viral nucleoprotein (H) were measured by western blot. (I and J) One-step growth curve was observed in miR340-knockout cells and control knockout cells following infection with H5N1/HM (I) or vesicular stomatitis virus (J). Data represent the mean ± SD from triplicate independent experiments. p value was calculated by using Student’s t test or ANOVA (*p < 0.05, **p < 0.01). ns, not significant; RIOD, relative integrated optical density.
Furthermore, we infected miR340-knockout or control cells with H5N1/HM virus at an MOI of 0.1, and then we quantified the viral load and RIG-I expression. As expected, viral M1 and NP mRNA were considerably less abundant in miR340-deficient cells (Figure 7F), whereas RIG-I mRNA showed increased expression (Figure 7G). The mRNA levels of RIG-I and NP are consistent with their protein levels, as verified by western blot (Figure 7H). In addition, the growth curves of H5N1/HM and vesicular stomatitis viruses were measured in both miR340-knockout and control cell lines. Expectedly, the viruses proliferated more slowly in miR340-knockout cells, indicating restricted virus growth in comparison to that in the control cells (Figures 7I and 7J). Taken together, these results indicate that endogenous miR340 is a key immune regulator under normal conditions or during viral infection.
Discussion
Humans and animals are susceptible to the influenza A virus, which may then cause serious sequelae. Notably, miRNAs, which are crucial post-transcriptional regulators, are also believed to be involved in virus-host interactions, although the underlying mechanisms continue to be a very active area of research. miR340 was largely revealed as a tumor suppressor by targeting key regulators of tumor cell migration and invasion.29, 30, 31 We now report that miR340 suppresses RIG-I and OAS2 by directly binding to their 3ʹ UTRs, thereby attenuating immunity. As a result, cells appear to downregulate miR340 in response to the influenza A virus to boost immunity. Indeed, our data not only indicate that miR340 is a negative regulator of the RIG-I-mediated innate antiviral response but also suggest a novel mechanism regarding host control of viral infections.
Viral infection is important to drive differential expression of cellular miRNA. Initially, we observed that miR340 levels decreased upon infection with influenza A virus in either A549 cells or 293T cells. In addition, miR340 expression was correlated with other RNA virus infections, since Sendai virus and vesicular stomatitis virus can also reduce the abundance of cellular miR340. Likewise, a previous report suggested that the expression of miR340 is downregulated in hepatitis B virus-infected cells.32 miR340 may thus play an even more profound role in the host antiviral response. Moreover, we speculate that miR340 is regulated at the transcriptional level, because miR340 precursor levels were correspondingly reduced following influenza virus infection. Many studies have already demonstrated that miRNA promoters are controlled by classic transcription factors, such as E2F1, NF-κB, and/or Sp1,33, 34, 35 which are associated with viral infections. Still, miRNA stability can be occasionally regulated by viral infections.36 Nonetheless, the precise mechanisms by which the influenza A virus regulates miR340 warrant further investigation.
Given that the host strategically reduces miR340 abundance to respond to viral infections, it may be responsible for fine-tuning virus-induced gene expression in infected cells. Accordingly, the ectopic expression of miR340 in A549 cells markedly enhances the proliferation of influenza A virus, while inhibiting or knocking out endogenous miR340 significantly inhibits infection. We found that miR340 targets serial tumor-associated genes, including tissue plasminogen activator (PLAT),29 phosphoserine aminotransferase 1 (PSAT1),30 DNA-directed polymerase zeta catalytic subunit (REV3L),31 and signal transducer and activator of transcription 3 (STAT3).32 We attempted to investigate such mechanisms, although discrepancies in results among different cell lines discouraged us from further pursuing these studies. Instead, using transcriptomic analyses in A549 cells, we found that the overexpression of miR340 may diminish antiviral responses. Several downregulated genes, including OAS2, RIG-I, IFNL1, and CCL4L2, were identified as potential targets of miR340, indicating that these regulatory relationships may play key roles during viral infection. Our data, together with those of others, imply that miR340 acts as a multifunctional regulator in different signaling pathways and in different cell types.
RIG-I is a crucial cytosolic pattern recognition receptor that triggers antiviral cascades primarily in response to RNAs from viruses, such as the influenza virus, Sendai virus, and vesicular stomatitis virus.20, 21 Accordingly, RIG-I is strictly controlled not only to prevent infection but also to avoid immunotoxicity. RIG-I regulators include miRNAs such as miR545, which directly targets RIG-I in pancreatic ductal adenocarcinoma.37 Similarly, miR146a was demonstrated to suppress RIG-I, thereby impeding the clearance of hepatitis B virus,38 while miR485 was recently shown to target RIG-I mRNA for degradation, resulting in an impaired antiviral response and enhanced replication of the influenza A virus.13 In the present study, two miR340 target sites were identified in the RIG-I 3ʹ UTR, and cellular RIG-I expression levels were clearly reduced by miR340. Expectedly, inhibition or deletion of endogenous miR340 can lead to elevated RIG-I expression. This regulatory pattern was also observed during influenza virus infection, indicating that miR340 plays a crucial role in immunity adjustment. Collectively, these studies suggest that miRNAs, apart from post-translational modifications, are key regulators of RIG-I in different cell types.
In parallel, we determined that miR340 can target the OAS2 3ʹ UTR by using luciferase assays. A follow-up study showed that miR340 suppressed OAS2 expression with or without influenza virus infection. Although both RIG-I and OAS2 are believed to be important antiviral genes, we focused more on the alteration of RIG-I mediated by miR340, since its activation could have more profound effects on the release of interferon-stimulated genes (ISGs) during virus infection. In fact, we found that miR340 mitigated the secretion of cytokines downstream of RIG-I, such as CCL5, CXCL10, IL-6, and IFNβ, in influenza virus-infected cells. Accordingly, we inferred that miR340 simultaneously degrades RIG-I and OAS2 transcripts and that this dual effect likely results in the facilitation of viral replication. However, in this study, we did not exclude the possibility that miR340 targets other cellular factors identified by RNA sequencing, including IFNL1 and CCL4L2, which may have effects on viral replication. Hence, the intrinsic relationships whereby miR340 modulates cellular signaling networks during viral infection still need to be clarified further in future studies.
Unfortunately, in vivo studies were not attempted since miR340-binding sites were not identified in the 3ʹ UTRs of mouse RIG-I and OAS2, suggesting distinctive functions of miR340 in different species. Whether there are conserved miR340-targeting sites among different species requires further investigation. Encouragingly, miR144-deficient mice were reported to be more resistant to influenza virus because of a decrease in immunosuppression.14 Indeed, stimulating immunity via RIG-I is an effective strategy to combat the influenza A virus,39, 40 and experimental miRNA-dependent strategies have been developed against hepatitis C and influenza A viruses.41, 42 In the present study, the antiviral factors RIG-I and OAS2 were identified as bona fide targets of miR340, supporting the concept that targeting host miRNAs will become novel therapeutic approaches against influenza virus infection, considering the pleiotropic function of miRNAs in the virus-host interplay.
Materials and Methods
Cells and Viruses
Madin-Darby canine kidney (MDCK), human A549 lung epithelial (A549), and HEK293T cells were cultured at 37°C and 5% CO2 in DMEM (Invitrogen, Carlsbad, CA, USA), F12 medium, and RPMI 1640 medium (Invitrogen), respectively, supplemented with 10% fetal bovine serum (Gibco, Auckland, NZ). The highly pathogenic avian influenza H5N1 strain designated H5N1/HM was isolated from duck brain tissues in our laboratory. H5N6 strain A/duck/Hubei/WH18/2015 was isolated and propagated by our laboratory. A/PR/8/34 (H1N1) (PR8) was acquired from the State Key Laboratory of Agricultural Microbiology. Sendai virus was kindly provided by Professor Zhengfan Jiang at the Institute of Life & Science, Peking University, China, while vesicular stomatitis virus encoding EGFP was provided by the Harbin Veterinary Research Institute. H5N1 and H5N6 viruses were handled in an animal Biosafety Level 3 laboratory. The titers of influenza A virus and vesicular stomatitis virus in culture media were measured using the TCID50 and plaque assays in MDCK cells.
Vector Construction and Reagents
The hsa-mir-340 precursor was PCR amplified from genomic DNA and subcloned into pcDNA3.1+ (Invitrogen) between the XhoI and BamHI restriction sites. Similarly, the 3′ UTRs of putative miR340 targets were PCR amplified from genomic DNA and cloned into the luciferase reporter vector pmirGLO (Promega, Madison, WI, USA). These targets were computationally identified using miRanda and TargetScan, both of which are available online. Variants of the 3′ UTRs in RIG-I and OAS2 were constructed by PCR-mediated mutagenesis. All DNA constructs were validated by sequencing. miR340 mimetics, negative control mimetics, miR340 inhibitors, and inhibitor controls were purchased from GenePharma (Shanghai, China), while poly(I:C) and Lipofectamine 2000 were purchased from Sigma and Invitrogen, respectively.
Lentivirus Production
A lentiviral vector encoding EGFP-Cas9, the packaging plasmid pMD2.G, and the envelope plasmid psPAX2 were kindly provided by Lisheng Zhang. Oligonucleotides, containing a U6 promoter and a miR340 guide RNA at each end, were chemically synthesized and subcloned into the BsmBI site in the EGFP-Cas9 lentiviral vector. A control vector was also constructed via insertion of a nonsense guide RNA. To produce lentiviruses, 293T cells in a 6-well plate were cotransfected with 10 μg EGFP-Cas9 lentiviral vector, psPAX2, and pMD2.G at a ratio of 4:3:1. The medium was collected 36 h post-transfection and centrifuged for 15 min at 3,000 × g and 4°C to remove cell debris. The resulting supernatant was directly added to A549 cells in a 12-well plate, which were then incubated for 24 h, and sorted by flow cytometry to isolate cells expressing EGFP. Subsequently, purified cells were diluted and seeded in a 96-well plate to generate clonal cell lines. miR340 loci in clones were amplified by PCR and inserted into the pClone007 simple vector (TsingKe Biotech, Beijing, China) for sequencing. The forward primer (5′-TCCTTTCCCTACTCCTTTCCCTACTC-3′) and reverse primer (5′-TTACTACCTTACTCAAAATTGTTACA-3′) were used to detect miR340 mutations. miR340 expression was also measured by RT-PCR. The miR340 guide RNAs were 5′-GATTGTCATATGTCGTTTG-3′ and 5′-TACCTGGTATCTTATACCACT-3′, while the control guide RNA was 5′-GTCAAAGTGCTTACAGTGC-3′.
High-Throughput RNA Sequencing
A549 cells seeded in 12-well plates were transfected with 1 μg miR340 expression plasmid or empty vector, and total RNA was isolated after 24 h using TRIzol (Invitrogen). RNA degradation and contamination were assessed by electrophoresis on 1% agarose gels. Purity was assessed on a NanoPhotometer (Implen, Munich, Germany), while RNA integrity was assessed using the RNA Nano 6000 Assay Kit for the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA), following the manufacturer’s recommendations, and index codes were added to attribute sequences to a sample. Index-coded samples were then clustered on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. Finally, libraries were sequenced on an Illumina Hiseq platform to generate 125- or 150-bp paired-end reads. Differential expression was determined in DESeq using a model based on a negative binomial distribution, and the resulting p values were adjusted using the Benjamini-Hochberg approach to control false discovery rates. Genes with adjusted p values < 0.05 were considered significant. The raw data and processed files were submitted to NCBI’s GEO and are accessible through GEO: GSE115361.43
qRT-PCR
Total RNA was isolated using TRIzol (Invitrogen) in accordance with the manufacturer’s instructions. Genomic DNA was removed with DNase I, and 2 mg RNA was reverse transcribed using avian myeloblastosis virus reverse transcriptase (TaKaRa Biotechnology, DaLian, China) and an oligo (dT)18 primer. GAPDH was used as a reference to normalize the amount of mRNA in each sample. Mature miR340 was reverse transcribed using specific stem-loop primers, quantified by SYBR-based (Roche, Basel, Switzerland) qRT-PCR on an ABI ViiA7 instrument (Applied Biosystems, Foster City, CA, USA) and normalized to U6 RNA. miR340 precursors were transcribed using specific primers, and GAPDH was used as a reference gene. The sequences of all primers used for qRT-PCR are available from the corresponding author upon request.
Cytokine and Chemokine Measurements
The 12-well-plated A549 cells were mock transfected or transfected with miR340 or control mimetics for 24 h, and then they were infected with or without influenza A virus at an MOI of 0.1. At 12 h later, the medium of these cell cultures was harvested, and the cytokine and chemokine concentrations (CCL5, CXCL10, IL-6, and IFNβ) were assessed using the Human Premixed Multi-Analyte Kit (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s protocol.
Western Blot
The cells were washed and lysed on ice in Tris Lysis Buffer (Cell Signaling Technology, Danvers, MA, USA) containing 1% EDTA-free protease inhibitors (Roche). Lysates were collected, quantified, and then separated by 10% SDS-PAGE. The proteins were transferred to nitrocellulose membranes. Each membrane was blocked in 5% nonfat milk and then incubated with anti-RIG-I, anti-OAS2, anti-virus NP, or anti-GAPDH antibody. After washing, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody. Finally, the signals were detected using an Immobilon Western Chemiluminescent HRP Substrate kit (Thermo Fisher Scientific) and ChemBis (Eastwin). The antibodies against RIG-I, OAS2, and GAPDH were purchased from ABclonal (Woburn, MA, USA), and the anti-rabbit and anti-mouse immunoglobulin G (IgG) conjugated to HRP were obtained from GE Healthcare (Chicago, IL, USA). The antibodies specific for influenza virus NP, PA and M1, were obtained from GeneTex (Irvine, CA, USA). All blots were quantified by calculating the relative integrated optical density (RIOD) in Image-Pro Plus software; the relative ratio is given below each blot.
Dual Luciferase Assays
293T cells were seeded into 24-well plates, and they were transfected for 24 h with 6 μL Lipofectamine 2000 (Invitrogen) containing 0.5 μg pGLO encoding 3′ UTRs of interest, as well as 60 nM miR340 mimetics, control mimetics, miR340 inhibitors, or inhibitor controls. Luciferase was then quantified according to standard protocols using the Dual-Luciferase Reporter Assay System Kit (Promega) and normalized to Renilla luciferase activity.
Statistical Analysis
Groups were compared using Student’s t tests or ANOVA in GraphPad Prism (San Diego, CA, USA). Data were collected from triplicates, are representative of at least three independent experiments, and are reported as means ± SD; p values less than 0.05 were considered statistically significant.
Author Contributions
M.J., H.C., and L.Z. conceived and designed the experiments. L.Z., Z.W., and X.Z. performed the experiments. L.Z. and X.S. supervised the study design and analyzed the data. X.Z. and K.H. contributed reagents and materials. L.Z. and M.J. wrote the paper. All authors reviewed the manuscript.
Conflicts of Interest
The authors have no conflicts to disclose.
Acknowledgments
This work was supported by funding from the National Key Research and Development Program of China (2016YFD0500205), National Natural Science Foundation of China (31702251), and the China Postdoctoral Science Foundation (2016M602325). We acknowledge Zhengfan Jiang and Hualan Chen for kindly providing the SEV and VSV, respectively. We thank Lisheng Zhang for providing the lentiviral CRISPR-Cas9 system.
References
- 1.World Health Organization (2017). Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003-2017. https://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/
- 2.Korteweg C., Gu J. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am. J. Pathol. 2008;172:1155–1170. doi: 10.2353/ajpath.2008.070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Ro S., Park C., Young D., Sanders K.M., Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Pedersen I.M., Cheng G., Wieland S., Volinia S., Croce C.M., Chisari F.V., David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli R., Paone A., Fabbri M., Zanesi N., Calore F., Cascione L., Acunzo M., Stoppacciaro A., Tubaro A., Lovat F. Toll-like receptor 3 (TLR3) activation induces microRNA-dependent reexpression of functional RARβ and tumor regression. Proc. Natl. Acad. Sci. USA. 2013;110:9812–9817. doi: 10.1073/pnas.1304610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trobaugh D.W., Klimstra W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loveday E.K., Svinti V., Diederich S., Pasick J., Jean F. Temporal- and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J. Virol. 2012;86:6109–6122. doi: 10.1128/JVI.06892-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buggele W.A., Johnson K.E., Horvath C.M. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J. Biol. Chem. 2012;287:31027–31040. doi: 10.1074/jbc.M112.387670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buggele W.A., Krause K.E., Horvath C.M. Small RNA profiling of influenza A virus-infected cells identifies miR-449b as a regulator of histone deacetylase 1 and interferon beta. PLoS ONE. 2013;8:e76560. doi: 10.1371/journal.pone.0076560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarbrough M.L., Zhang K., Sakthivel R., Forst C.V., Posner B.A., Barber G.N., White M.A., Fontoura B.M. Primate-specific miR-576-3p sets host defense signalling threshold. Nat. Commun. 2014;5:4963. doi: 10.1038/ncomms5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingle H., Kumar S., Raut A.A., Mishra A., Kulkarni D.D., Kameyama T., Takaoka A., Akira S., Kumar H. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015;8:ra126. doi: 10.1126/scisignal.aab3183. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberger C.M., Podyminogin R.L., Diercks A.H., Treuting P.M., Peschon J.J., Rodriguez D., Gundapuneni M., Weiss M.J., Aderem A. miR-144 attenuates the host response to influenza virus by targeting the TRAF6-IRF7 signaling axis. PLoS Pathog. 2017;13:e1006305. doi: 10.1371/journal.ppat.1006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song L., Liu H., Gao S., Jiang W., Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010;84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y.J., Yang J., Fan X.L., Zhao H.B., Hu W., Li Z.P., Yu G.C., Ding X.R., Wang J.Z., Bo X.C. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell. Mol. Med. 2012;16:2539–2546. doi: 10.1111/j.1582-4934.2012.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf S., Wu W., Jones C., Perwitasari O., Mahalingam S., Tripp R.A. MicroRNA regulation of human genes essential for influenza A (H7N9) replication. PLoS ONE. 2016;11:e0155104. doi: 10.1371/journal.pone.0155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y., Jiang L., Lai W., Qin Y., Zhang T., Wang S., Ye X. MicroRNA-33a disturbs influenza A virus replication by targeting ARCN1 and inhibiting viral ribonucleoprotein activity. J. Gen. Virol. 2016;97:27–38. doi: 10.1099/jgv.0.000311. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 21.Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 22.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 23.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 24.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Hou J., Wang P., Lin L., Liu X., Ma F., An H., Wang Z., Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Fan X., He X., Sun H., Zou Z., Yuan H., Xu H., Wang C., Shi X. MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-alpha. Cell. Mol. Immunol. 2012;9:497–502. doi: 10.1038/cmi.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L., Zhu J., Zhou H., Zhao Z., Zou Z., Liu X., Lin X., Zhang X., Deng X., Wang R. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci. Rep. 2015;5:14991. doi: 10.1038/srep14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita D., Kondo T., Ohue S., Takahashi H., Ishikawa M., Matoba R., Suehiro S., Kohno S., Harada H., Tanaka J., Ohnishi T. miR340 suppresses the stem-like cell function of glioma-initiating cells by targeting tissue plasminogen activator. Cancer Res. 2015;75:1123–1133. doi: 10.1158/0008-5472.CAN-14-0938. [DOI] [PubMed] [Google Scholar]
- 30.Yan S., Jiang H., Fang S., Yin F., Wang Z., Jia Y., Sun X., Wu S., Jiang T., Mao A. MicroRNA-340 inhibits esophageal cancer cell growth and invasion by targeting phosphoserine aminotransferase 1. Cell. Physiol. Biochem. 2015;37:375–386. doi: 10.1159/000430361. [DOI] [PubMed] [Google Scholar]
- 31.Arivazhagan R., Lee J., Bayarsaikhan D., Kwak P., Son M., Byun K., Salekdeh G.H., Lee B. MicroRNA-340 inhibits the proliferation and promotes the apoptosis of colon cancer cells by modulating REV3L. Oncotarget. 2017;9:5155–5168. doi: 10.18632/oncotarget.23703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong Q., Wu S., Wang J., Zeng X., Chen J., Wei M., Guan H., Fan C., Chen L., Guo D., Sun G. Hepatitis B virus promotes cancer cell migration by downregulating miR-340-5p expression to induce STAT3 overexpression. Cell Biosci. 2017;7:16. doi: 10.1186/s13578-017-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woods K., Thomson J.M., Hammond S.M. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 34.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Z., Xiao S.B., Xu P., Xie Q., Cao L., Wang D., Luo R., Zhong Y., Chen H.C., Fang L.R. miR-365, a novel negative regulator of interleukin-6 gene expression, is cooperatively regulated by Sp1 and NF-kappaB. J. Biol. Chem. 2011;286:21401–21412. doi: 10.1074/jbc.M110.198630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buck A.H., Perot J., Chisholm M.A., Kumar D.S., Tuddenham L., Cognat V., Marcinowski L., Dölken L., Pfeffer S. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song B., Ji W., Guo S., Liu A., Jing W., Shao C., Li G., Jin G. miR-545 inhibited pancreatic ductal adenocarcinoma growth by targeting RIG-I. FEBS Lett. 2014;588:4375–4381. doi: 10.1016/j.febslet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Hou Z., Zhang J., Han Q., Su C., Qu J., Xu D., Zhang C., Tian Z. Hepatitis B virus inhibits intrinsic RIG-I and RIG-G immune signaling via inducing miR146a. Sci. Rep. 2016;6:26150. doi: 10.1038/srep26150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin L., Liu Q., Berube N., Detmer S., Zhou Y. 5′-Triphosphate-short interfering RNA: potent inhibition of influenza A virus infection by gene silencing and RIG-I activation. J. Virol. 2012;86:10359–10369. doi: 10.1128/JVI.00665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martínez-Gil L., Goff P.H., Hai R., García-Sastre A., Shaw M.L., Palese P. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J. Virol. 2013;87:1290–1300. doi: 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 42.Peng S., Wang J., Wei S., Li C., Zhou K., Hu J., Ye X., Yan J., Liu W., Gao G.F. Endogenous cellular microRNAs mediate antiviral defense against influenza A virus. Mol. Ther. Nucleic Acids. 2018;10:361–375. doi: 10.1016/j.omtn.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]