Abstract
Clostridium thermocellum is a good candidate organism for producing cellulosic biofuels due to its native ability to ferment cellulose, however its maximum biofuel titer is limited by tolerance. Wild type C. thermocellum is inhibited by 5 g/L n-butanol. Using growth adaptation in a chemostat, we increased n-butanol tolerance to 15 g/L. We discovered that several tolerant strains had acquired a D494G mutation in the adhE gene. Re-introducing this mutation recapitulated the n-butanol tolerance phenotype. In addition, it increased tolerance to several other primary alcohols including isobutanol and ethanol. To confirm that adhE is the cause of inhibition by primary alcohols, we showed that deleting adhE also increases tolerance to several primary alcohols.
Introduction
Metabolic engineering has been widely applied to different hosts for the production of pharmaceuticals, biofuels and bulk chemicals. Producing chemicals at high titer often results in inhibition of the host organism. Therefore, improving tolerance is an essential step in engineering microorganisms to maximize their productivity and develop economically feasible processes1–3. Previous studies looking at strategies that microbes use to increase their inhibitor tolerance have focused on changes in the cell membrane, including: lipid composition, membrane fluidity and changes to specific efflux pumps4–9. Other strategies include upregulation of chaperones to increase protein stability10–12 and mutations in transcription factors that regulate the cellular response to environmental stress13–15.
Clostridium thermocellum is a good candidate organism for the production of biofuels by consolidated bioprocessing due to its ability to rapidly ferment cellulosic biomass16,17. It can natively produce ethanol, and some strains have been engineered to produce ethanol at a titer of 25–30 g/L18,19. Currently, tolerance appears to be the main cause of the titer limitation19,20. Besides ethanol, C. thermocellum has the potential to produce a variety of other products, including lactate21, amino acids19,22 and several advanced biofuel and chemical products23. Recently, a heterologous pathway was introduced into C. thermocellum to increase its isobutanol production24.
Historically, n-butanol has been produced by fermentations involving the related organism, Clostridium acetobutylicum25, and for this reason, we think n-butanol may be a good candidate biofuel molecule for production in C. thermocellum. In this work, we applied adaptive laboratory evolution to isolate a strain of C. thermocellum tolerant to increased concentrations of n-butanol. To understand the genotype-phenotype relationship of this mutant, we re-sequenced the genomes of tolerant mutants, and re-introduced observed mutations to recapitulate the n-butanol tolerance phenotype.
Results and Discussion
Isolation and characterization of a strain with improved n-butanol tolerance
First, we tested the tolerance to n-butanol in batch culture and found that 4 g/L inhibited but did not completely eliminate growth (Figure S1). Adaptive laboratory evolution experiments for n-butanol tolerance were carried out in chemostat bioreactors. After more than 2000 hours, butanol tolerance had improved to 10 g/L (Fig. 1), and we purified 12 isolates.
Figure 1.
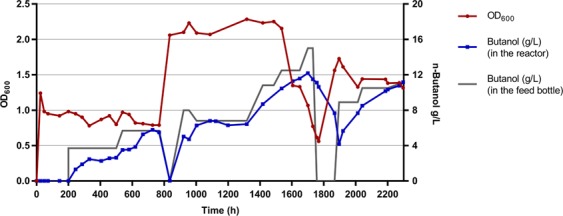
Chemostat culture of the C. thermocellum for selection of improved n-butanol tolerance. The strains were grown on MTC-5 minimal medium in a bioreactor with pH regulation at 55 °C. From time 0 to 800 hours and 1700 to 2300 hours, the concentration of cellobiose in the feed was 5 g/L . From 800 to 1700 hours, the concentration of cellobiose in the feed was 10 g/L. The red line represents the biomass concentration (as measured by OD600, using standard absorbance units). The blue line represents the n-butanol concentration in the bioreactor and the grey line represents the n-butanol concentration in the feed bottle.
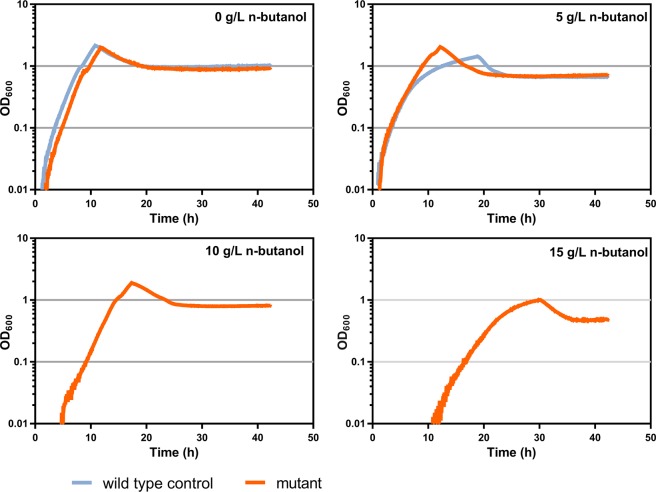
To evaluate the n-butanol tolerance of the 12 isolates, we tested the growth of the isolates and the wild type strain in the presence of 0, 5, 10, and 15 g/L n-butanol. All 12 isolates showed increased growth compared with the wild type control with 5 g/L n-butanol, while maintaining similar growth in the absence of n-butanol (Fig. 2). The wild type strain did not grow in the presence of 10 and 15 g/L n-butanol; however, all of the selected isolates were still able to grow. The maximum OD600 was slightly lower for 10 g/L n-butanol condition and 50% lower with 15 g/L n-butanol condition, compared to the no-butanol control, and the lag phase was about 2 and 15 hours longer for 10 g/L n-butanol and 15 g/L n-butanol respectively (Fig. 2). These results indicate that evolutionary selection by chemostat was successful in isolating n-butanol tolerant strains of C. thermocellum.
Figure 2.
Improved n-butanol tolerance of C. thermocellum mutants. Growth rate was measured in MTC-5 medium at 55 °C. Growth comparison of wild type and butanol-tolerant mutant in different concentrations of n-butanol. Cells were grown in a 96-well plate and absorbance at 600 nm was measured at 3-minute intervals. Of the 12 isolates selected, all showed similar improvements in n-butanol tolerance, and the results from a single representative isolate are presented. The OD600 values are plotted on a semi-logarithmic scale. The orange line represents the OD600 value of the mutant strain and the light blue line represents the OD600 value of wild type strain. The wild type strain did not grow at n-butanol concentrations above 5 g/L.
Whole-genome sequencing of n-butanol tolerant strains and identification of key mutations
To identify specific mutations in the genome of the n-butanol tolerant strains, we re-sequenced the genomes of 6 isolates. Common mutations among these 6 isolates are listed in Table 1.
Table 1.
Common mutations among the 6 selected n-butanol tolerant strains.
| Regiona | C. thermocellum DSM 1313 genome (CDS) | Type | Ref | Allele | Gene product |
|---|---|---|---|---|---|
| 309131 | Clo1313_0284 upstream | SNV | G | A | hypothetical protein |
| 493829^493830 | Clo1313_0438 | Insertion | — | T | transcription elongation factor GreA |
| 590805^590806 | Clo1313_0538 upstream | Insertion | — | C | hypothetical protein |
| 904318 | Clo1313_0785 | SNV | G | T | transposase mutator type |
| 989039^989040 | Clo1313_0853 | Insertion | — | A | phospholipase D/Transphosphatidylase |
| 2097217 | Clo1313_1798 | SNV | T | C | iron-containing alcohol dehydrogenase |
aGenome coordinates based on NC_017304.1 sequence from Genbank.
Among the 6 common mutations, the mutations in coding sequences of Clo1313_0853 and Clo1313_1798 were found in a previous study looking at ethanol tolerance in C. thermocellum26. The gene Clo1313_0853 is annotated to encode a phospholipase D enzyme (PLD), which catalyzes the hydrolysis of phosphatidylcholine and other phospholipids to generate phosphatidic acid (PA), a necessary structural element of membranes. However, in the presence of a primary alcohol, PLD activity can generate phosphatidyl alcohol instead of PA27,28, and the resulting PA deficiency may be toxic. The mutation in the Clo1313_0853 gene truncates the protein by frameshift, which should eliminate activity. This mutation could protect the membrane when ethanol or n-butanol is present. The mutation in the Clo1313_1798 gene, the bifunctional alcohol dehydrogenase (adhE) generates a D494G amino acid change. This mutation has been previously shown to increase the ability of the alcohol dehydrogenase reaction to use NADPH as a cofactor29. It has also been shown to increase ethanol yield and titer30, however its effect on ethanol tolerance has not been studied.
Reconstruction of n-butanol tolerance
To determine whether mutations in Clo1313_0853 and Clo1313_1798 were the cause of the observed increase in n-butanol tolerance, we set out to reconstruct the phenotype by introducing specific mutations in the wild type strain. For Clo1313_0853, we inactivated the gene by deleting it (strain LL1636, Table 3). This strain did not show any obvious improvement in n-butanol or ethanol tolerance compared to the control. For the adhED494G mutation, we have a pair of strains that differ only by its absence or presence (strains LL1160 and LL1161 respectively, Table 3), whose construction is described elsewhere31.
Table 3.
Strains used in this work.
| Organism | Strain number | Description | Accession number | Source or reference |
|---|---|---|---|---|
| C. thermocellum | LL1004 | wild type strain DSM 1313 | CP002416 | DSMZ |
| LL1599 | n-butanol tolerant mutant isolate 1 | SRP163075 | this work | |
| LL1600 | n-butanol tolerant mutant isolate 2 | SRP163076 | this work | |
| LL1601 | n-butanol tolerant mutant isolate 3 | SRP163072 | this work | |
| LL1603 | n-butanol tolerant mutant isolate 4 | SRP163070 | this work | |
| LL1604 | n-butanol tolerant mutant isolate 5 | SRP163069 | this work | |
| LL1605 | n-butanol tolerant mutant isolate 6 | SRP163071 | this work | |
| LL1111 | LL1004 Δhpt ΔadhE ldh(S161R) | SRX744221 | 21 | |
| LL1160 | LL1111 adhE ldh(S161R) | SRA273168 | 31 | |
| LL1161 | LL1111 adhED494G ldh(S161R) | SRA273169 | 31 | |
| AG929 | DSM1313 Δhpt ΔClo1313_0478 | SRP097241 | 41 | |
| LL1636 | AG929 ΔClo1313_0853 | this work | ||
| E. coli | BL21(DE3) | T7 Express lysY/lq Used for heterologous protein expression | New England Biolabs | |
| DH5α | DH5α Used for plasmid screening and propagation | New England Biolabs | ||
| BL21(DE3) overexpressing C. thermocellum adhE | 39 | |||
| BL21(DE3) overexpressing C. thermocellum adhED494G | 39 | |||
| BL21(DE3) overexpressing C. thermocellum gapdh | 20 |
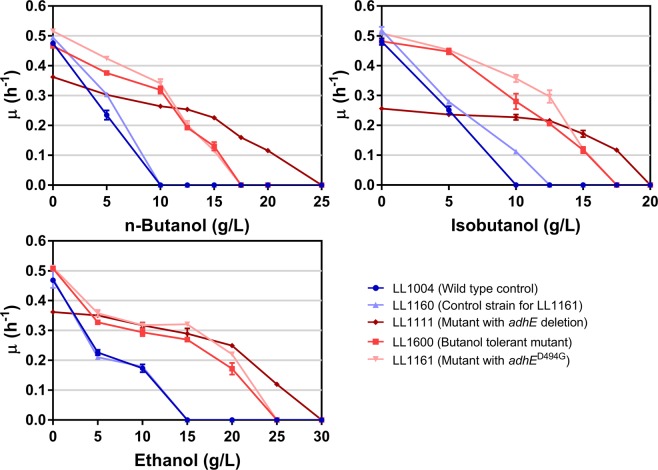
For the strain with the mutant adhE gene (LL1161), n-butanol tolerance was similar to that of the n-butanol-adapted strain (LL1600, Table 3) (Fig. 3). Since we know that mutations in the AdhE protein have been shown to increase ethanol tolerance26,32, we hypothesized that this mutation might increase tolerance to other primary alcohols. This is, in fact, what we found when we tested tolerance to isobutanol (Fig. 3).
Figure 3.
Tolerance of the reconstructed C. thermocellum mutant to ethanol, n-butanol and isobutanol. Growth rate was measured in MTC-5 medium at 55 °C. Maximum specific growth rate is plotted against the concentration of the alcohol. The data represents the average of three biological replications. Error bars represent one standard deviation. Strains colored in shades of blue are wild type for the adhE gene. Strains colored in shades of red have mutations or deletion in the adhE gene. The dark blue circle represents the wild type strain LL1004; the light blue triangle (up-direction) represents the control strain, LL1160; The dark red diamond represents the adhE deletion strain, LL1111; the red square represents the selected n-butanol tolerant strain, LL1600; the light red triangle (down-direction) represents the strain with the mutant adhE (D494G mutation, strain LL1161).
Effect of AdhE mutations
Ethanol production in C. thermocellum involves successive reduction of acetyl-CoA and acetaldehyde with electrons provided by NADH (i.e., the ALDH and ADH reactions). These two reactions are both catalyzed by the bifunctional AdhE enzyme. Previously we have shown that as ethanol concentrations increase, the NADH/NAD+ ratio also increases20. In the case of added ethanol, this may be due to reverse flux through the ADH and ALDH reactions, however even with produced ethanol, when net flux is in the direction of ethanol formation, the NADH/NAD+ ratio may need to increase to maintain a negative Gibbs free energy change for the ALDH and ADH reactions. High NADH/NAD+ ratios have been shown to inhibit the GAPDH reaction33,34 which inhibits the carbon flux of glycolysis in C. thermocellum20. The increased NADPH-linked activity associated with the AdhED494G mutation provides a potential explanation for this. In this mutant, reverse flux through the ADH reaction can affect both the NADH/NAD+ ratio and the NADPH/NADP+ ratio. Changes in the NADPH/NAD+ ratio, however, will not affect the GAPDH reaction, which is strictly NADH-linked in C. thermocellum (Table S1).
To apply the same explanation for n-butanol and isobutanol tolerance, we need to demonstrate that AdhE can also use butyryl-CoA/isobutyryl-CoA and butyraldehyde/isobutyraldehyde as substrates. Both the wild type AdhE and mutant AdhE were cloned and purified in E. coli and their activities were measured (Table 2). From these results, we confirmed that AdhE has the capacity to catalyze both butyryl-CoA conversion to butyraldehyde and isobutyryl-CoA conversion to isobutyraldehyde. In addition to lending support for our hypothesis that isobutanol and n-butanol inhibit C. thermocellum metabolism by inhibiting the GAPDH reaction, results from enzyme assays provide insight into the native pathway for isobutanol production in this organism, the final steps of which were previously unknown23.
Table 2.
Kinetic parameters of purified C. thermocellum wild type and D494G mutant AdhE proteins.
| Substrate | Cofactor | AdhE | AdhE D494G | ||||
|---|---|---|---|---|---|---|---|
| Km (µM) | Vmax (U/mg)a | kcat/Km (M−1s−1) | Km (µM) | Vmax (U/mg) | kcat/Km (M−1s−1) | ||
| Forward Direction | |||||||
| acetyl-CoA | NADH | 38.3 ± 1.5b | 1.84 ± 0.13 | 7.69E + 04 | 45.8 ± 2.1 | 1.51 ± 0.11 | 5.28E + 04 |
| acetaldehyde | 4658 ± 594 | 1.21 ± 0.09 | 4.16E + 02 | 5136 ± 485 | 1.04 ± 0.08 | 3.24E + 02 | |
| butyryl-CoA | 75.7 ± 3.7 | 3.08 ± 0.21 | 6.51E + 04 | 84.5 ± 5.5 | 2.88 ± 0.19 | 5.45E + 04 | |
| butyraldehyde | 4152 ± 485 | 6.14 ± 0.42 | 2.37E + 03 | 4351 ± 407 | 5.37 ± 0.34 | 1.98E + 03 | |
| Iso-butytyl-CoA | 54.5 ± 2.8 | 4.18 ± 0.23 | 1.23E + 05 | 65.5 ± 4.8 | 3.78 ± 0.19 | 9.24E + 04 | |
| Iso-butyradehyde | 3578 ± 485 | 5.84 ± 0.32 | 2.61E + 03 | 3897 ± 414 | 5.78 ± 0.34 | 2.37E + 03 | |
| acetaldehyde | NADPH | NDc | 4754 ± 214 | 1.45 ± 0.21 | 4.88E + 02 | ||
| butyraldehyde | 3457 ± 248 | 8.51 ± 0.41 | 3.94E + 03 | ||||
| Iso-butyradehyde | 3789 ± 234 | 6.25 ± 0.21 | 2.64E + 03 | ||||
| Reverse Direction | |||||||
| Substrate | Cofactor | AdhE | AdhE D494G | ||||
| K m (mM) | V max (U/mg) | k cat /K m (M −1 s 1 ) | K m (mM) | V max (U/mg) | k cat /K m (M −1 s −1 ) | ||
| ethanol | NAD + | 59.06 ± 6.01 | 3.79 ± 0.60 | 1.03E + 02 | 57.21 ± 5.01 | 3.14 ± 0.42 | 8.78E + 01 |
| n-butanol | 62.21 ± 5.54 | 2.98 ± 0.54 | 7.67E + 01 | 58.32 ± 6.75 | 2.18 ± 0.23 | 5.98E + 01 | |
| Isobutanol | 65.7 ± 6.12 | 2.58 ± 0.46 | 6.28E + 01 | 61.40 ± 4.54 | 2.94 ± 0.32 | 7.67E + 01 | |
| ethanol | NADP+ | ND | 48.51 ± 6.32 | 3.52 ± 0.63 | 1.16E + 02 | ||
| n-butanol | 53.14 ± 4.58 | 2.59 ± 0.68 | 7.80E + 01 | ||||
| Isobutanol | 51.15 ± 3.65 | 2.77 ± 0.59 | 8.67E + 01 | ||||
a1U = 1 µmol/min.
bThe data represents the average of three individual rounds of assays.
cND = not detected.
To further confirm our hypothesis that reverse flux through AdhE is the mechanism for inhibition by primary alcohols, we tested the tolerance of a strain with an adhE deletion (strain LL1111). As shown in Fig. 3, the adhE deletion strain shows tolerance to all three primary alcohols and demonstrated the highest level of tolerance among the strains tested.
Conclusions
After isolating a butanol tolerant mutant of C. thermocellum, we discovered that the strain had acquired a D494G mutation in the adhE gene. We showed that this mutation is sufficient to recapitulate the n-butanol tolerance phenotype. We further showed that this is applicable to other primary alcohols including ethanol and isobutanol. To show that reverse flux through AdhE is the mechanism of inhibition, we confirmed that it has NADH-linked activity with both isobutyryl-CoA and butyryl-CoA. Furthermore, deletion of the adhE gene increases tolerance for all three primary alcohols tested. The mechanism found in this study can be widely applied to other organisms.
Methods
Bacterial strains, media and cultivation
Strains used in this study are listed in Table 3. All chemicals were reagent grade and obtained from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) unless indicated otherwise. CTFUD rich medium35 and MTC-5 defined medium36 were used for routine strain maintenance and strain evolution.
Strain evolution
Serum bottles cultures were incubated at 55 °C and shaken at 180 rpm. Serum bottles were purged with N2 and sealed with butyl rubber stoppers. In batch cultures, pH was regulated with 40 mM MOPS buffer. Chemostat bioreactor fermentations were carried out in 0.5 L (100 ml working volume) bioreactors (NDS Technologies Ins, Vineland NJ) in modified MTC-5 medium without MOPS buffer and with 2 g/L urea as the nitrogen source, with the temperature maintained at 55 °C and stirred at 150 rpm. The dilution rate was set to 0.04 h−1. The pH was controlled at 7.0 with a Mettler-Toledo pH probe (Columbus, OH) by the addition of 8 N KOH. The bioreactor was inoculated with 5 mL fresh culture grown on 5 g/L cellobiose in MTC-5. The headspace of the bioreactor was flushed with N2 gas prior to inoculation. The feed bottle was continuously purged with N2 gas to maintain anaerobic conditions. Appropriate amounts of n-butanol were added to the feed bottle. To avoid selecting an inducible mutation (as opposed to a constitutive mutation), the n-butanol concentration was reduced to 0 g/L twice during the selection process. 16 s rRNA gene sequences of cell pellets from the fermentation were used to verify culture purity. The adapted culture was first grown on CTFUD agar plates and then 12 isolates were selected and inoculated into liquid CTFUD medium.
Tolerance test
Tolerance was measured in a COY (Ann Arbor, MI) anaerobic chamber (85% N2, 10% CO2, and 5% H2). 200 µl cultures were grown in 96-well pre-sterilized polystyrene plates in an anaerobic chamber (85% N2, 10% CO2, and 5% H2). Absorbance measurements at 600 nm (OD600) were taken every 10 minutes for 36 hours using a BioTek plate reader (BioTek Instruments Inc., Winooski, VT). Growth rates were determined based on the slope of blank-subtracted, log-transformed absorbance data in the range of 0.1 to 1. Averages were based on at least three independent biological replicates.
Protein purification
For expression and purification of proteins in E. coli, cell preparation and cell free extract were prepared as described previously37. Cells were grown aerobically in TB medium at 37 °C with a stirring speed of 225 rpm. When the OD600 reached 0.6, 4 mM rhamnose was added to induce expression of the target gene. Cells were then grown aerobically for 4 h before harvesting by centrifugation. Cell pellets were washed with buffer (50 mM Tris-HCl, pH 7.5 and 0.5 mM DTT) and stored at −80 °C.
The cell pellet was resuspended in lysis buffer (1X BugBuster reagent (EMD Millipore, Darmstadt, Germany) with 0.2 mM dithiothreitol). The cells were lysed with Ready-Lyse lysozyme (Epicentre, Madison, WI, USA), and DNase I (New England Biolabs, Ipswich, MA, USA) was added to reduce the viscosity. After incubation for 30 min at room temperature, the resulting solution was centrifuged at 10,000 X g for 5 min. The supernatant was used as cell free extract for enzyme assays or protein purification.
All purification steps were performed at room temperature as described previously38. His-tag affinity spin columns (His SpinTrap; GE Healthcare BioSciences, Pittsburgh, PA, USA) were used to purify the protein. The column was first equilibrated with binding buffer (50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.5). Cell free extracts (in 50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.5) were applied to the column, and then the column was washed twice with wash buffer (50 mM sodium phosphate, 500 mM NaCl, 50 mM imidazole, 20% ethanol, pH 7.5). The His-tagged protein was eluted with elution buffer (50 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, pH 7.5).
Enzyme assays
Enzyme assays were performed in a COY (Ann Arbor, MI) anaerobic chamber (85% N2, 10% CO2, and 5% H2, <5 ppm O2). ALDH and ADH activities were measured as described previously39. The reaction mix contained: 50 mM pH 7.0 Tris-HCl buffer (The pH was adjusted at 55 °C to avoid changes in pH with changes in temperature), 2 mM MgCl2, 0.5 mM DTT, 5 µM FeSO4, 0.3 mM NADH or NADPH. For ALDH activity, six different substrate concentrations between 10 and 500 µM of acetyl-CoA, butyryl-CoA or isobutyryl-CoA were used as substrates to start the reaction. For ADH activity, six different substrate concentrations between 0.25 to 20 mM acetaldehyde, butyraldehyde or isobutyraldehyde were used. The consumption of NADH or NADPH was followed spectrophotometrically at 340 nm (molar extinction coefficient ε of NADH/NADPH = 6.22 mM−1cm−1) in a BioTek PowerWave XS plate reader (BioTek Instruments Inc., Winooski, VT, USA).
The activity of the glyceraldehyde-3-phosphate dehydrogenase enzyme (GAPDH EC 1.2.1.12) was measured at 55 °C as previously described40. The standard assay (200 µl working volume) contained 50 mM Tris-HCl pH 7.0, 10 mM sodium arsenate, 10 mM glyceraldehyde-3-phosphate, and 0.5 mM NAD+. To avoid thermal destruction of glyceraldehyde-3-phosphate, this substrate was added to the mixture immediately before starting the enzyme reaction. The formation NADH or NADPH were followed by photometric observation at 340 nm (ε = 6.2 mM−1 cm−1) in a BioTek PowerWave XS plate reader (BioTek Instruments Inc., Winooski, VT, USA).
The protein concentration was determined using the Bradford protein reagent with bovine serum albumin as the standard (BioRad, Hercules, CA).
Supplementary information
Table S1 : Kinetic parameters of C. thermocellum GAPDH enzyme.
Figure S1: Growth of C. thermocellum in thepresence of different concentrations of n-butanol from 0 to 8 g/L.
Acknowledgements
The Center for Bioenergy Innovation is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Resequencing was performed by the Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, and is supported by the Office of Science of the U.S. Department of Energy under contract number DE-AC02–05CH11231.
Author Contributions
L.T. and D.G.O. designed experiments, performed experiments, analyzed data and wrote the manuscript. L.A. carried out the chemostat fermentations. N.C. carried out the tolerance test assays. L.R.L. designed experiments and wrote the manuscript. All authors read and approved the manuscript.
Competing Interests
Lee R. Lynd is a founder of the Enchi Corporation, which has a financial interest in Clostridium thermocellum.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37979-5.
References
- 1.Atsumi S, et al. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 2010;6:1–11. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghiaci P, Norbeck J, Larsson C. Physiological adaptations of Saccharomyces cerevisiae evolved for improved butanol tolerance. Biotechnol. Biofuels. 2013;6:101. doi: 10.1186/1754-6834-6-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherkhanov S, Korman TP, Bowie JU. Improving the tolerance of Escherichia coli to medium-chain fatty acid production. Metab. Eng. 2014;25:1–7. doi: 10.1016/j.ymben.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Segura A, et al. Solvent tolerance in Gram-negative bacteria. Curr. Opin. Biotechnol. 2012;23:415–421. doi: 10.1016/j.copbio.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Pini CV, Bernal P, Godoy P, Ramos JL, Segura A. Cyclopropane fatty acids are involved in organic solvent tolerance but not in acid stress resistance in Pseudomonas putida DOT-T1E. Microb. Biotechnol. 2009;2:253–261. doi: 10.1111/j.1751-7915.2009.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Royce LA, Liu P, Stebbins MJ, Hanson BC, Jarboe LR. The damaging effects of short chain fatty acids on Escherichia coli membranes. Appl. Microbiol. Biotechnol. 2013;97:8317–8327. doi: 10.1007/s00253-013-5113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunlop MJ, et al. Engineering microbial biofuel tolerance and export using efflux pumps. Mol. Syst. Biol. 2011;7:1–7. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minty JJ, et al. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb. Cell Fact. 2011;10:1–38. doi: 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D, et al. Identification of ethanol tolerant outer membrane proteome reveals OmpC-dependent mechanism in a manner of EnvZ/OmpR regulation in Escherichia coli. J. Proteomics. 2018;179:92–99. doi: 10.1016/j.jprot.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Tomas CA, Welker NE, Papoutsakis ET. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Appl. Environ. Microbiol. 2003;69:4951–4965. doi: 10.1128/AEM.69.8.4951-4965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zingaro KA, Terry Papoutsakis E. GroESL overexpression imparts Escherichia coli tolerance to i-, n-, and 2-butanol, 1,2,4-butanetriol and ethanol with complex and unpredictable patterns. Metab. Eng. 2013;15:196–205. doi: 10.1016/j.ymben.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Zingaro KA, Papoutsakis ET. Toward a semisynthetic stress response system to engineer. MBio. 2012;3:1–9. doi: 10.1128/mBio.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duque E, et al. The RpoT regulon of Pseudomonas putida DOT-T1E and its role in stress endurance against solvents. J. Bacteriol. 2007;189:207–219. doi: 10.1128/JB.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez-Herva JJ, García V, Hurtado A, Segura A, Ramos JL. The ttgGHI solvent efflux pump operon of Pseudomonas putida DOT-T1E is located on a large self-transmissible plasmid. Environ. Microbiol. 2007;9:1550–1561. doi: 10.1111/j.1462-2920.2007.01276.x. [DOI] [PubMed] [Google Scholar]
- 15.Duque E, Segura A, Mosqueda G, Ramos JL. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 2001;39:1100–1106. doi: 10.1046/j.1365-2958.2001.02310.x. [DOI] [PubMed] [Google Scholar]
- 16.Lynd L, van Zyl WH, McBride J, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 2012;23:396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 18.Hon S, et al. Expressing the Thermoanaerobacterium saccharolyticum pforA in engineered Clostridium thermocellum improves ethanol production. Biotechnol. Biofuels. 2018;11:242. doi: 10.1186/s13068-018-1245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian L, et al. Simultaneous achievement of high ethanol yield and titer in Clostridium thermocellum. Biotechnol. Biofuels. 2016;9:116. doi: 10.1186/s13068-016-0528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian L, et al. Metabolome analysis reveals a role for glyceraldehyde 3-phosphate dehydrogenase in the inhibition of C. thermocellum by ethanol. Biotechnol. Biofuels. 2017;10:1–11. doi: 10.1186/s13068-017-0961-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo J, Zheng T, Hon S, Olson DG, Lynd LR. The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J. Bacteriol. 2015;197:JB.02450–14. doi: 10.1128/JB.02450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veen, D. va.d. et al. Characterization of Clostridium thermocellum strains with disrupted fermentation end-product pathways. Journal of Industrial Microbiology & Biotechnology.10.1007/s10295-013-1275-5. 40(7), 725–34 (July 2013). [DOI] [PubMed]
- 23.Holwerda EK, et al. The exometabolome of Clostridium thermocellum reveals overflow metabolism at high cellulose loading. Biotechnol. Biofuels. 2014;7:155. doi: 10.1186/s13068-014-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin PP, et al. Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum. Metab. Eng. 2015;31:44–52. doi: 10.1016/j.ymben.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Lütke-Eversloh T, Bahl H. Metabolic engineering of Clostridium acetobutylicum: Recent advances to improve butanol production. Curr. Opin. Biotechnol. 2011;22:634–647. doi: 10.1016/j.copbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Shao X, et al. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl. Microbiol. Biotechnol. 2011;92:641–652. doi: 10.1007/s00253-011-3492-z. [DOI] [PubMed] [Google Scholar]
- 27.Banno Y. Regulation and possible role of mammalian phospholipase D in cellular functions. J Biochem. 2002;131:301–6. doi: 10.1093/oxfordjournals.jbchem.a003103. [DOI] [PubMed] [Google Scholar]
- 28.Kolesnikov YS, et al. Molecular structure of phospholipase D and regulatory mechanisms of its activity in plant and animal cells. Biochem. Biokhimii͡a. 2012;77:1–14. doi: 10.1134/S0006297912010014. [DOI] [PubMed] [Google Scholar]
- 29.Zheng T, et al. Both adhE and a separate NADPH-dependent alcohol dehydrogenase, adhA, are necessary for high ethanol production in Thermoanaerobacterium saccharolyticum. J. Bacteriol. 2017;199:1–10. doi: 10.1128/JB.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas R, Zheng T, Olson DG, Lynd LR, Guss AM. Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol. Biofuels. 2015;8:20. doi: 10.1186/s13068-015-0204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hon S, et al. Development of a Plasmid-Based Expression System in Clostridium thermocellum and its use to Screen Heterologous Expression of bifunctional alcohol dehydrogenases (adhEs) Metab. Eng. Commun. 2016;3:120–129. doi: 10.1016/j.meteno.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown SD, et al. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc. Natl. Acad. Sci. USA. 2011;108:13752–7. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Copeland L. Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean root nodules. Archives of biochemistry and biophysics. 1994;312:107–113. doi: 10.1006/abbi.1994.1287. [DOI] [PubMed] [Google Scholar]
- 34.Even S, Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. Pyruvate metabolism in Lactococcus lactis is dependent upon glyceraldehyde-3-phosphate dehydrogenase activity. Metab. Eng. 1999;1:198–205. doi: 10.1006/mben.1999.0120. [DOI] [PubMed] [Google Scholar]
- 35.Olson, D. G. & Lynd, L. R. Transformation of Clostridium thermocellum by electroporation. Methods in enzymology510, (Elsevier Inc. 2012). [DOI] [PubMed]
- 36.Hon, S., et al. The ethanol pathway from Thermoanaerobacterium saccharolyticum improves ethanol production in Clostridium thermocellum. Metabolic Engineering42, 175–84 (2017). [DOI] [PubMed]
- 37.Tian L, et al. Ferredoxin:NAD+ oxidoreductase of Thermoanaerobacterium saccharolyticum and its role in ethanol formation. Appl. Environ. Microbiol. 2016;82:7134–7141. doi: 10.1128/AEM.02130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Liang, P. S. J., et al. Enhanced ethanol formation by Clostridium thermocellum via pyruvate decarboxylase. Microb. Cell Fact. 10.1186/s12934-017-0783-9 (2017). [DOI] [PMC free article] [PubMed]
- 39.Zheng T, et al. Cofactor specificity of the bifunctional alcohol and aldehyde dehydrogenase (AdhE) in wild-type and mutants of Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J. Bacteriol. 2015;197:JB.00232–15. doi: 10.1128/JB.00232-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zwickl P, Fabry S, Bogedain C, Haas A, Hensel R. Glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaebacterium Pyrococcus woesei: Characterization of the enzyme, cloning and sequencing of the gene, and expression in Escherichia coli. J. Bacteriol. 1990;172:4329–4338. doi: 10.1128/jb.172.8.4329-4338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hon S, et al. The ethanol pathway from Thermoanaerobacterium saccharolyticum improves ethanol production in Clostridium thermocellum. Metab. Eng. 2017;42:175–184. doi: 10.1016/j.ymben.2017.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 : Kinetic parameters of C. thermocellum GAPDH enzyme.
Figure S1: Growth of C. thermocellum in thepresence of different concentrations of n-butanol from 0 to 8 g/L.