Abstract
The zinc finger protein Snail is a master regulator of epithelial–mesenchymal transition (EMT) and a strong inducer of tumor metastasis, yet the signal cascades triggered by Snail have not been completely revealed. Here, we report the discovery of the sulfation program that can be induced by Snail in breast cancer cells, and which plays an essential role in cell migration and metastasis. Specifically, Snail induces the expression of PAPSS2, a gene that encodes a rate-limiting enzyme in sulfation pathway, and VCAN, a gene that encodes the chondroitin sulfate proteoglycan Versican in multiple breast cancer cells. Depletion of PAPSS2 in MCF7 and MDA-MB-231 cells results in reduced cell migration, while overexpression of PAPSS2 promotes cell migration. Moreover, MDA-MB-231-shPAPSS2 cells display a significantly lower rate of lung metastasis and lower number of micrometastatic nodules in nude mice, and conversely, MDA-MB-231-PAPSS2 cells increase lung metastasis. Similarly, depletion of VCAN dampens the cell migration activity induced by Snail or PAPSS2 in MCF 10A cells. Moreover, PAPSS inhibitor sodium chlorate effectively decreases cell migration induced by Snail and PAPSS2. More importantly, the expression of Snail, PAPSS2, and VCAN is positively correlated in breast cancer tissues. Together, these findings are important for understanding the genetic programs that control tumor metastasis and may identify previously undetected therapeutic targets to treat metastatic disease.
Subject terms: Metastasis, Tumour biomarkers
Introduction
The transcriptional factor Snail is a critical regulator of cell migration in normal embryonic development and diseases such as tumor metastasis and fibrosis [1–3]. Genetic deletion of the snail gene in mice results in embryonic lethality due to defects in gastrulation [2, 3]. Snail is also highly expressed in the invasive cells of various types of tumors including ductal breast carcinomas, colorectal cancer, prostate cancer, and hepatocellular carcinomas and serves as an early marker for the malignant phenotype and prognosis [4–7]. Forced expression of Snail in various types of epithelial cells induces mesenchymal phenotype accompanied by increased cell survival, migration, stemness, invasiveness, and chemoresistance [4, 8, 9]. Most recently, Snail was found to be essential for cancer-associated fibroblast activation and promote tumor-initiating cell expansion in mouse breast [10, 11]. These studies collectively demonstrate that Snail plays critical roles in both tumor metastasis and recurrence.
Snail belongs to the C2H2 superfamily of transcription factors containing C-terminal tandem zinc finger motifs and an N-terminal SNAG repression domain [1]. The zinc finger motifs can recognize the E-box DNA sequences of the target genes, whereas the SNAG domain is a potent, highly conserved, and transferable repression motif and recruits various repressive cofactors. The transcriptional repressive function of Snail has been extensively interrogated and various proteins involved in gene silencing were identified as Snail interacting cofactors such as histone deacetylases, mSin3A, Ezh2, LSD1, and Ajuba/Prmt5/14-3-3 ternary complex [12–17]. We further found that Snail, Ring1B, and EZH2 form distinct protein complexes, which are cooperatively recruited to the target promoter to repress Snail target gene expression [18]. Notably, several studies have shown that Snail can directly activate gene expression. For example, Snail can directly activate genes during mesoderm development of Drosophila by potentiating Twist-mediated enhancer activation [19]; in HepG2 cells, Snail associates with SP-1 and EGR-1 to induce transcription of p15INK4b [20]; MMP9 and Fibronectin are also transcriptionally activated by Snail [21, 22]. Genome-wide gene expression profiling analyses revealed that Snail can induce a large pool of gene expression in MCF 10A and MCF7 cells. However, the role of these Snail-activated genes in tumor development and metastasis remain elusive.
Sulfation is a process of transferring a sulfate group (SO4−2) from the universal sulfate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to appropriate acceptor molecules including xenobiotics, hormones, lipids, neurotransmitters, steroids, proteins, and proteoglycans. A major class of sulfation substrates is the carbohydrate side-chains of proteoglycans, which are important structural components of extracellular matrix (ECM) in various tissues. Sulfation involves in three indispensable steps: transport of inorganic sulfate into cytoplasm; synthesis of PAPS; the transfer of SO4− from PAPS to “acceptor” molecules by sulfotransferases (SULTs) [23, 24].
3′-Phosphoadenosine 5′-phosphosulfate synthase 2 (PAPSS2) catalyzes the PAPS synthesis with two sequential reactions: inorganic sulfate combines with ATP to form adenosine 5′-phosphosulfate (APS) and pyrophosphate catalyzed by the ATP sulfurylase domain on PAPSS2; in the second step, APS combines with another molecule of ATP to form PAPS and ADP catalyzed by the APS kinase domain [24–26]. Sulfation process is tightly controlled and alterations in any of these steps might result in impaired sulfation, leading to significant pathophysiological disorders and developmental consequences. For example, human mutations in the ATP sulfurylase domain of PAPSS2 induce defect in sulfation of the proteoglycans of the cartilage ECM, presenting with spondyloepimetaphyseal dysplasia involving the spine and long bones [27]. A spontaneous mutation in the APS kinase domain of PAPSS2 in mice results in decreased synthesis of chondroitin sulfate in cartilage, presenting with disproportionate short-limb dwarfism, a short spine, tail, and a domed skull [27, 28]. However, the role of PAPSS2 in tumor progression is poorly defined. Versican, a product of the gene VCAN and belonging to the chondroitin sulfate proteoglycan group, is one of the main components of ECM and plays important roles in cell adhesion, survival, proliferation, migration, and assembly of ECM by interaction with cells and molecules directly or indirectly [29–31]. As such, Versican induces tumor invasion and metastasis through forming a suitable extracellular microenvironment by interacting to cytokines, growth factors, cell surface, and matrix proteins with its protein core or chondroitin sulfate side-chains [32–34].
In this paper, we demonstrate that Snail induces the expression of PAPSS2 and VCAN by binding to their promoter sequences, elevated levels of PAPSS2 and Versican are essential for Snail-mediated breast cancer cell migration and metastasis, and clinically, expression of Snail is positively correlated to the expression of PAPSS2 and Versican in breast cancer tissue.
Results
Snail transactivates a large pool of gene expression including PAPSS2 and VCAN
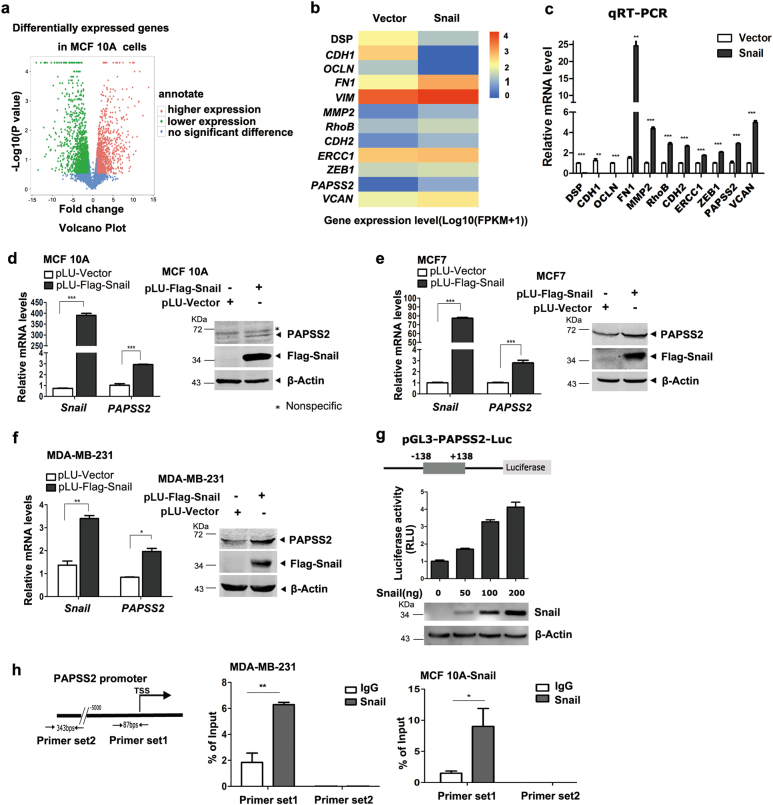
To define the genetic programs mediated by Snail, a genome-wide gene expression profiling was performed in MCF 10A cells stably expressing Snail by employing the RNA-seq approaches, which led to the identification of genes repressed or activated by Snail. Notably, the proportion of the genes activated by Snail was similar to that of the repressed (Fig. 1a). Functional category enrichment analysis of the RNA-seq data revealed that genes repressed by Snail mainly encode tight-junction-related proteins, while the activated genes encode ECM-related proteins (Supplemental Figure 1a, b). For example, the well-known Snail target genes E-cadherin and Claudins were markedly repressed by Snail, while the classical mesenchymal markers CDH2, FN1, and Vimentin were upregulated. Interestingly, PAPSS2 was induced by Snail (Fig. 1b, c, d). We further observed that Snail induced PAPSS2 expression in MCF7 and MDA-MB-231 cells, respectively (Fig. 1e, f).
Fig. 1.
Snail activates the transcription of PAPSS2 in breast cancer cells. a The Volcano Plot analysis of the differentially expressed genes in MCF 10A-Snail and -vector cells identified by the RNA-seq approach. Red dots represent genes induced by Snail, green dots represent genes repressed by Snail, and blue dots represent genes without significant difference. b Heat map showed the representatives of differentially expressed genes regulated by Snail expression in MCF 10A cells. c qRT-PCR validation of the known Snail target genes in MCF 10A cells. Data were shown as mean ± S.D. from three independent experiments. **P < 0.001, ***P < 0.0001, compared with Vector group. d–f Overexpression of Snail increased PAPSS2 mRNA level in MCF 10A cells (d), MCF7 cells (e), and MDA-MB-231 cells (f). Data were shown as mean ± S.D. from three independent experiments. ***P < 0.0001, **P < 0.001, *P < 0.05, compared with vector group (left panels). Western blots showed the protein levels of Snail and PAPSS2 (right panels). g Human PAPSS2 promoter-luc reporter activity was induced by Snail in 293T cells. Human PAPSS2 gene promoter (−138 +138) was subcloned into pGL3 basic luciferase vector to create a PAPSS2-Luc reporter (upper panel). Assays were performed in 293T cells and the luciferase activity was normalized to β-galactosidase activity. Error bars show standard deviations (bottom panel). h Snail bound to the proximal promoter region of PAPSS2. The ChIP assays were performed in MDA-MB-231 (middle panel) and MCF 10A-Snail cells (right panel) with specific antibody against Snail and the enriched DNA fragments were examined by qPCR. Error bars show standard deviations. **P < 0.001, *P < 0.05
To further examine the role of Snail in inducing the transcription of PAPSS2, we cloned the human PAPSS2 gene promoter (−138 to +138) into the pGL3 basic luciferase vector to make a PAPSS2-luc reporter (Fig. 1g, upper). The reporter plasmid was transiently co-transfected with various doses of Snail-expressing vectors into 293T cells and the luciferase activity was assayed and normalized to β-galactosidase activity. Snail induced the activity of PAPSS2-luc reporter in a dose-dependent manner (Fig. 1g, bottom). Moreover, the ChIP assays showed that DNA fragment flanking the proximal promoter of PAPSS2 was readily enriched by the antibody specifically against Snail (Fig. 1h). Collectively, these data indicate that Snail may function as a transcriptional activator to induce PAPSS2 expression in normal and cancerous breast epithelial cells.
PAPSS2 is critical for breast cancer cell migration and metastasis
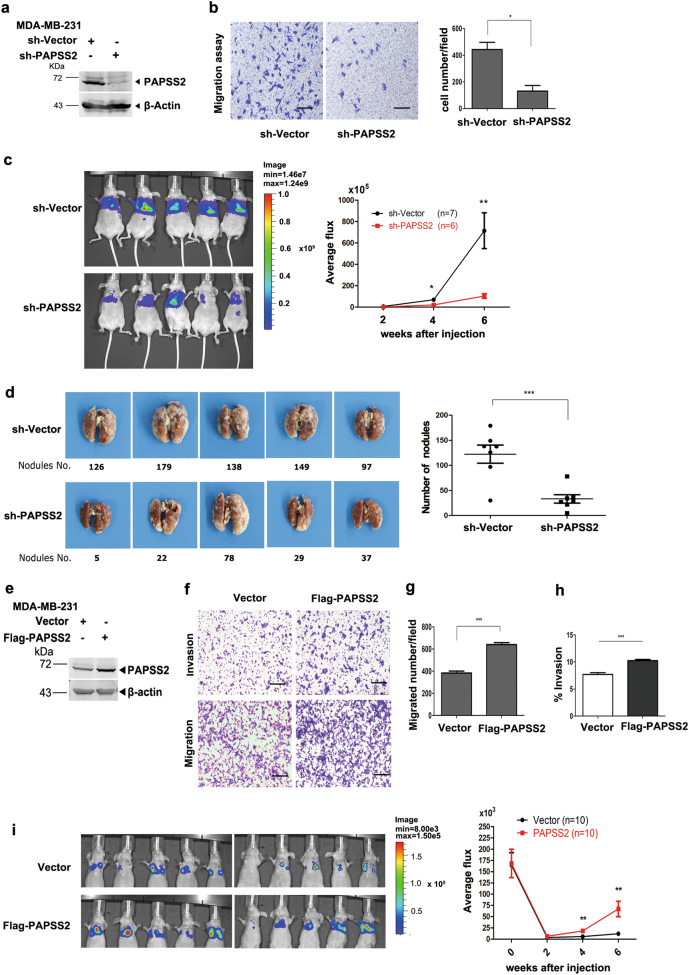
To determine the role of PAPSS2 in breast cancer cell migration and metastasis, we first performed western blot assays to examine a panel of breast cell lines for expression of PAPSS2 (Supplemental Figure 2a), MDA-MB-231 and MCF7 cells expressed higher levels of PAPSS2 than MCF 10A cells in which PAPSS2 was low and barely detectable. Next, we depleted PAPSS2 in MDA-MB-231 cells by using shRNA sequences specifically targeting PAPSS2 (Fig. 2a), which resulted in significant decrease in cell migration capability (Fig. 2b), as well as in cell proliferation ability (Supplemental Figure 2b). In addition, depletion of PAPSS2 in MCF7 cells also decreased cell migration and proliferation (Supplemental Figure 2c, d, e).
Fig. 2.
PAPSS2 is required for breast cancer cell migration and metastasis. a, e Protein levels of PAPSS2 were quantified by immunoblot analysis in MDA-MB-231 cells with PAPSS2 knocking-down (a) and overexpression (e), respectively. b Transwell assay shows migration abilities of MDA-MB-231-shPAPSS2 cells. Left panel: representative images of migrated cells; right panel: statistical analysis of the average number of migrated cells from the nine randomly chosen fields. Data were shown as mean ± S.D. from three independent experiments. *P < 0.05, compared with Vector groups, 100×, scale bar, 200 μm. c Knocking-down of PAPSS2 in MDA-MB-231 cells inhibited lung metastasis in vivo. Left panel: intensities of lung metastasis in mice at the 6th week were analyzed by in-vivo imaging. Right panel: quantification of lung photon flux at the 2nd, 4th, and 6th week. P value was determined using Student’s t-test, *P < 0.05,**P < 0.001, compared with vector group (n = 7 and n = 6, respectively). d Knocking-down of PAPSS2 decreased number of metastatic nodules on lungs. Left panel: Representative images of metastatic nodules are shown. Right panel: the average number of metastatic tumor nodules shows significant difference between the vector and PAPSS2 knocking-down group. Data were shown as mean ± S.D. ***P < 0.0001, compared with vector group. f–h Migration and invasion capabilities were elevated in PAPSS2-overexpressed MDA-MB-231 cells. f Representative images of invading and migrating cells, 100×, scale bar, 200 μm. g Statistic analysis of migrating cell number, ***P < 0.0001. h Statistic analysis of invasion percentage, % Invasion = (mean number of cell invading through Matrigel Matrix coated membrane/mean number of cells migrating through uncoated membrane) x 100. ***P < 0.0001. i Overexpression of PAPSS2 in MDA-MB-231 cells promoted lung metastasis in vivo. Left panel: intensities of lung metastasis in mice at the 4th week were analyzed by in-vivo imaging. Right panel: quantification of lung photon flux at the 0th, 2nd, 4th and 6th week. P value was determined using Student’s t-test, **P < 0.001, compared with vector group (n = 10)
To examine the role of PAPSS2 in breast cancer cell metastasis in vivo, luciferase-labeled MDA-MB-231-shPAPSS2 and MDA-MB-231-control cells were transplanted into BALB/C nude mice by tail vein injection. The mice were monitored biweekly using the Bioluminescence Imaging system and the total metastasized tumors in lung were reflected by the luciferase intensity. Indeed, a significantly lower rate of lung metastasis in mice injected with MDA-MB-231-shPAPSS2 cells was observed at the 4th and 6th week post-injection (Fig. 2c). Moreover, the number of micrometastatic nodules on lungs was much less in MDA-MB-231-shPAPSS2 mice, comparing with those of MDA-MB-231-shVector mice (Fig. 2d).
Notably, overexpression of PAPSS2 in MDA-MB-231 cells resulted in significant increase in migration and invasion abilities (Fig. 2f, g, h), but no effect on the proliferation capability and cell cycle (Supplemental Figure 2f, g, h). Similar results were obtained in MCF7 and MCF 10A cells stably expressing PAPSS2 (Supplemental Figure 2k, n, j, m). Further, luciferase-labeled MDA-MB-231-PAPSS2 and MDA-MB-231-vector cells were also transplanted into BALB/C nude mice by tail vein and a significantly higher rate of lung metastasis in mice injected with MDA-MB-231-PAPSS2 cells was observed at the 4th and 6th week post-injection (Fig. 2i). Collectively, these data demonstrate that PAPSS2 is required for breast cancer cell migration and elevated PAPSS2 can promote metastasis.
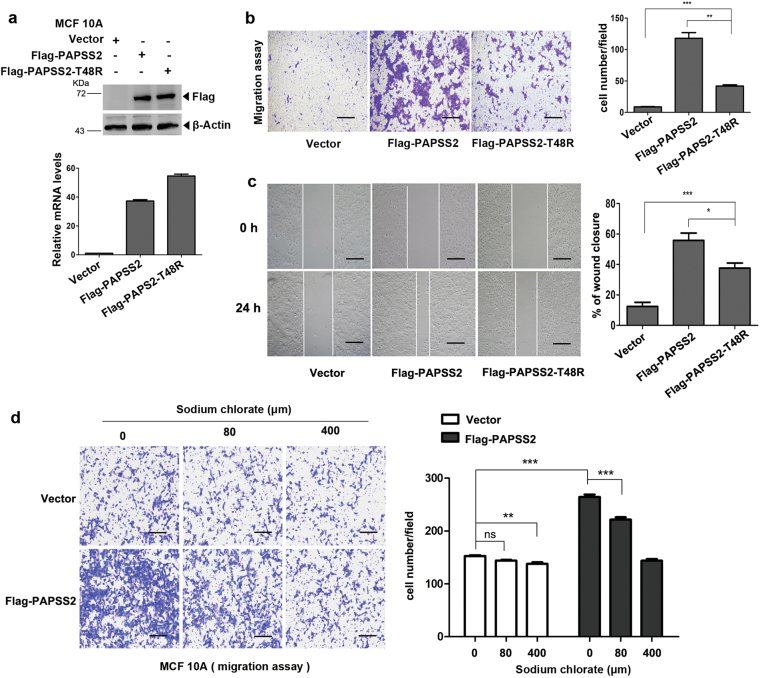
The enzymatic activity is required for PAPSS2 to induce cell migration
To determine if the enzymatic activity of PAPSS2 is required for cell migration, we stably expressed wild-type PAPSS2 and the mutant PAPSS2-T48R, which retains only partial enzymatic activity in MCF 10A cells via lentiviral infections [35]. The expression level of PAPSS2 wild type and the mutant PAPSS2-T48R was examined by western blotting and qRT-PCR assays (Fig. 3a). Overexpression of PAPSS2-T48R resulted in significant increase in cell migration rate compared with the vector control, and was remarkably lower than that of overexpression of wild-type PAPSS2 (Fig. 3b). These observations were further supported by the wound healing assays (Fig. 3c). Moreover, treatment of MCF 10A cells with sodium chlorate, a competitive inhibitor of ATP sulfurylase of PAPSS2 [36, 37], effectively inhibited sulfation reflected by the level of Versican (Supplemental Figure 3a), and markedly decreases MCF 10A-PAPSS2 cell migration (Fig. 3d) without apparently affecting the cell viabilities at concentration of 80 and 400 μM (Supplemental Figure 3b). Notably, MCF 10A-PAPSS2 cells were more sensitive to sodium chlorate treatment. Collectively, these data indicate that enzymatic activity of PAPSS2 is critical for PAPSS2 to induce cell migration.
Fig. 3.
The enzymatic activity is critical for PAPSS2 to induce cell migration. a Upper panel: western blots showed overexpression of PAPSS2 wild type and the mutant T48R in MCF 10A cells. Bottom panel: qRT-PCR analyses showed the mRNA levels of PAPSS2 and T48R. b Left panel: Transwell assays were performed in stable cell lines. Right panel: nine fields chosen randomly were counted for statistical analysis. Data were shown as mean ± S.D. from three independent experiments. **P < 0.001, ***P < 0.0001. c Left panel: Wound healing assay was performed to examine migratory ability, images were taken at 0th, 24th hour post-scratch separately, 40×, scale bar, 500 μm. Right panel: the percentage of wound closure were quantified, wound gaps in six fields chosen randomly were measured at the 0th, 12th, 24th, and 48th hour post-scratch separately. The percentage of wound closure = widths [(0th−24th)/0th * 100%], *P < 0.05, compared with Flag-PAPSS2 group. d Transwell assays showed migration capabilities of PAPSS2-overexpressed cell lines treated with different concentrations of sodium chlorate for 20 h, Right panel: nine fields chosen randomly were counted for statistical analysis. Data were shown as mean ± S.D. from three independent experiments, *P < 0.05, ***P < 0.0001, ns no significance
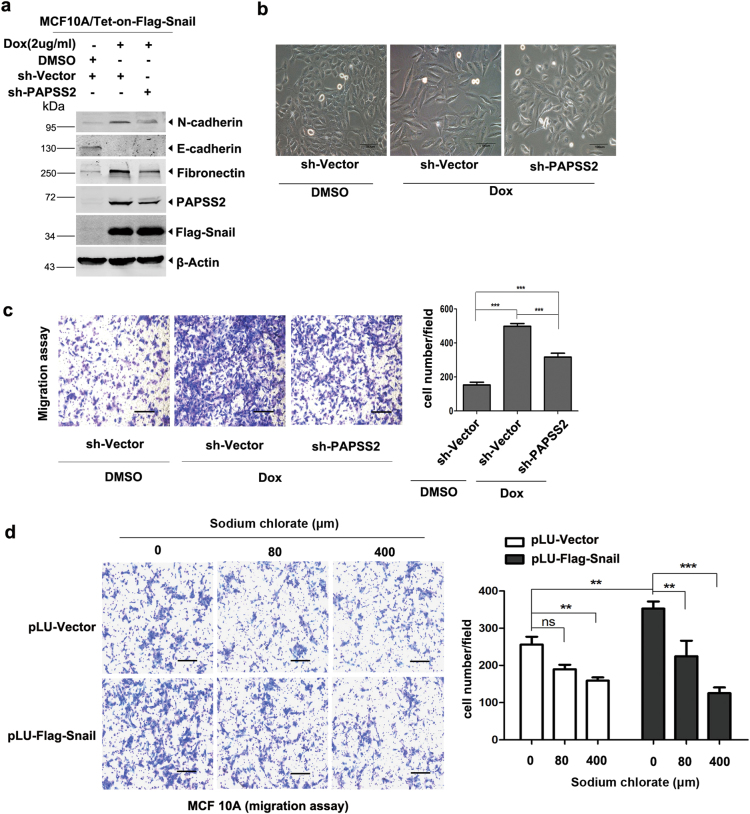
PAPSS2 is an obligate downstream target to mediate Snail-induced cell migration
To examine if PAPSS2 is a critical target gene participating in Snail-induced epithelial–mesenchymal transition (EMT) and cell migration processes, we depleted PAPSS2 in MCF 10A-tet-Snail cells using specific targeting shRNAs (Fig. 4a). Snail was markedly induced by adding Dox into the cell culture media, which resulted in drastic cell shape changes from epithelial to spindle-like mesenchymal phenotype (Fig. 4b) and promoted cell migration (Fig. 4c). Remarkably, reduced PAPSS2 expression in MCF 10A-tet-Snail cells resulted in reversed phenotypic changes from spindle-like to partial epithelial shape, and decreased the cell migration concomitantly (Fig. 4b, c). Similarly, depletion of PAPSS2 in MCF7-Snail cells also reduced cell migration (Supplemental Figure 4a, b). In addition, inhibition of PAPSS2 activity by sodium chlorate in MCF 10A-Snail cells also decreased cell migration rate significantly (Fig. 4d). Taken together, these data demonstrate that PAPSS2 is a critical downstream target of Snail and that elevated PAPSS2 expression is essential for maintaining the mesenchymal phenotype and cell migration induced by Snail.
Fig. 4.
PAPSS2 is an obligate target gene for Snail-induced cell migration. a Western blots showed the protein levels of Snail, PAPSS2, and EMT markers. Depletion of PAPSS2 in Snail-inducible MCF 10A cells. Expression of Snail was induced by Dox (2 μg/ml). b Images showed cellular morphology (200×). Snail induced cell morphological change to mesenchymal type, but knocking-down of PAPSS2 reversed the mesenchymal phenotype into epithelial type in MCF 10A cells. Scale bar, 100 μm. c Transwell assays in MCF 10A-tet-Snail cells with depletion of PAPSS2. Three independent experiments were performed, nine fields chosen randomly were counted for statistical analysis and data were shown as mean ± S.D., ***P < 0.0001. d Transwell assays in Snail-overexpressed MCF 10A cells treated with varied concentration of sodium chlorate for 20 h, **P < 0.001, ***P < 0.0001, ns no significance
Sulfation of Versican is elevated by Snail and PAPSS2
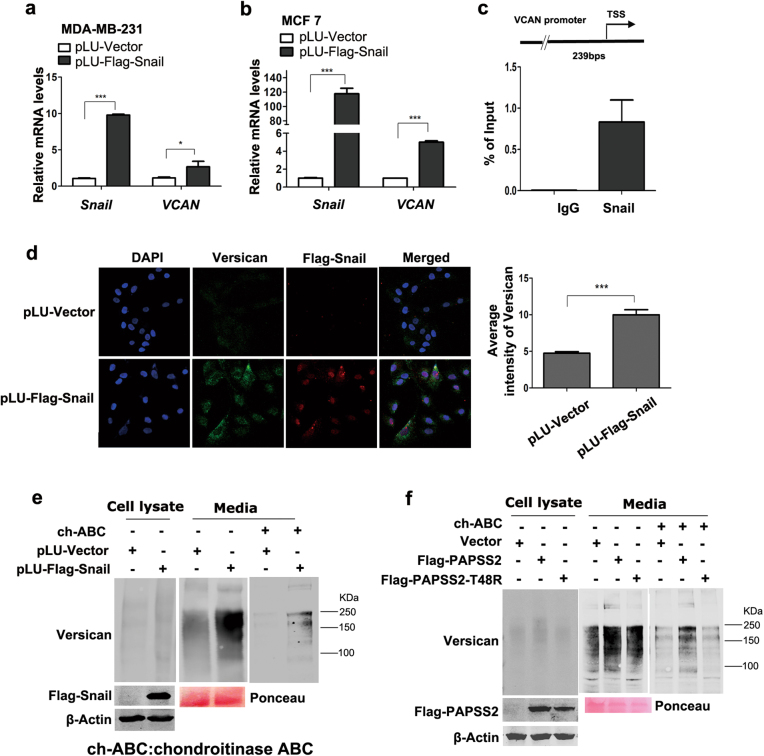
Analysis of the RNA-seq data showed that VCAN was also induced by Snail (Fig. 1b). We further showed that Snail induced expression of VCAN in MDA-MB-231 and MCF7 cells, respectively (Fig. 5a, b), and the ChIP assays found that Snail could bind the DNA fragment flanking the proximal promoter of VCAN (Fig. 5c). Moreover, indirect immunofluorescence assays showed that overexpression of Snail resulted in stronger staining of Versican localized mostly on the boundary of cells (Fig. 5d). Taken together, these observations demonstrate that VCAN is also a target gene of Snail.
Fig. 5.
Snail activates transcription of VCAN and increases sulfation of Versican. a, b Real-time PCR analysis showed increased transcription levels of VCAN in Snail-overexpressed MDA-MB-231 (a) and MCF7 cells lines (b), *P < 0.05, ***P < 0.0001, compared with the vector groups. c Upper: diagram illustrates the human VCAN promoter and the PCR primers used for ChIP. Bottom: examination of the enriched DNA fragments by the ChIP assays using qPCR in MDA-MB-231 cells. Error bars show standard deviations, IgG immunoglobulin. d Snail increased the expression of Versican in MCF 10A cells. Left panel: indirect immunofluorescent assays were performed to examine the subcellular localization of Versican in Snail-overexpressed MCF 10A and MCF 10A-Vector cell lines. Right panel: Average intensity of Versican on every cell in six fields chosen randomly was measured, ***P < 0.0001, compared to the vector group. The average number of cells located in every vision was counted, No = 24/Flag-vector; No = 13/Flag-Snail. e Western blots showed that expression levels of Versican in cell lysate and extracellular matrix, as well as sulfation level in Snail-overexpressed MCF 10A cells. ch-ABC chondroitinase ABC. f Western blots showed that expression levels of Versican in cell lysate and extracellular matrix, as well as sulfation level in PAPSS2-overexpressed MCF 10A cells. Note: Ponceau staining is used as loading control for proteins collected from media
To examine whether the sulfated Versican is increased by Snail and PAPSS2, we established cell lines in MCF 10A stably expressing Snail or PAPSS2, respectively. Culture media and whole cell lysates were prepared respectively and subjected to western blotting assays using specific antibody against Versican. Consistent with the prior reports, Versican molecules were found predominantly in the culture media, and were detected as smear with several major bands. Indeed, higher level of the sulfated Versican was observed in Snail over-expressing MCF 10A cell lysates and the media. The smear was diminished by pre-treating the samples with the Chondroitinase ABC to remove the sulfated glycosaminoglycan side chains (Fig. 5e). Similar patterns of Versican expression were observed in PAPSS2-overexpressing MCF 10A cells (Fig. 5f). Collectively, these data demonstrate that sulfation of Versican is elevated by enhanced expression of Snail or PAPSS2.
Versican is essential for Snail/PAPSS2-induced cell migration
To determine the role of Versican in Snail/PAPSS2-induced cell migration, we effectively depleted VCAN expression by stably expressing the specific targeting shRNAs in Snail- or PAPSS2-overexpressing MCF 10A cells, respectively (Fig. 6a, c).
Fig. 6.
Versican is essential for Snail/PAPSS2-induced cell migration. a Western blot showed the protein levels of Versican and Snail when VCAN was knocked down in Snail-overexpressed MCF 10A cells. b Migration capabilities were tested by Transwell assays, depletion of VCAN inhibited Snail-induced cell migration in MCF 10A cells. ***P < 0.0001, compared with vector group. c Western blot showed the protein levels of Versican and PAPSS2 when VCAN was knocked down in PAPSS2-overexpressed MCF 10A cells. d Migration capabilities were tested by Transwell assays, depletion of VCAN inhibited PAPSS2-induced cell migration in MCF 10A cells. ***P < 0.0001, compared with vector group
Knocking-down VCAN markedly decreased cell migration rate in MCF 10A-Snail cells (Fig. 6b). Similarly, depletion of Versican resulted in reduced cell migration capability of MCF 10A-PAPSS2 cells (Fig. 6d). Together, these data demonstrate that Versican, an important downstream substrate of Snail and PAPSS2, is necessary for cell migration.
Expression of Snail is positively correlated with the expression of PAPSS2 and Versican in breast cancer tissues
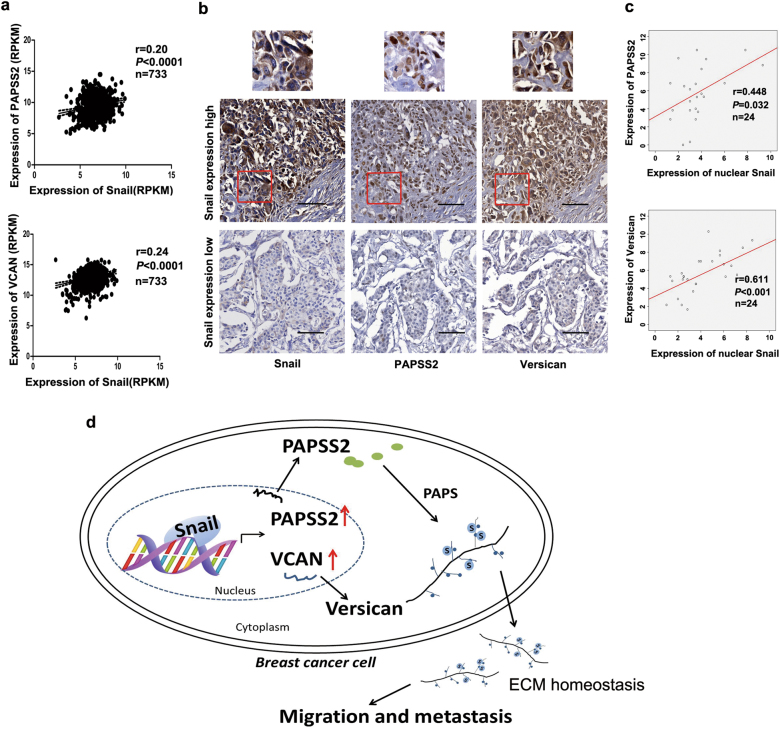
To examine the clinical correlation between Snail, PAPSS2, and Versican in breast cancer samples, we first analyzed their expression in 733 breast carcinomas samples found in The Cancer Genome Atlas (TCGA-BRCA-exp-HiSeqV2-2015-02-24). Notably, Snail expression was positively associated with the expression of PAPSS2 (P < 0.0001, r = 0.20) (Fig. 7a, upper panel), and VCAN (P < 0.0001, r = 0.24) (Fig. 7a, lower panel).
Fig. 7.
Expression of Snail is positively correlated with the expression of PAPSS2 and Versican in breast cancer specimens. a Scatter plots showed the correlation of Snail expression with PAPSS2 (upper panel) and VCAN (lower panel) expression in 733 breast tumor samples from a TCGA dataset. The r value was calculated via Pearson’s ranking correlation coefficient analysis. b Representative IHC images showed the protein levels of Snail, PAPSS2, and Versican in TMA. Red box indicates cells with amplification. The original magnifications, 200×, scale bar, 100 μm. c Scatter plots showed the correlation of Snail expression with PAPSS2 and Versican expression in TMA samples with positive Snail-nuclear staining. n = 24, the r value was calculated via Pearson’s ranking correlation coefficient analysis. d Schematic model for the role of Snail/PAPSS2/VCAN sulfation axis in breast cancer cell migration and metastasis. Snail activated transcription of PAPSS2 and VCAN via binding to promoter sequence, and simultaneously, induced-PAPSS2 increased sulfation level of Versican, which enhances migration and metastasis of breast cancer cells
To further validate the observations obtained from TCGA database, we performed IHC assays on tissue array chips containing 60 paired human breast ductal tumor specimens and peri-tumor tissue to examine the expression of Snail, PAPSS2, and Versican. The staining for PAPSS2 was stronger in the nuclei of tumor cells around duct and acinus (Fig. 7b). In tumor tissue, 38 cases showed high expression of PAPSS2 and 22 cases showed low expression; while in peri-tumor tissue, four cases showed high expression and 56 cases showed low expression. Statistical analysis indicated that there was a positive correlation between the expression level of PAPSS2 and breast tumor lymph node metastasis (P < 0.0001, Supplemental Figure 5a, b). Similarly, 43 cases were scored high for Versican in tumor specimens, comparing to 31 cases scored high in peri-tumor specimens. A positive correlation between expression level of Versican and metastatic lymph nodes, or tumor volumes was also observed (P < 0.05, Supplemental Figure 5d, e, f). Notably, Snail was detected in nuclei of 24 (40%) cases, and nuclear Snail expression was generally accompanied with cytoplasm expression (Fig. 7b). Next, we examined the correlation between the expression level of nuclear Snail, PAPSS2, and Versican in breast tumors. Consistent with the observations obtained from TCGA dataset, among the 24 cases showing high level of Snail in the nuclei, Snail protein level was positively associated with the expression of PAPSS2 (P = 0.032) and Versican (P < 0.001), respectively (Fig. 7b, c). Taken together, these data indicate that Snail expression is positively associated with the expression of its target genes PAPSS2 and Versican in breast cancer specimens and all three can be diagnostic markers.
Discussion
In this paper, we describe a novel pathway controlled by Snail through the activation of gene expression program to regulate EMT and metastasis in breast cancer cells (Fig. 7d). Specifically, Snail induces the expression of PAPSS2 and VCAN in multiple breast cancer cells, and elevated PAPSS2 results in increased sulfation modification in the cells such as Versican, which is one of the main components of ECM and plays important roles in maintaining ECM homeostasis [38]. The imbalanced ECM may help to trigger EMT program and promote cell migration and metastasis [39]. This hypothesis is supported by the data obtained from the genetic manipulations of Snail, PAPSS2, and VCAN expression in breast cancer cells, and is further strengthened by clinical findings that expressions of Snail, PAPSS2, and VCAN are positively correlated in breast cancer tissues.
It was reported that depletion of PAPSS2 expression inhibits cell proliferation and induces senescence in liver cancer cells [40]. We found that knockdown of PAPSS2 reduces both cell proliferation and migration in breast cancer cells, suggesting that the decreased metastasis potential in vivo may be due to decrease in cell migration and proliferation. Surprisingly, we found that overexpression of PAPSS2 in MDA-MB-231 cells has no effect on cell proliferation, but results in greater metastatic capability in vivo. Moreover, MDA-MB-231-Snail or PAPSS2 cell are more sensitized to sodium chlorate treatment than the vector control cells. These data collectively suggest that promoting cell migration and metastasis requires higher level of PAPSS2 expression, although the underlying mechanism is not clear. Nevertheless, these observations further strengthen our initial claim that enhanced PAPSS2 is essential to promote breast cancer cell migration and metastasis.
We identified Versican as an example of ultimate sulfated substrates to promote cell migration and metastasis and as a diagnostic marker for breast cancer, however, we believe that other substrates existed to mediate Snail-induced EMT and metastasis. Sulfated biomolecules comprise a remarkable array of substances, including xenobiotics, neurotransmitters, hormones, glycosaminoglycans (GAG) and proteoglycans (PG) with molecular weight ranging from less than 103 to greater than 106 Dalton [27]. Of those substrates, Sulfated GAGs and PGs are major components of the cell surface, basement membrane, and ECM, and play critical roles in the regulation of signal transduction, cell–cell and cell–ECM communications, cell movement both in normal embryonic development and tumor metastasis [41–43]. Aggrecans, Versicans, Syndecans, Glypicans, NG2, Perlecan, and CD44 are the major PG sulfates that have been extensively studied in tumorigenesis and metastasis [44, 45]. However, depending on the type, their localization, and the accessibility of their ligands, PGs can either promote or inhibit tumors. Therefore, a comprehensive study to identify Snail-regulated PGs will be necessary.
In animals, two functionally identical PAPS synthases PAPSS1 and PAPSS2 are found. However, the expression of PAPSS1 is not affected by Snail. The catalytic activity of PAPSS2 is 10–15-fold higher than that of PAPSS1 [46], and moreover, the subcellular localization is different: PAPSS1 is located predominantly in the nucleus, whereas PAPSS2 shows pronounced heterogeneity in sub-cellular localization. PAPSS2 contains a conserved N-terminal nuclear localization signal and can shuttle between cytoplasm and nucleus. Schroder et al. reported that PAPSS2 shows pronounced heterogeneity in sub-cellular localization in HeLa cells [47]. Consistently, we observed remarkable heterogeneity of the localization for PAPSS2 in breast cancer samples: positive staining both in the nucleus and the cytoplasm, nucleus only, and predominantly cytoplasm. We speculate that when PAPSS2 is depleted in cells, PAPSS1 can maintain a certain level of sulfation in cells. To fully evaluate the biological significance of sulfation in cells, double knockout of PAPSS1 and PAPSS2 should be generated by employing shRNA approach or CRISPR system.
Although Snail has been extensively interrogated as a transcriptional repressor, several studies have shown that Snail can directly activate gene expression [19, 21]. Here, we also demonstrated that Snail can bind to the promoter regions of PAPSS2 and VCAN, and induces their expression in breast cancer cells, and moreover, genome-wide gene expression profiling analyses revealed that Snail can induce a large pool of gene expression in MCF 10A cells. However, the molecular mechanism by which Snail activates gene expression still remains elusive and need to be explored further.
In summary, these studies mechanistically link the transcription factor Snail, which plays vital roles in both normal development and tumor metastasis to the biologically important, but less well-appreciated sulfation pathway, and lead to a new and exciting direction for mechanistic research in EMT and metastasis. Clinically, this enzyme-rich sulfation pathway provides ample targets for breast cancer therapy development and stands a high chance for possible rapid translation of existing drugs to the clinic.
Material and methods
Plasmids
The human PAPSS2 cDNA was subcloned into pCDH-CMV-MCS-EF1-Puro-vector by PCR method between Nhe l and Not I restriction enzyme sites. The pCDH-Flag-PAPSS2-T48R were cloned from pCDH-Flag-PAPSS2 by point mutation of the base 143C into G. The PAPSS2 gene promoter from −138 to +138 was amplified by PCR and subcloned into pGL3.0-Luc basic vector to create the pGL3-PAPSS2-Luc reporter. The pLU-Flag-Snail has been previously described [16]. The plasmids of PLKO.1-shPAPSS2 and shVCAN were obtained from GE-Lifescience and the short hairpin RNA sequences are listed in Table 1.
Table 1.
Primer list
| Primers | Sequences | |
|---|---|---|
| Sense sequences in shRNA | PAPSS2 shRNA-1 | TTTGGCTTTGTTGTAGGCAGC |
| PAPSS2 shRNA-2 | AATCCGAGATTTCTGTTAAGG | |
| VCAN shRNA-1 | AATACTCTCTATCAATATCGG | |
| VCAN shRNA-2 | ATCTTAGACCATTTGATGCGG | |
| Primers for ChIP | PAPSS2-set1-Forward | GCCTTTTGGTGATAGGGTCA |
| PAPSS2-set1-Reverse | GGGCTGCATGAGGTATTCAC | |
| PAPSS2-set2-Forward | GTGGCGGGAGGAGGAGTAG | |
| PAPSS2-set2-Reverse | CTCCCGGGAAGGAGGTATAA | |
| VCAN- Forward | AGAGCGATGTTACTGAGTG | |
| VCAN-Reverse | GGTTCATTCACAGGCTCC | |
| Primers for qRT-PCR | Snail-84-Forward | TTTACCTTCCAGCAGCCCTA |
| Snail-291-Reverse | CCCACTGTCCTCATCTGACA | |
| PAPSS2-1488-Forward | CAGTTGCGCAATCCTGTCCACAAT | |
| PAPSS2-1785-Reverse | AGAAATTGGCACCCGCAATCATCC | |
| VCAN-947-Forward | GAGGTGGTCTACTTGGGGTGA | |
| VCAN-1203-Reverse | ACAAGTGGCTCCATTACGAC |
Cell culture and luciferase reporter assay
The human breast cell lines MCF 10A, MCF7, and MDA-MB-231 were originally obtained from the American Type Culture Collection (ATCC) and were authenticated by DNA fingerprinting in our lab. MCF7, MDA-MB-231, and 293T were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM L-glutamine and penicillin (50 U/ml)/streptomycin (50 μg/ml) at 37 ℃ under 5% CO2 in a humidified chamber. MCF 10A cells were maintained in DMEM/F12 supplemented with 5% horse serum, EGF (20 ng/ml), insulin (10 μg/ml), hydrocortisone (0.5 μg/ml), cholera toxin (100 ng/ml), and penicillin/streptomycin. Transfection and luciferase reporter assays in 293T cells were performed as described [48].
Western blot, Immunofluorescence, and antibodies
The western blot and immunofluorescent assays were described previously [16]. The antibodies used are as follows: anti-PAPSS2 (sc-100801, Santa Cruz), anti-Flag (F3165, F7425, Sigma), anti-Versican (sc-25831, Santa Cruz), anti-β-Actin (60008-1-lg, Proteintech), anti-Snail (sc-28199, Santa Cruz), and normal rabbit IgG (sc-2027, Santa Cruz).
Chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR)
ChIP assays were carried out in MDA-MB-231 and MCF 10A-Snail cells as described previously [16]. The chromatin was sonicated into fragments ranging from 500 to 800 bps in size and the immunoprecipitation was performed using the antibody against Snail or equal amount of rabbit normal IgG as control. The precipitated DNA fragments were detected by qPCR with primer sets listed in Table 1.
Total RNA was isolated from cells with Trizol reagent (Invitrogen) and the reverse transcription (RT) procedure was described previously [49]. qPCR was performed on a 7500 Fast Realtime PCR system (Applied Biosystem) using SYBR Green reagent. The primer sets are listed in Table 1.
Migration and in vitro invasion assay
Transwell assays were performed to assess cell migration and invasion. To evaluate migration, MCF 10A cells were harvested after serum-free starvation for 12 h, and then resuspended in plain DMEM/F12 media. 5 × 104 cells were applied into the upper 8-μm pore Transwell filters (Corning). Complete DMEM/F12 medium was added to the bottom chamber as attractants. After incubation for 20 h, cells were fixed with 4% paraformaldehyde and stained with crystal violet. The non-migrated cells on the top of the chamber were removed gently with cotton swabs, the migrated cells at the bottom of filter were quantified by counting nine randomly chosen fields (100×) using an inverted phase contrast microscope in each experiment. Experiments were repeated in triplicates. Scale bar, 200 μm. The procedure of migration assays in MCF7 and MDA-MB-231 cells was essentially the same as that in MCF 10A cells, but with 4 × 104 MCF7 cells and 2 × 104 MDA-MB-231 cells and complete DMEM media as attractants instead. In invasion assays, diluted Matrigel matrix (Corning, #354234) was coated onto Transwell filters firstly, subsequent procedure was same to that in migration assays [50].
In vivo metastasis assay
Female BALB/c nude mice were purchased from SLAC laboratory Co. Ltd. (Shanghai, China) and divided into two groups randomly. Exponentially growing MDA-MB-231 cells and derivatives labeled with luciferase were harvested and resuspended in PBS buffer. Cells (MDA-MB-231-shPAPSS2/Vector: 5 × 105 cells, MDA-MB-231-Flag-PAPSS2/Vector: 2 × 105 cells) in 100 μl of PBS suspension were injected into the tail vein of 8-week-old mice (n = 10 for each group). Metastatic tumors were examined using Xenogen IVIS Imaging System biweekly. All animal experiments were approved by the Institutional Animal Care and Use Committee of Shanghai in accordance with the National Research Council Guide for Care and Use of Laboratory Animals (SCXK, Shanghai 2007-0005).
Cell proliferation and viability assays
Cell proliferation and viability were evaluated using the cell counting kit-8 (CCK-8, Dojindo) according to the instructions from the manufacturer. Highly water-soluble tetrazolium salt, WST-8, is reduced by dehydrogenase activities in cells to generate a yellow-color formazan dye, which is soluble in the tissue culture media. The amount of the formazan dye is directly proportional to the number of living cells and viability. Absorbance at 450 nm was detected every other day for proliferation assay, and cell viability was detected at the 24th hour post-addition of sodium chlorate.
Wound healing assays
MCF 10A cells were overexpressed with Flag-PAPSS2 and Flag-PAPSS2-T48R individually, cells were trypsinized and 2.3 × 105 cells were reseeded on a 6-well tissue culture plate. After 12 h, the attached cells were scratched with a 10 μl pipette tip and 0 h images were captured using an inverted phase contrast microscope. The plates were placed back at 37 ℃ under 5% CO2 in a humidified chamber. Other sets of images were captured of the same wounds at 12, 24, 48 h separately. The wound widths were measured, normalized, and presented as the percentage of wound measured at time 0 h (mean ± S.D.). The percent of migration was calculated as the width difference of a scratch divided by the initial width. At least five fields were analyzed for each scratch and each sample was performed in triplicate.
Immunohistological chemistry (IHC) and tissue microarray (TMA)
Protein levels of Snail, PAPSS2, and Versican were examined by IHC staining using polyclonal antibodies to Snail (sc28199, Santa Cruz, dilution 1:100), PAPSS2 (sc100801, Santa Cruz, dilution 1:100), Versican (sc-25831, Santa Cruz, dilution 1:50). TMA chips containing 60 cases of paired tumor and peri-tumor specimens were purchased (HBre-Duc060CS-03 and HBre-Duc060CS-04, Shanghai Outdo Biotech Co., Ltd.). All tumor specimens were evaluated according to the tumor size and number of metastatic lymph nodes. After IHC staining, TMA chips were scanned by Aperio ImageScope and every tissue spot was scored by a senior pathologist. The immunoreactive score system (IRS) [51, 52] was adopted to assess the staining of every sample as follows: negative, 0–1 point; mild(+), 2–3 points; moderate(++), 4–8 points; and strongly positive(+++), 9–12 points. The IHC staining of more than 4 points was considered as high.
Statistical analysis
Data presented as mean ± S.D. or mean ± SEM was analyzed by the independent Student t test. The distribution of the IHC scoring results on TMA chips was analyzed by the Pearson Chi-square test. The correlation between the expression of Snail and PAPSS2 or Versican in breast tumor was evaluated by the Pearson rank correlation coefficient test. Values of P < 0.05 were considered statistically significant. Statistical analysis was performed using SPSS software (SPSS 13; SPSS lnc., Chicago, IL, USA).
Electronic supplementary material
Acknowledgments
We thank Dr. Frank J. Rauscher III for his critical reading and insightful suggestions.
This work was supported by the Ministry of Science and Technology of China (No. 2014DFA32120); the National Science Foundation of China (Grant Nos. 81372309, 31671415, 81471000); the Shanghai Committee of Science and Technology (15410724200); and the Natural Science Foundation of Liaoning (No. 2014023042).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by B. Dynlacht
These authors contributed equally: Yihong Zhang, Xiuqun Zou.
Contributor Information
Gang Wei, Email: weigang@picb.ac.cn.
Yingjie Wu, Email: yingjiewu@dmu.edu.cn.
Zhaoyuan Hou, Email: houzy@sjtu.edu.cn.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0147-y) contains supplementary material, which is available to authorized users.
References
- 1.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of Snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013;13:963–72. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson P. Maternal–zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics. 1983;105:615–32. doi: 10.1093/genetics/105.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–92. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 4.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Brzozowa M, Michalski M, Wyrobiec G, Piecuch A, Dittfeld A, Harabin-Slowinska M, et al. The role of Snail1 transcription factor in colorectal cancer progression and metastasis. Contemp Oncol. 2015;19:265–70. doi: 10.5114/wo.2014.42173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baulida J, Garcia de Herreros A. Snail1-driven plasticity of epithelial and mesenchymal cells sustains cancer malignancy. Biochim Biophys Acta. 2015;1856:55–61. doi: 10.1016/j.bbcan.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Kaufhold S, Bonavida B. Central role of Snail1 in the regulation of EMT and resistance in cancer: a target for therapeutic intervention. J Exp Clin Cancer Res. 2014;33:62. doi: 10.1186/s13046-014-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen WJ, Wang H, Tang Y, Liu CL, Li HL, Li WT. Multidrug resistance in breast cancer cells during epithelial–mesenchymal transition is modulated by breast cancer resistant protein. Chin J Cancer. 2010;29:151–7. doi: 10.5732/cjc.009.10447. [DOI] [PubMed] [Google Scholar]
- 9.Deep G, Jain AK, Ramteke A, Ting H, Vijendra KC, Gangar SC, et al. SNAI1 is critical for the aggressiveness of prostate cancer cells with low E-cadherin. Mol Cancer. 2014;13:37. doi: 10.1186/1476-4598-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, Grither WR, Van Hove S, Biswas H, Ponik SM, Eliceiri KW, et al. Mechanical signals regulate and activate SNAIL1 protein to control the fibrogenic response of cancer-associated fibroblasts. J Cell Sci. 2016;129:1989–2002. doi: 10.1242/jcs.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2003;24:306–19. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–81. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–16. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–15. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 16.Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou Z, Peng H, White DE, Wang P, Lieberman PM, Halazonetis T, et al. 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial–mesenchymal differentiation. Cancer Res. 2010;70:4385–93. doi: 10.1158/0008-5472.CAN-10-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Xu H, Zou X, Wang J, Zhu Y, Chen H, et al. Snail recruits Ring1B to mediate transcriptional repression and cell migration in pancreatic cancer cells. Cancer Res. 2014;74:4353–63. doi: 10.1158/0008-5472.CAN-14-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rembold M, Ciglar L, Yanez-Cuna JO, Zinzen RP, Girardot C, Jain A, et al. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 2014;28:167–81. doi: 10.1101/gad.230953.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu CT, Chang TY, Cheng CC, Liu CS, Wu JR, Li MC, et al. Snail associates with EGR-1 and SP-1 to upregulate transcriptional activation of p15INK4b. FEBS J. 2010;277:1202–18. doi: 10.1111/j.1742-4658.2009.07553.x. [DOI] [PubMed] [Google Scholar]
- 21.Jorda M, Olmeda D, Vinyals A, Valero E, Cubillo E, Llorens A, et al. Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J Cell Sci. 2005;118(Pt 15):3371–85. doi: 10.1242/jcs.02465. [DOI] [PubMed] [Google Scholar]
- 22.Stanisavljevic J, Porta-de-la-Riva M, Batlle R, de Herreros AG, Baulida J. The p65 subunit of NF-kappaB and PARP1 assist Snail1 in activating fibronectin transcription. J Cell Sci. 2011;124(Pt 24):4161–71. doi: 10.1242/jcs.078824. [DOI] [PubMed] [Google Scholar]
- 23.Klaassen CD, Boles JW. Sulfation and sulfotransferases 5: the importance of 3’-phosphoadenosine 5’-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 1997;11:404–18. doi: 10.1096/fasebj.11.6.9194521. [DOI] [PubMed] [Google Scholar]
- 24.Venkatachalam KV. Human 3’-phosphoadenosine 5’-phosphosulfate (PAPS) synthase: biochemistry, molecular biology and genetic deficiency. IUBMB Life. 2003;55:1–11. doi: 10.1080/1521654031000072148. [DOI] [PubMed] [Google Scholar]
- 25.Grum D, van den Boom J, Neumann D, Matena A, Link NM, Mueller JW. A heterodimer of human 3’-phospho-adenosine-5’-phosphosulphate (PAPS) synthases is a new sulphate activating complex. Biochem Biophys Res Commun. 2010;395:420–5. doi: 10.1016/j.bbrc.2010.04.039. [DOI] [PubMed] [Google Scholar]
- 26.Venkatachalam KV, Fuda H, Koonin EV, Strott CA. Site-selected mutagenesis of a conserved nucleotide binding HXGH motif located in the ATP sulfurylase domain of human bifunctional 3’-phosphoadenosine 5’-phosphosulfate synthase. J Biol Chem. 1999;274:2601–4. doi: 10.1074/jbc.274.5.2601. [DOI] [PubMed] [Google Scholar]
- 27.Faiyaz ul Haque M, King LM, Krakow D, Cantor RM, Rusiniak ME, Swank RT, et al. Mutations in orthologous genes in human spondyloepimetaphyseal dysplasia and the brachymorphic mouse. Nat Genet. 1998;20:157–62. doi: 10.1038/2458. [DOI] [PubMed] [Google Scholar]
- 28.Lane PW, Dickie MM. Three recessive mutations producing disproportionate dwarfing in mice: achondroplasia, brachymorphic, and stubby. J Hered. 1968;59:300–8. doi: 10.1093/oxfordjournals.jhered.a107725. [DOI] [PubMed] [Google Scholar]
- 29.Kinsella MG, Bressler SL, Wight TN. The regulated synthesis of versican, decorin, and biglycan: extracellular matrix proteoglycans that influence cellular phenotype. Crit Rev Eukaryot Gene Expr. 2004;14:203–34. doi: 10.1615/CritRevEukaryotGeneExpr.v14.i3.40. [DOI] [PubMed] [Google Scholar]
- 30.Rahmani M, Wong BW, Ang L, Cheung CC, Carthy JM, Walinski H, et al. Versican: signaling to transcriptional control pathways. Can J Physiol Pharmacol. 2006;84:77–92. doi: 10.1139/y05-154. [DOI] [PubMed] [Google Scholar]
- 31.Foulcer SJ, Day AJ, Apte SS. Isolation and purification of versican and analysis of versican proteolysis. Methods Mol Biol. 2015;1229:587–604. doi: 10.1007/978-1-4939-1714-3_46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricciardelli C, Sakko AJ, Ween MP, Russell DL, Horsfall DJ. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev. 2009;28:233–45. doi: 10.1007/s10555-009-9182-y. [DOI] [PubMed] [Google Scholar]
- 33.Cattaruzza S, Schiappacassi M, Kimata K, Colombatti A, Perris R. The globular domains of PG-M/versican modulate the proliferation-apoptosis equilibrium and invasive capabilities of tumor cells. FASEB J. 2004;18:779–81. doi: 10.1096/fj.03-0660fje. [DOI] [PubMed] [Google Scholar]
- 34.Zheng PS, Wen J, Ang LC, Sheng W, Viloria-Petit A, Wang Y, et al. Versican/PG-M G3 domain promotes tumor growth and angiogenesis. FASEB J. 2004;18:754–6. doi: 10.1096/fj.03-0545fje. [DOI] [PubMed] [Google Scholar]
- 35.Noordam C, Dhir V, McNelis JC, Schlereth F, Hanley NA, Krone N, et al. Inactivating PAPSS2 mutations in a patient with premature pubarche. N Engl J Med. 2009;360:2310–8. doi: 10.1056/NEJMoa0810489. [DOI] [PubMed] [Google Scholar]
- 36.Baeuerle PA, Huttner WB. Chlorate—a potent inhibitor of protein sulfation in intact cells. Biochem Biophys Res Commun. 1986;141:870–7. doi: 10.1016/S0006-291X(86)80253-4. [DOI] [PubMed] [Google Scholar]
- 37.Mencio C, Garud DR, Kuberan B, Koketsu M. Synthesis of selective inhibitors of heparan sulfate and chondroitin sulfate proteoglycan biosynthesis. Methods Mol Biol. 2015;1229:69–78. doi: 10.1007/978-1-4939-1714-3_8. [DOI] [PubMed] [Google Scholar]
- 38.Wight TN. Versican: a versatile extracellular matrix proteoglycan in cell biology. Curr Opin Cell Biol. 2002;14:617–23. doi: 10.1016/S0955-0674(02)00375-7. [DOI] [PubMed] [Google Scholar]
- 39.Schedin P, Keely PJ. Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression. Cold Spring Harb Perspect Biol. 2011;3:a003228. doi: 10.1101/cshperspect.a003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung SH, Lee HC, Yu DM, Kim BC, Park SM, Lee YS, et al. Heparan sulfation is essential for the prevention of cellular senescence. Cell Death Differ. 2016;23:417–29. doi: 10.1038/cdd.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malavaki C, Mizumoto S, Karamanos N, Sugahara K. Recent advances in the structural study of functional chondroitin sulfate and dermatan sulfate in health and disease. Connect Tissue Res. 2008;49:133–9. doi: 10.1080/03008200802148546. [DOI] [PubMed] [Google Scholar]
- 42.Morgenstern DA, Asher RA, Fawcett JW. Chondroitin sulphate proteoglycans in the CNS injury response. Prog Brain Res. 2002;137:313–32. doi: 10.1016/S0079-6123(02)37024-9. [DOI] [PubMed] [Google Scholar]
- 43.Trowbridge JM, Gallo RL. Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology. 2002;12:117R–25R. doi: 10.1093/glycob/cwf066. [DOI] [PubMed] [Google Scholar]
- 44.Venero Galanternik M, Kramer Kenneth L, Piotrowski T. Heparan Sulfate Proteoglycans Regulate Fgf Signaling and Cell Polarity during Collective Cell Migration. Cell reports 2015;10:414-28. [DOI] [PMC free article] [PubMed]
- 45.Baghy K, Tatrai P, Regos E, Kovalszky I. Proteoglycans in liver cancer. World J Gastroenterol. 2016;22:379–93. doi: 10.3748/wjg.v22.i1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuda H, Shimizu C, Lee YC, Akita H, Strott CA. Characterization and expression of human bifunctional 3’-phosphoadenosine 5’-phosphosulphate synthase isoforms. Biochem J. 2002;365(Pt 2):497–504. doi: 10.1042/bj20020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroder E, Gebel L, Eremeev AA, Morgner J, Grum D, Knauer SK, et al. Human PAPS synthase isoforms are dynamically regulated enzymes with access to nucleus and cytoplasm. PLoS One. 2012;7:e29559. doi: 10.1371/journal.pone.0029559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–8. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- 49.Hou Z, Peng H, White DE, Negorev DG, Maul GG, Feng Y, et al. LIM protein Ajuba functions as a nuclear receptor corepressor and negatively regulates retinoic acid signaling. Proc Natl Acad Sci USA. 2010;107:2938–43. doi: 10.1073/pnas.0908656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marshall J. Transwell((R)) invasion assays. Methods Mol Biol. 2011;769:97–110. doi: 10.1007/978-1-61779-207-6_8. [DOI] [PubMed] [Google Scholar]
- 51.Remmele W, Stegner HE. [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue] Pathologe. 1987;8:138–40. [PubMed] [Google Scholar]
- 52.Maacke H, Opitz S, Jost K, Hamdorf W, Henning W, Kruger S, et al. Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int J Cancer. 2000;88:907–13. doi: 10.1002/1097-0215(20001215)88:6<907::AID-IJC11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.