Abstract
The transcription factor c-Myc is an important regulator of cellular proliferation, differentiation and embryogenesis. While c-Myc can inhibit myoblast differentiation, the underlying mechanisms remain poorly understood. Here, we found that c-Myc does not only inhibits myoblast differentiation but also promotes myoblast proliferation and muscle fibre hypertrophy. By performing chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq), we identified the genome-wide binding profile of c-Myc in skeletal muscle cells. c-Myc achieves its regulatory effects on myoblast proliferation and differentiation by targeting the cell cycle pathway. Additionally, c-Myc can regulate cell cycle genes by controlling miRNA expression of which dozens of miRNAs can also be regulated directly by c-Myc. Among these c-Myc-associated miRNAs (CAMs), the roles played by c-Myc-induced miRNAs in skeletal muscle cells are similar to those played by c-Myc, whereas c-Myc-repressed miRNAs play roles that are opposite to those played by c-Myc. The cell cycle, ERK–MAPK and Akt-mediated pathways are potential target pathways of the CAMs during myoblast differentiation. Interestingly, we identified four CAMs that can directly bind to the c-Myc 3' UTR and inhibit c-Myc expression, suggesting that a negative feedback loop exists between c-Myc and its target miRNAs during myoblast differentiation. c-Myc also potentially regulates many long intergenic noncoding RNAs (lincRNAs). Linc-2949 and linc-1369 are directly regulated by c-Myc, and both lincRNAs are involved in the regulation of myoblast proliferation and differentiation by competing for the binding of muscle differentiation-related miRNAs. Our findings do not only provide a genome-wide overview of the role the c-Myc plays in skeletal muscle cells but also uncover the mechanism of how c-Myc and its target genes regulate myoblast proliferation and differentiation, and muscle fibre hypertrophy.
Subject terms: Epigenetics, Epigenetics
Introduction
c-Myc is a multifunctional transcription factor that regulates various processes including development and differentiation. One of its regulatory functions involved genes transcription where it binds to E-box sequences (CANNTG) located on the gene promoter regions. The c-Myc target gene network has been estimated to comprise ~15% of all genes from flies to humans [1]. A c-Myc gene knock-out is lethal in mice and results in smaller embryos and retarded development [2]. c-Myc is also an important regulator of the cell cycle and its knock-out or knockdown severely affects cell proliferation [3–5]. It also plays a role in cell differentiation. c-Myc is induced by the differentiation of long-term haematopoietic stem cells and balances stem cell self-renewal and differentiation [6]. In mouse embryo stem cells, c-Myc inhibits the expression of differentiation marker genes and affects stem cell differentiation by inducing miRNA expression [7].
Myoblasts are mononuclear myogenic precursor cells that can be induced to form multinuclear myotubes under differentiation conditions. c-Myc expression is downregulated during myoblast differentiation into myotubes [8], while its overexpression induces myoblast proliferation but has no effect on myoblast differentiation [8, 9]. However, the full-length c-Myc protein can be cleaved by calpain and forms a cytoplasmic protein called Myc-nick [8]. The ectopic expression of Myc-nick promotes myogenic marker expression and accelerates myoblast differentiation and fusion [8]. The deficient cleavage of c-Myc to Myc-nick can reduce myoblast differentiation [9], suggesting the full-length of c-Myc protein can inhibit myoblast differentiation. But this inhibition role can be repressed by Myc-nick. The detailed mechanism underlying the regulation of myoblast differentiation by the full-length c-Myc remains unknown.
Many miRNAs are regulated by c-Myc [10]. In addition, long noncoding RNAs (lncRNAs) are important regulatory targets of c-Myc [11]. Most c-Myc functions involving cell development are achieved via miRNAs and lncRNAs [7, 10, 12]. Both miRNAs and lncRNAs play critical roles in myoblast proliferation and differentiation [13, 14]. Interestingly, many muscle differentiation-related miRNAs (MyomiRs) and lncRNAs are regulated by c-Myc [7, 15–20], such as miR-135, miR-124, miR-199 and H19 [21–25]. Therefore, we hypothesized that the regulatory roles played by c-Myc in myoblast proliferation and differentiation, at least partially, may be achieved via its transcriptional regulation of miRNAs and lncRNAs. A detailed verification of this hypothesis is needed.
Here, we used chicken primary myoblasts and skeletal muscle to study the functions and regulatory mechanisms of c-Myc during skeletal muscle development. Using chromatin immunoprecipitation-sequencing (ChIP-seq) and expression profiling data from RNA sequencing, we found that several genes including miRNAs and lincRNAs, can be directly regulated by c-Myc and that these c-Myc-regulated targets are important for the functions of c-Myc in myoblast proliferation, differentiation and muscle fibre hypertrophy.
Results
c-Myc expression during chicken skeletal muscle development
To study the c-Myc gene, we isolated its full-length cDNA from chicken embryonic skeletal muscle. We obtained a 2111 bp length of chicken c-Myc cDNA (Fig. 1a; Supplementary Figure S1, accession number KU981087 in the NCBI database). c-Myc is widely expressed in various tissues (Fig. 1b). We observed that during chicken skeletal muscle development, c-Myc showed increasing and then decreasing trends in expression (Fig. 1c) and that c-Myc expression was downregulated during chicken primary myoblast differentiation (Fig. 1d). In the mouse cell lines, the full-length of c-Myc protein was cleaved to generate Myc-nick of which the major c-Myc cleavage site is conserved in vertebrates (Fig. 1e). Notably, a potential Myc-nick band was expressed in the chicken myotubes but not in the myoblasts, and the expression of the full-length c-Myc is reduced in the myotubes (Fig. 1f). The expression of c-Myc was mainly observed in the nuclear region of the mononuclear myoblasts but it increased in the cytoplasm as the myoblasts differentiated into multinuclear myotubes (Fig. 1g).
Fig. 1.
c-Myc expression during chicken skeletal muscle development. a Genomic structure of the chicken c-Myc gene. Black boxes indicate the coding sequence regions, and the white boxes indicate the UTRs. b Relative c-Myc mRNA expression in chicken tissues. c Relative c-Myc mRNA expression in chicken embryonic leg muscle. d Relative c-Myc mRNA expression during chicken primary myoblast differentiation. e The major c-Myc calpain cleavage site is conserved among vertebrates. f Chicken primary myoblasts cultured in growth medium (GM) and differentiation medium for 4 days (DM). Total cell extracts were immunoblotted to detect c-Myc. g Chicken primary myoblasts cultured in DM4 were stained with anti-c-Myc. The results are shown as the mean ± standard error of mean (sem) of three independent experiments. One-way analysis of variance (ANOVA) followed by Dunnett’s test was performed to determine the significant differences between the groups. Different letters (a), (b) above the bars indicate significant differences (p < 0.05) by Duncan’s multiple range test. *p < 0.05; ns, no significant difference
c-Myc regulates myoblast proliferation and differentiation in vitro and induces muscle fibre hypertrophy in vivo
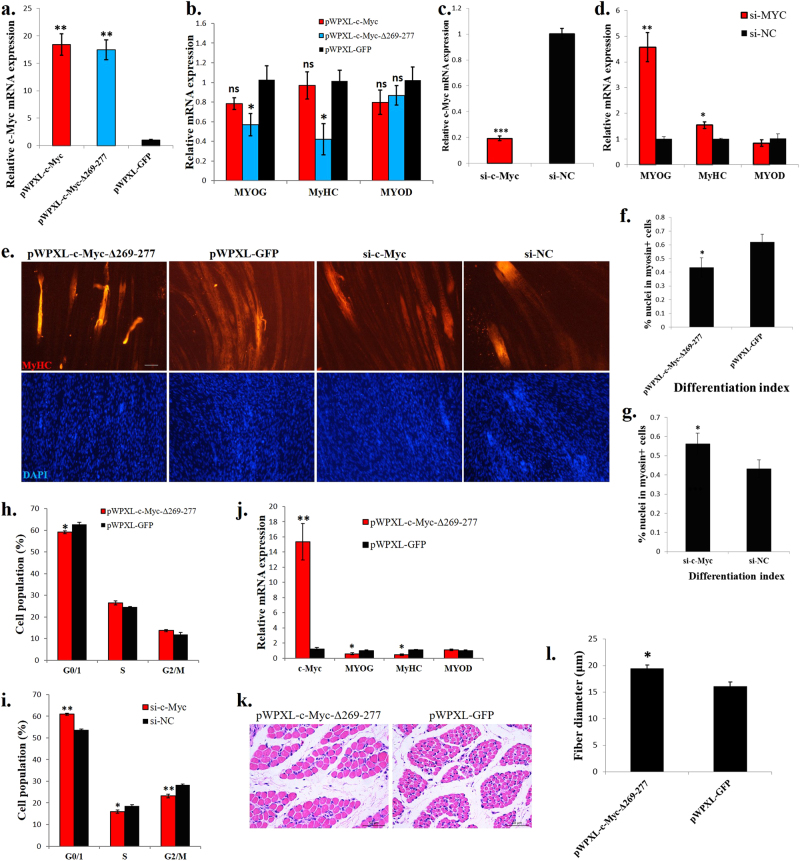
To determine the function of c-Myc in skeletal muscle cells, we constructed a full-length c-Myc lentiviral vector and a mutant c-Myc (c-Myc-Δ269–77) that is unable to generate Myc-nick. In chicken primary myoblasts, the ectopic expression of the full-length c-Myc had no effect on the expression of muscle differentiation marker genes whereas, the c-Myc-Δ269–77 could significantly reduce the marker genes expression (Fig. 2a, b). Myoblasts transfected with siRNA specifically designed for c-Myc showed increased marker genes expression (Fig. 2c, d). During myotube formation, the myotube-specific structural gene, myosin heavy chain (MyHC), would increase its expression, and the mononuclear myoblast will fuse to form multinuclear myotube. These two characteristics are widely used in judging the degree of myotube formation. c-Myc-Δ269–277 overexpression inhibited myoblast differentiation, as indicated by decreased myotube number (Fig. 2e). Quantification of the myotubes revealed significant decreases in differentiation index (percentage of nuclei in MyHC-positive cells, Fig. 2f), indicating that c-Myc-Δ269–277 inhibited myotube formation. c-Myc-Δ269–277 overexpression also promoted myoblast proliferation by increasing the cell population in the S phase and decreasing it in the G1/0 phase (Fig. 2h). Furthermore, the c-Myc loss-of-function promoted myotube formation (Fig. 2e), increased the differentiation index (Fig. 2g) and inhibited myoblast proliferation (Fig. 2i). Therefore, c-Myc promotes myoblast proliferation and inhibits myoblast differentiation in chicken.
Fig. 2.
c-Myc regulates myoblast proliferation and differentiation in vitro and induces muscle fibre hypertrophy in vivo. a Relative c-Myc mRNA expression after introducing c-Myc, c-Myc-Δ269–277 and GFP into chicken primary myoblasts. b Muscle differentiation marker genes expression after introducing c-Myc, c-Myc-Δ269–277 and GFP into chicken primary myoblasts. c Relative c-Myc mRNA expression after c-Myc knockdown in chicken primary myoblasts. d Muscle differentiation marker gene expression after c-Myc knockdown in chicken primary myoblasts. e MyHC immunostaining of primary myoblasts transduced with indicated vectors or siRNAs. Cells were differentiated for 72 h after transfection. The nuclei were visualized with DAPI. Bar, 100 µm. f Differentiation index of cells expressing c-Myc-Δ269–277 or GFP. g Differentiation index of cells transfected with si-c-Myc or si-NC. h Primary myoblasts expressing c-Myc-Δ269–277 or GFP were cultured in GM, and the cell cycle phase was analysed after 2 days. i Primary myoblasts transfected with si-c-Myc and si-NC were cultured in GM, and the cell cycle phase was analysed after 2 days. j Relative mRNA expression of the indicated genes in chicken breast muscles infected with a c-Myc-Δ269–277-expressing lentivirus or control (GFP). k H–E staining of a breast muscle fibre cross section from chickens infected with a c-Myc-Δ269–277-expressing lentivirus or control (GFP). l Fibre diameter of chicken breast muscle infected with a c-Myc-Δ269–277-expressing lentivirus or control (GFP). The results are shown as the mean ± sem of three independent experiments. In a, b, ANOVA followed by Dunnett’s test was used. In c, d, f–j, l, independent sample t-test was used. *p < 0.05; **p < 0.01; ns, no significant difference
To investigate the function of c-Myc in skeletal muscle in vivo, we introduced lentiviral vectors carrying c-Myc-Δ269–277 or GFP into the breast muscle of 1-day-old chicks. After 14 days and two introductions, the c-Myc-Δ269–277 transfection increased the c-Myc expression but decreased the muscle differentiation marker genes expression (Fig. 2j). Notably, the c-Myc-Δ269–277 transfection induced muscle fibre hypertrophy (Fig. 2k). Compared to the control group, the fibre diameter significantly increased after the c-Myc-Δ269–277 introduction (Fig. 2l).
Genome-wide mapping of c-Myc binding during myoblast differentiation by ChIP-seq
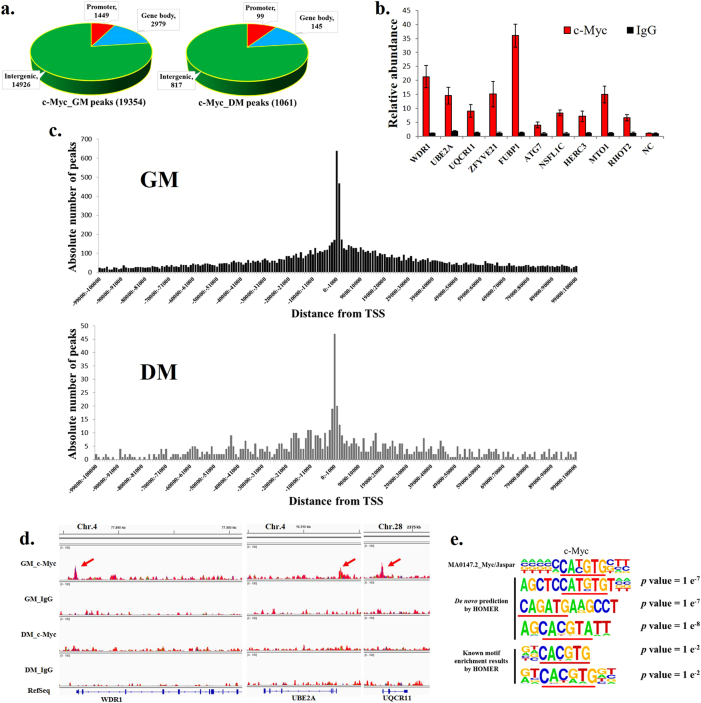
To obtain global insight into the roles played by c-Myc in myoblast differentiation, we performed ChIP-seq to determine the c-Myc occupancy in myoblasts and myotubes. In total, 19,354 c-Myc-binding sites were identified with high confidence in chicken primary myoblasts, while only 1061 binding sites were identified in myotubes (Supplementary file 1); 77.1% were found in intergenic regions, 15.4% were found in the gene body of annotated RefSeq genes, and 7.5% were found in the promoter region (±2 kb from the transcription start site (TSS)) of the annotated RefSeq genes (Fig. 3a). The distribution of the c-Myc binding sites in the myotubes was similar to the one in the myoblasts (Fig. 3a). To validate the ChIP-seq data, ten binding sites were randomly selected for ChIP-qPCR (Fig. 3b). The results obtained from ChIP-qPCR were concordant to the one from ChIP-seq, demonstrating the high quality of the ChIP-seq data. The c-Myc-binding sites were concentrated around the TSS, their number decreased as the distance from the TSS increased (Fig. 3c) and from the myoblasts to the myotubes (Fig. 3d; Supplementary file 2). c-Myc has been shown to regulate gene transcription by binding to the E-box motif [1, 26]. Subsequently, we performed a de novo prediction of the highly enriched DNA sequences from the c-Myc-binding sites. It revealed that a Myc-bound motif among the top 10 scored motifs, and certain canonical and non-canonical E-box sequences were significantly enriched (Fig. 3e; Supplementary Figure S2). HOMER known motif enrichment results revealed that Myc-bound motif was also significantly enriched and this motif occupied 25.88% of the target motifs (Fig. 3e; Supplementary Figure S3). In addition, 19,828 E-boxes were observed in the 19,354 c-Myc peak sequences and ~65.15% of the peak sequences contained at least one E-box motif (Supplementary file 3). Therefore, the c-Myc genome-wide association with muscle cell chromatin is directly mediated by its DNA-binding activity.
Fig. 3.
Genome-wide mapping of c-Myc binding during myoblast differentiation by ChIP-seq. a Distribution of c-Myc-binding peaks in myoblasts and myotubes. Promoter = TSS ± 2 kb. b Results of the ChIP-qPCR validation of 10 selected c-Myc-binding sites associated with RefSeq gene promoters. NC represents a genomic region without an identified c-Myc-binding peak. c Absolute number of peaks in myoblasts and myotubes distributed between −100 kb and +100 kb from the TSS. Each bin represents 1 kb. d Genomic snapshots depicting the ChIP-seq results of c-Myc and IgG at the promoter regions of the selected genomic loci. e De novo motif prediction and known motif enrichment results by HOMER of DNA sequences enriched in the c-Myc-binding regions. The c-Myc JASPAR matrix is presented for comparison. The results are shown as the mean ± sem of three independent experiments
Subsequently, we examined the function of the c-Myc target genes that were bound to c-Myc in their gene bodies and promoter regions. The target genes in the myoblasts were significantly enriched in cellular metabolic process and cell development-related processes, such as cell proliferation and cell migration (Supplementary Figure S4). In the myotubes, they were also enriched in cell developmental processes (Supplementary Figure S5), suggesting that c-Myc plays a role in the regulation of skeletal muscle development.
Cell cycle pathway as important regulatory target of c-Myc during myoblast differentiation
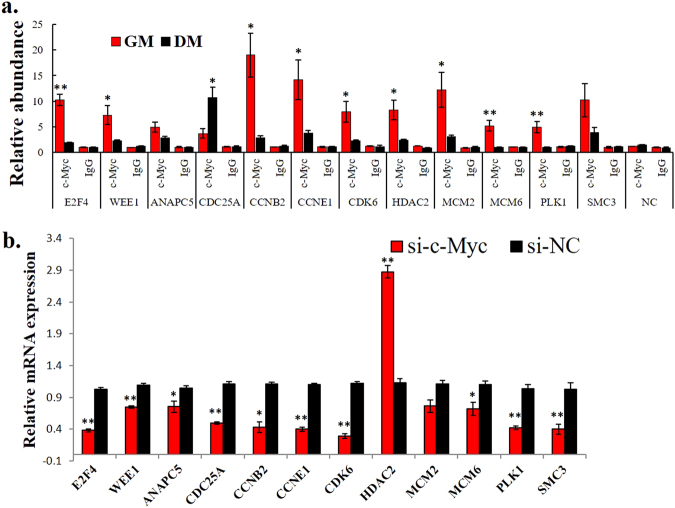
To determine the number of genes that are differentially regulated by c-Myc during myoblast differentiation, we identified the genes that do not only show differential c-Myc promoter binding between myoblasts and myotubes (Supplementary file 2) but also showed differential expression (p < 0.05) (Supplementary file 4). We then performed GO and KEGG pathway and found that these genes were enriched in cell proliferation-related processes, such as the cell cycle and DNA replication (Supplementary Figure S6 and S7). Following the KEGG analysis, 12 c-Myc-bound genes were enriched in the cell cycle pathway (Supplementary Figure S7 and S8), suggesting that c-Myc may promote the cell cycle by directly binding and regulating the transcription of the genes involved in this pathway. By performing ChIP-qPCR, we validated that most of the 12 cell cycle-related genes differentially bound c-Myc in the myoblasts and myotubes (Fig. 4a). Additionally, the c-Myc knockdown in the chicken primary myoblasts led to a downregulation of the 11 cell cycle genes and upregulation of 1 cell cycle gene (Fig. 4b), demonstrating that the cell cycle pathway is an important regulatory target of c-Myc during myoblast differentiation.
Fig. 4.
Cell cycle pathway is an important regulatory target of c-Myc during myoblast differentiation. a Results of the ChIP-qPCR validation of the 12 c-Myc-bound genes shown in Fig. 4c. NC represents a genomic region without an identified c-Myc-binding peak. b Relative mRNA expression of the indicated genes after c-Myc knockdown in primary myoblasts. The results are shown as the mean ± sem of three independent experiments. Independent sample t-test was performed to determine the significant differences between the groups. *p < 0.05; **p < 0.01
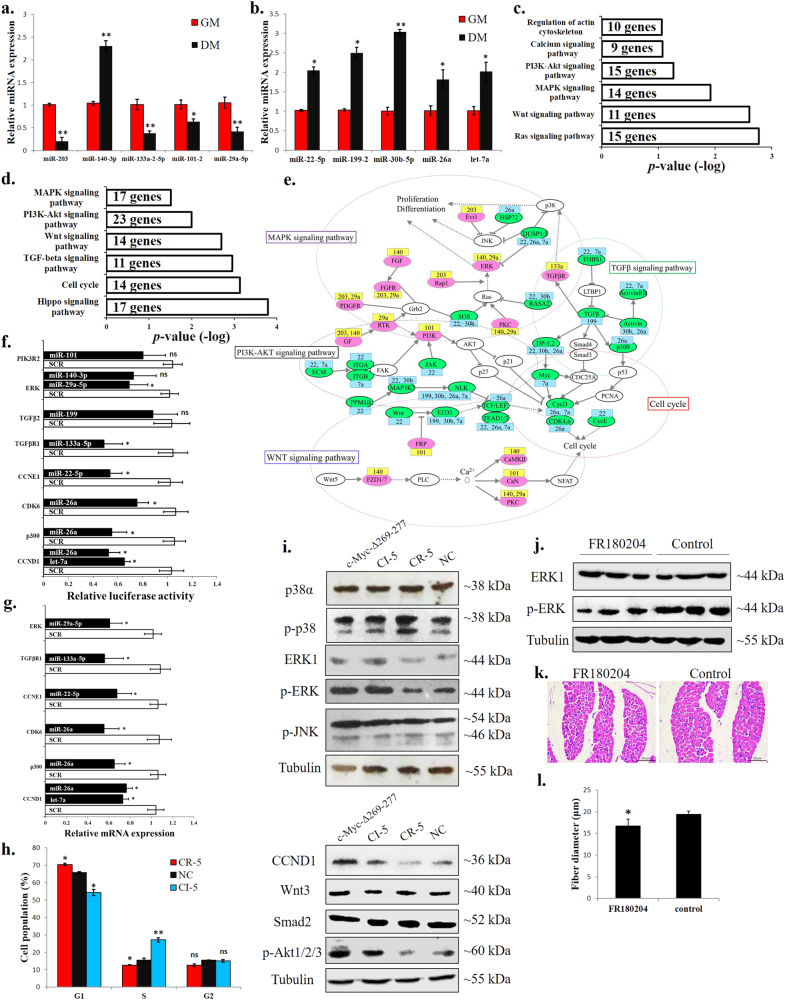
miRNAs are associated with c-Myc and are important for c-Myc’s regulation of myoblast proliferation, differentiation and muscle fibre hypertrophy
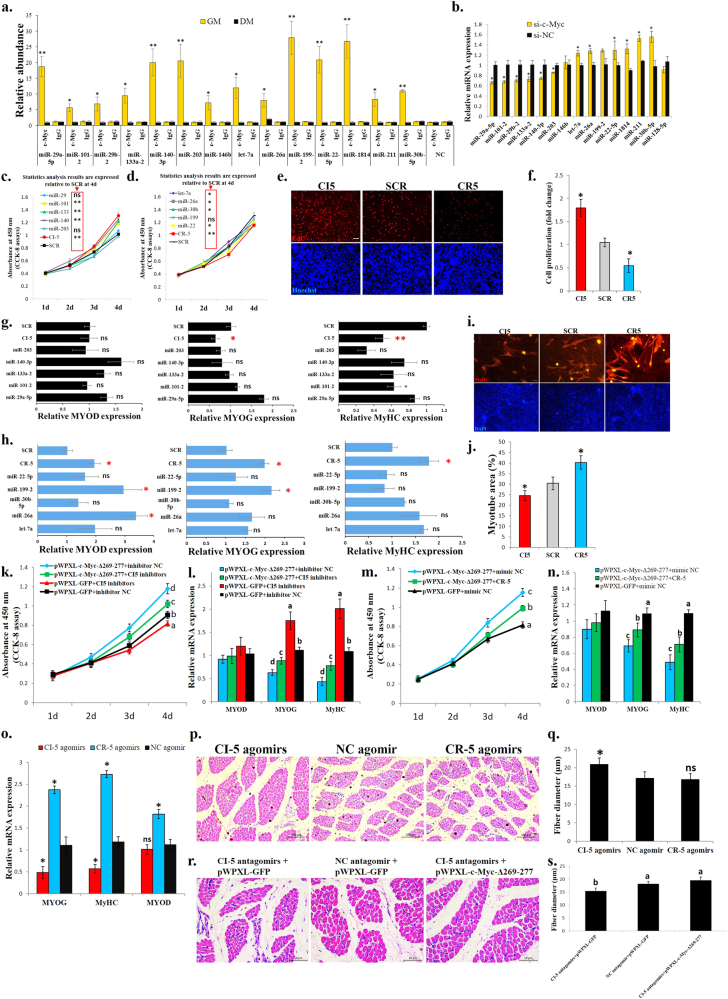
Among the differential c-Myc binding sites between myoblasts and myotubes, 97 peaks are localized in miRNA promoter regions (−5000 bp to +2000 bp from the mature miRNA gene loci) (Supplementary file 5). We selected 14 miRNAs that have been reported to play roles in muscle development for the ChIP-qPCR validation. The ChIP-qPCR results validated our ChIP-seq findings (Fig. 5a), with downregulation of six miRNAs and upregulation of six miRNAs with the c-Myc knockdown (Fig. 5b).
Fig. 5.
miRNAs are associated with c-Myc and are important for c-Myc’s roles in the regulation of myoblast proliferation and differentiation and muscle fibre hypertrophy. a Results of the ChIP-qPCR validation of 14 c-Myc-bound miRNAs with different peak values between GM and DM. NC represents a genomic region without an identified c-Myc-binding peak. b Relative miRNA expression after c-Myc knockdown in primary myoblasts. c CCK-8 assay was performed to assess the effect of the CI-5 miRNAs on myoblast proliferation. SCR, scramble miRNA. d CCK-8 assay was performed to assess the effect of the CR-5 miRNAs on myoblast proliferation. SCR, scramble miRNA. e EdU staining after transfection of CI-5s and CR-5s. Bar, 50 μm. f Proliferation rate of myoblasts transfected with CI-5s and CR-5s. g Relative mRNA expression of the differentiation marker genes after transfection with the CI-5 miRNAs. h Relative mRNA expression of the differentiation marker genes after transfection with the CR-5 miRNAs. i MyHC staining of primary myoblasts 72 h after transfection with CI-5s and CR-5s. j Myotube area (%)72 h after transfection with CI-5s and CR-5s. k CCK-8 assay was performed to assess the effect of c-Myc and the CI-5 inhibitors on myoblast proliferation. l Primary myoblasts were co-transfected with mixtures of the indicated vectors and miRNA inhibitors, and the expression of the muscle differentiation marker genes was then analysed. m CCK-8 assay was performed to assess the effect of c-Myc and the CR-5 mimics on myoblast proliferation. n Primary myoblasts were co-transfected with mixtures of the indicated vectors and miRNA mimics, and the expression of the muscle differentiation marker genes was then analysed. o Chicken breast muscles were injected with CI-5, CR-5 or NC agomirs, and the expression of the muscle differentiation marker genes was then analysed. p H–E staining of breast muscle fibre cross sections from chickens injected with CI-5, CR-5 or NC agomirs. q Fibre diameter of chicken breast muscles injected with CI-5, CR-5 or NC agomirs. r H–E staining of breast muscle fibre cross sections from chickens injected with CI-5 antagomirs + pWPXL-GFP, NC antagomir + pWPXL-GFP and CI-5 antagomirs + pWPXL-c-Myc-Δ269–277. s Fibre diameter of chicken breast muscles injected with CI-5 antagomirs + pWPXL-GFP, NC antagomir + pWPXL-GFP and CI-5 antagomirs + pWPXL-c-Myc-Δ269–277. The results are shown as the mean ± sem of three independent experiments. In k, m, different letters a–d indicate values of each group at 4 d are significantly different at p < 0.05 by Duncan’s multiple range test. In l, n, s, different letters a–d above the bars indicate significant differences (p < 0.05) by Duncan’s multiple range test. In c, d, f–h, j, o, q, ANOVA followed by Dunnett’s test was used. In a–d, independent sample t-test was performed to determine the significant differences between the groups. *p < 0.05; **p < 0.01; ns, no significant difference
Among these 12 miRNAs 5 c-Myc-induced miRNAs (CI-5s) and 5 c-Myc-repressed miRNAs (CR-5s) were selected to test their roles in chicken primary myoblast proliferation and differentiation because they had been reported to play similar roles previously [23, 27–35]. Most CI-5s can promote cell proliferation (Fig. 5c), while CR-5s can repress cell proliferation (Fig. 5d). Interestingly, combinations of CI-5s and CR-5s exert stronger effects on cell proliferation than individual CI-5 and CR-5 miRNAs (Fig. 5c, d). Based on the EdU staining, the CI-5s can induce cell proliferation, while the CR-5s repressed cell proliferation (Fig. 5e, f). To determine the functions of the CI-5s and CR-5s in myoblast differentiation, we transfected the CI-5s and/or CR-5s into myoblasts and induced cell differentiation. The individual CI-5s and CR-5s had a weak effect on myoblast differentiation, but the combinations of CI-5s and CR-5s could significantly affect muscle differentiation marker genes expression (Fig. 5g, f). The CI-5s repressed and CR-5s promoted myoblast differentiation and fusion (Fig. 5g, j).
To determine whether these c-Myc-associated miRNAs (CAMs) are necessary for c-Myc function, we performed rescue assays. The c-Myc-Δ269–277 overexpression could induce cell proliferation but repressed myoblast differentiation. However, the co-transfection of the c-Myc-Δ269–277 vector with the CI-5 inhibitors (mixtures of the individual CI-5 miRNAs) (Fig. 5k, i), and the co-transfection of the c-Myc-Δ269–277 vector with the CR-5s (Fig. 5m, n) weakened the effects of c-Myc on myoblast proliferation and differentiation.
Subsequently, we introduced the CI-5s and CR-5s into skeletal muscle of 1-day-old chicks. After 14 days and 2 introductions, the CI-5s inhibited muscle differentiation marker genes expression, while the CR-5s promoted the expression of these genes (Fig. 5o). The skeletal muscle fibres were induced to hypertrophy after the CI-5s were introduced, but the introduction of the CR-5s had no significant effect (Fig. 5p, q). Additionally, CI-5s inhibition reduced muscle fibre diameter, and the mutant c-Myc introduction rescued the effect of CI-5 antagomirs on muscle fibre hypertrophy (Fig. 5r, s). Altogether, these results indicate that c-Myc performs its functions in skeletal muscle development, at least partially, by regulating miRNA expression.
c-Myc-associated miRNAs are involved in the regulation of the cell cycle, ERK–MAPK and Akt-mediated pathways
To further study the CAM regulatory pathways, we detected their expression levels from myoblasts to myotubes. Except for miR-140-3p, the other individual CI-5s were downregulated during myoblast differentiation (Fig. 6a). However, the CR-5 miRNAs are all upregulated (Fig. 6b). We then used TargetScan to predict the potentiality of CI-5 and CR-5 target genes. The genes that were downregulated from the myoblasts to myotubes and predicted to be potential target genes of the CI-5s were selected as CI-5 target genes. The genes that were upregulated from the myoblasts to myotubes and predicted to be potential target genes of the CR-5s were selected as CR-5 target genes. A KEGG analysis was then performed. The CI-5 and CR-5 target genes were all enriched in many muscle development-related pathways, such as the MAPK signalling, PI3K–Akt and cell cycle (Fig. 6c, d). To better understand the roles played by the CAMs and their target genes in the muscle development-related pathways, we generated an interaction network consisting of the CAMs and their target genes using the KEGG results and Cytoscape (Fig. 6e). The CI-5s and their target genes were mainly enriched in the MAPK pathway, whereas the CR-5s and their target genes were mainly enriched in the cell cycle and PI3K–Akt pathways (Fig. 6c, e).
Fig. 6.
c-Myc-associated miRNAs are involved in the regulation of the cell cycle, ERK–MAPK and Akt-mediated pathways. a Relative miRNA expression of the CI-5s between GM and DM. b Relative miRNA expression of the CR-5s between GM and DM. c KEGG analysis of the CI-5 target genes. d KEGG analysis of the CR-5 target genes. e Gene-miRNA network consisting of 57 proteins (from the CI-5 and CR-5 target genes enriched in the indicated pathways) and their connections (grey lines). miRNA targets are displayed in green and pink. Targets in pink indicate genes upregulated from GM to DM. Targets in green indicate genes downregulated from GM to DM. miRNA names are reported in the yellow and blue boxes. CI-5s are shown in yellow, and CR-5s are shown in blue. The dotted line marks the areas with nodes belonging to canonical pathways. f Relative luciferase activity of 3’ UTR reporter constructs in certain genes shown in figure (e) after transfection with selected miRNA mimics. g Relative mRNA expression after transfection of individual CR-5 miRNAs in chicken primary myoblast. h Primary myoblasts expressing CR-5s, CI-5s or NC were cultured in GM, and the cell cycle phase was analysed after 2 days. i Primary myoblasts were transfected with c-Myc-Δ269–277, CI-5s, CR-5s or NC, and the protein levels of the indicated proteins were analysed after 3 d. j Chicken breast muscles were injected with FR180204 or control, and the protein levels of the ERK1 and p-ERK were analysed. k H–E staining of breast muscle fibre cross sections from chickens injected with FR180204 or control. l Fibre diameter of chicken breast muscles injected with FR180204 or control. The results are shown as the mean ± sem of three independent experiments. In (f (ERK and CCND1), g (CCND1) and h), ANOVA followed by Dunnett’s test was used. In (a,b,h,l), independent sample t-test was performed to determine the significant differences between the groups. *p < 0.05; **p < 0.01; ns, no significant difference
Subsequently, we examined the function of the CI-5s and CR-5s in these pathways. According to a dual-luciferase reporter gene assay, many of our analysed CR-5 target genes can directly interact with CR-5 miRNAs (Fig. 6f), and the introduction of these CR-5 miRNAs can inhibit target gene expression (Fig. 6g). However, the CR-5s can increase the cell populations in the G1 phase and decrease the cell populations in the S phase (Fig. 6h), and CR-5s transfection repressed Cyclin D1 protein expression (Fig. 6i), suggesting that the CR-5s may inhibit cell proliferation by repressing cell cycle-related genes. Interestingly, the CI-5s can also regulate the cell cycle. The CI-5s decreased the cell populations in the G1 phase and increased the cell populations in the S phase (Fig. 6h). Additionally, the myoblasts transfected with c-Myc and CI-5 showed increased ERK1 and phosphorylated ERK (p-ERK) protein levels, but no significant changes were observed in the other MAPK proteins (Fig. 6i). The p-Akt1/2/3 protein level was also showed increased when the cells transfected with c-Myc and CI-5 (Fig. 6i). However, the CR-5 transfection had no significant effects on the expression of ERK1, p-ERK and p-Akt1/2/3 (Fig. 6i). Notably, ERK is important for muscle growth. The p-ERK specific inhibition prevented muscle fibre hypertrophy (Fig. 6j–l). We therefore conclude that the CAMs are involved in the regulation of the cell cycle, ERK–MAPK and Akt-mediated pathways.
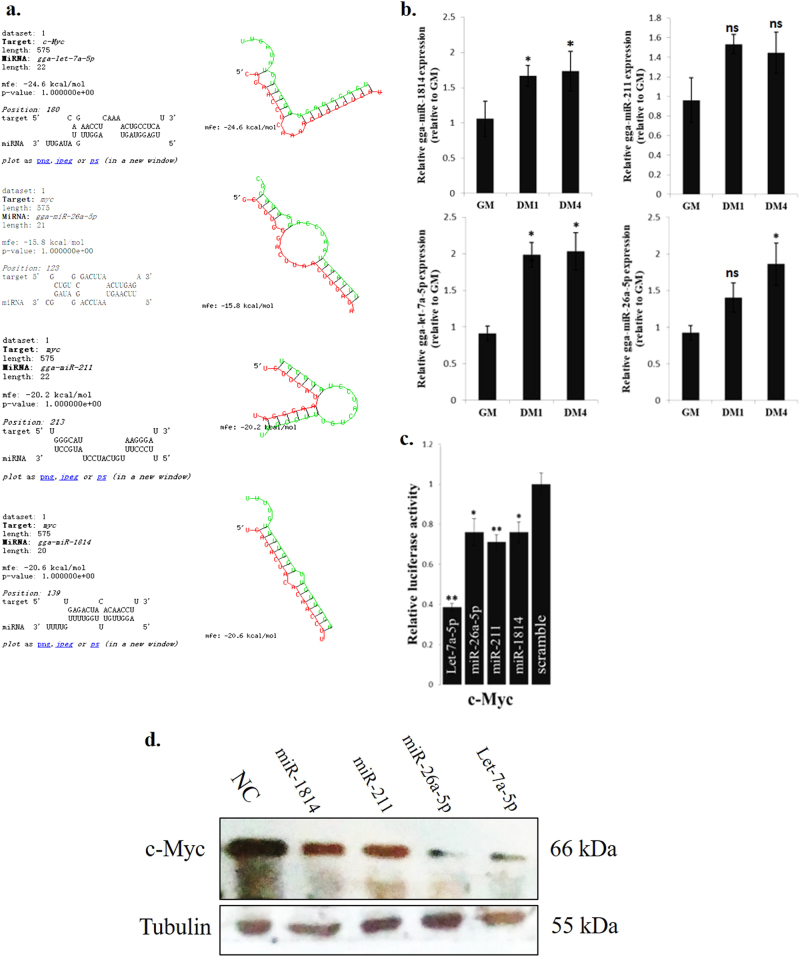
c-Myc-repressed miRNAs are involved in the regulation of c-Myc expression during myoblast differentiation
c-Myc expression is downregulated during myoblast differentiation, but the underlying mechanism remains unknown. Here, we found that 4 c-Myc-repressed miRNAs (CRM-4s) have potential binding sites in the c-Myc mRNA 3' UTR (Fig. 7a). The expression of the CRM-4s was upregulated during myoblast differentiation (Fig. 7b). Notably, these CRM-4s can directly interact with the c-Myc 3' UTR sequence (Fig. 7c). Additionally, the transfection of the CRM-4s reduced the c-Myc protein levels in the myoblasts (Fig. 7d). Therefore, a negative feedback loop likely exists between c-Myc and its associated miRNAs during myoblast differentiation.
Fig. 7.
c-Myc-repressed miRNAs are involved in the regulation of c-Myc expression during myoblast differentiation. a Schematic representation of four miRNAs and c-Myc 3' UTR target region duplexes. b Relative miRNA expression of 4 miRNAs during myoblast differentiation. c Relative luciferase activity of the c-Myc 3' UTR reporter constructs after transfection with selected miRNA mimics. d c-Myc protein expression after transfection with selected miRNA mimics in primary myoblasts. The results are shown as the mean ± sem of three independent experiments. One-way analysis of variance (ANOVA) followed by Dunnett’s test was performed to determine the significant differences between the groups. *p < 0.05; **p < 0.01; ns, no significant difference
c-Myc is associated with lincRNA loci and certain lincRNAs regulate myoblast proliferation and differentiation
Based on our ChIP-seq data, most c-Myc-binding sites are localized in intergenic regions. To determine whether c-Myc is associated with long intergenic noncoding RNA (lincRNA) loci and regulates lincRNA expression, we analysed and identified 1104 lincRNAs with promoters that display c-Myc binding peaks (Supplementary file 6). To determine whether the c-Myc-associated lincRNAs (CALs) are involved in myoblast differentiation, we performed RNAhybrid to predict the CALs that can interact with MyomiRs. In total, 5476 lincRNA-MyomiR target pairs containing 1104 lincRNAs and 23 MyomiRs were predicted (Supplementary file 7). Then, we selected the lincRNAs that can interact with multi-MyomiRs (Supplementary file 8) and used Cytoscape to construct lincRNA-MyomiR interaction networks. Based on the numerous interaction networks, certain lincRNAs mainly interacted with MyomiRs that played similar roles in muscle development (Fig. 8a, b). From Fig. 8a we selected NONGGAT012949.1 (named linc-2949) as candidate for further study, because this lincRNA not only have numerous target MyomiRs, but also have at least two target sites for muscle-specific miRNAs (miR-206 and miR-1) (Fig. 8c). From Fig. 8b we selected NONGGAT001369.2 (named linc-1369) because it has the most target MyomiRs from the lincRNAs that can interact with muscle-specific miRNAs (Fig. 8c). The promoter regions of these two lincRNAs differentially bound c-Myc during myoblast differentiation (Fig. 8d); the c-Myc knockdown led to a downregulation of linc-2949 and an upregulation of linc-1369 (Fig. 8e). During myoblast differentiation, linc-2949 expression was downregulated, while the expression of linc-1369 was upregulated (Fig. 8f). Both lincRNAs were abundantly expressed in the chicken muscles (Fig. 8g).
Fig. 8.
c-Myc is associated with lincRNA loci, and certain lincRNAs regulate myoblast proliferation and differentiation. a, b MyomiR-lincRNA interaction target networks. MyomiRs shown in red play positive roles in myoblast differentiation. MyomiRs shown in green play negative roles in myoblast differentiation. c Two selected lincRNAs and their target MyomiRs. d Results of the ChIP-qPCR validation of c-Myc binding to the linc-2949 and linc-1369 promoters between GM and DM. e Relative lincRNA expression after c-Myc knockdown in primary myoblasts. f Relative expression of linc-2949 and linc-1369 during myoblast differentiation. g Relative linc-2949 and linc-1369 expression in chicken tissues. h Relative luciferase activity of linc-2949 and linc-1369 reporter constructs after transfection with the indicated miRNA mimics. i CCK-8 assay was performed to assess the effect of si-linc-2949 and si-linc-1369 on myoblast proliferation. j Relative mRNA expression of the differentiation marker genes after transfection with si-linc-2949 and si-linc-1369. k Schematic of c-Myc-induced regulatory network in myoblast proliferation and differentiation and muscle fibre hypertrophy. The results are shown as the mean ± sem of three independent experiments. In f, h–j, ANOVA followed by Dunnett’s test was used. In e, independent sample t-test was performed to determine the significant differences between the groups. *p < 0.05; **p < 0.01; ns, no significant difference
Using dual-luciferase reporter gene assays, we validated that three MyomiRs can directly interact with linc-2949 and that two MyomiRs can directly interact with linc-1369 (Fig. 8h). According to a CCK-8 assay, linc-2949 promoted myoblast proliferation, whereas linc-1369 repressed myoblast proliferation (Fig. 8i). The knockdown of linc-2949 in myoblasts repressed muscle differentiation marker genes expression, and the linc-1369 knockdown led to an upregulation of marker genes expression (Fig. 8j). We therefore conclude that c-Myc-associated lincRNAs are involved in the regulation of myoblast proliferation and differentiation.
Discussion
In this study, we identified genome-wide c-Myc-binding sites in skeletal muscle cells by performing ChIP-seq and computational analyses and experimental validation. c-Myc is associated with dozens of genes, miRNAs and lincRNAs, and many c-Myc targets play regulatory roles in myoblast proliferation and differentiation and muscle fibre hypertrophy. Our results suggest that the c-Myc associated targets are critical for c-Myc’s function and that the c-Myc-induced regulatory network plays important roles in myoblast proliferation and differentiation and muscle fibre hypertrophy (Fig. 8k).
c-Myc has been shown to repress myoblast differentiation in C2C12 [10], but the main regulation pathway remains poorly understood. Here, we showed that the cell cycle pathway is a potential target of c-Myc during myoblast differentiation and that the inhibitory role played by c-Myc in myoblast differentiation may be due to its positive effects on the cell cycle. Cell cycle arrest is essential for myoblast differentiation. Myoblasts permanently retract from the cell cycle during terminal differentiation [36]. c-Myc is a oncogene that plays a profound role in the promotion of the cell cycle [1]. The promoting effects of c-Myc on the cell cycle may lead to the prevention of cell cycle arrest, therefore resulting myoblast differentiation inhibition.
In total, 19,354 c-Myc binding peaks were identified in myoblasts, while only 1061 c-Myc binding peaks were identified in myotubes, which is consistent with the gradual decline in the c-Myc protein levels in the nuclei during myoblast differentiation. Detailed analysis of c-Myc binding revealed that E-box is the core motif of c-Myc-bound sequences. This is because, we do not only find a Myc-bound motif among the top 10 de novo scored motifs, but also the motifs of HIF-1b and Tal1, which belong to bHLH proteins and preferred to bind E-box region like c-Myc, were among the top 10 scored motifs (Supplementary Figure S2). Additionally, the known motif enrichment results revealed that motifs containing E-box occupy at least 29.82% of target motifs (Myc- and Usf2-bound motifs, Supplementary Figure S3). These results are consistent with previous c-Myc ChIP-seq results in mouse fibroblasts [26]. Alternatively, many of the transcription factors among the de novo prediction list, such as Mad, Nrf2, Smad2, BRCA1 and RUNX1, have been previously shown to be interacting with proteins of c-Myc [37–40]. These results may reveal some potential interaction partners of c-Myc during myogenesis.
miRNAs are important regulators and mediators of c-Myc function [41]. Based on our results, c-Myc regulates myoblast differentiation, at least, by directly regulating the transcription of many MyomiRs. Interestingly, we found that a combination of CRMs and CIMs have more significant biological effects than the individual miRNAs alone. This was also observed in previous studies [40], suggesting that the joint regulation of different miRNAs has stronger activity than the regulation of individual miRNAs [42, 43]. Additionally, c-Myc could bind and regulate dozens of lincRNAs. Recent studies have identified diverse functions performed by lincRNAs in cells, such as local gene regulation, protein and RNA decoys, encoding micropeptides and enhancer RNAs [44]. Our results do not only elucidate the functionality of linc-1369 and linc-2949 in skeletal muscle cells but also revealed their interaction with c-Myc, demonstrating that lincRNAs are important components of the c-Myc-induced regulatory network that controls myogenic differentiation. More importantly, many lincRNAs are known to be involved in the regulation of muscle development. Certain myogenic lincRNAs can encode micropeptides and function in muscle development and/or regeneration [44–46], while other myogenic lincRNAs can function as competing endogenous RNAs that sponge miRNAs to regulate the miRNA target gene expression, thus controlling muscle differentiation [22]. Our study suggests that linc-1369 and linc-2949 act as MyomiR sponges and regulate myoblast proliferation and differentiation. However, the roles and mechanisms of other CALs still remain unknown. Altogether, our results reveal novel facets of c-Myc regulation in myogenic differentiation: in addition to regulating protein-coding genes, c-Myc controls noncoding RNA expression to regulate skeletal muscle development.
Materials and methods
Cell culture
Chicken primary myoblasts were isolated from chicken leg muscle of day 10 embryo as previous described [29]. The obtained cells were cultured in growth medium (GM) with RPMI-1640 (Gibco, Grand Island, NY, USA), 15% FBS (ExCell, Shanghai, China), 10% chicken embryo extract and 0.2% penicillin/streptomycin at 37 °C in 5% CO2, or induced to differentiation by culturing the cells in DM (RPMI-1640 without FBS containing 2% horse serum) when 90% confluent.
RNA extraction, cDNA synthesis and quantitative real-time PCR
The total RNA was extracted from tissues or cells using RNAiso reagent (Takara, Otsu, Japan). The reverse transcription reaction for mRNA was performed with PrimeScriptTM RT reagent Kit (Perfect Real Time) (Takara) according to manufacturer’s manual. The reverse transcription reaction and quantitation for miRNA were using All-in-OneTM miRNA qRT-PCR Detection Kit (GeneCopoeia). The specific miRNA PCR Primers were designed and provided in Supplementary file 9. qPCR program was carried out in Bio-rad CFX96 Real-Time Detection system (Bio-rad, CA, USA), and the method was as described [29]. All reactions were run in triplicate.
RNA sequencing
The proliferating myoblasts (GM, 50% confluence, cultured in GM) and differentiated myotubes (DM, 100% confluence, cultured in DM for 4 d) were harvested and total RNA was extracted using RNAiso reagent (Takara). Then the RNA samples were sent to Beijing Novogene Bioinformation Technology Co., Ltd. for RNA sequencing. cDNA libraries were constructed according to Illumina’s protocols and then each library was sequenced on a single line of Illumina Hi-seq 2000 (Illumina, San Diego, CA, USA) to obtain paired-end 101-bp reads. All the sequence data have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE109225.
Immunoblotting and immunofluorescence
Western blot was performed as previously described [47]. The following antibodies were used: anti-c-Myc (sc-42, Santa Cruz Biotechnology, CA, USA), anti-p38α (sc-271120, Santa Cruz Biotechnology), anti-p-p38 (sc-166182, Santa Cruz Biotechnology), anti-ERK (sc-376852, Santa Cruz Biotechnology), anti-p-ERK (sc-7383, Santa Cruz Biotechnology), anti-p-JNK (sc-6254, Santa Cruz Biotechnology), anti-Cyclin D1 (BS2436, Bioworld, St Louis Park, MN, USA), anti-Smad2 (ab228807, Abcam, Cambridge, USA), anti-p-Akt1/2/3 (sc-271966, Santa Cruz Biotechnology), anti-Wnt3 (ab50341, Abcam) and anti-Tubulin (BS1482M, Bioworld). The immunofluorescence was performed using anti-c-Myc (Santa Cruz Biotechnology, 1:150) or anti-MyHC (B103, DSHB, Iowa City, IA, USA, 1:100). The cell nuclei were stained for DAPI (Beyotime, Jiangsu, China). The total myotube area was calculated as the percentage of the total image area covered by myotubes, and the measurement was performed using ImageJ sofware (National Institutes of Health) on cells labelled with anti-MyHC.
The 5'- and 3'-RACE
The 5'- and 3'-rapid amplification of cDNA ends (RACE) were performed as previously described [28]. Both 5'- and 3'-RACE PCR products were cloned and sequenced. All the primers used in RACE were summarized in Supplementary file 9.
Plasmid construction
pmirGLO Dual-Luciferase Reporters: The partial 3'UTR fragment of c-Myc, PIK3R2, ERK, TGFβ2, TGFβR1, CCNE1, CDK6, p300 and CCD1 containing the indicated miRNA-binding sites were amplified by PCR from chicken embryonic skeletal muscle cDNA and then cloned into pmirGLO vector. To construct lincRNA reporter plasmids, 672-bp and 452-bp lincRNA gene fragments encompassing the predicted miRNAs binding sites were cloned into pmirGLO vector between XhoI and SalI sites.
c-Myc overexpression lentivirus vector: c-Myc coding sequence (NCBI Reference Sequence: KU981087) was amplified from chicken embryonic leg muscle cDNA by PCR. The PCR product was cloned into the pWPXL vector (Addgene, Cambridge, MA, USA) between BamHI and EcoRI sites. The successful c-Myc overexpression lentivirus vector was confirmed by DNA sequencing. A mutant c-Myc overexpression lentivirus plasmid (mutant the 269–277 region which is the putative c-Myc calpain cleavage region in chicken, Supplementary Figure S9) was generated by changing the cleavage site from TGGTCCTCAAGCG to CGCAGGCCCACAT.
Dual-luciferase reporter assay
Target genes 3' UTR dual-luciferase reporter (partial 3'UTR containing the indicated miRNA-binding sites, 200 ng) and miRNA mimic or NC duplexes (50 nM) were co-transfected into primary myoblasts using the Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) in 48-well plates. After 48 h transfection, cells were washed by PBS twice and the activities of Firefly and Renilla luciferase were measured according to the manual of Dual-luciferase reporter assay system (Promega, Madison, WI, USA). All the data were acquired by averaging the results from three independent repeats.
Transfection and RNA oligonucleotides
Transfection was carried out using Lipofectamine 3000 reagent (Invitrogen). Cells were transfected with 50 nM miRNA mimics (RiboBio, Guangzhou, China), 100 nM miRNA inhibitors (RiboBio) or 100 nM c-Myc specific siRNA (GenePharma, Suzhou, China). Chicken breast muscle was treated with 20 nM agomir or 40 nM antagomir each miRNA (RiboBio). Lipofectamine 3000 and nucleic acids were diluted in OPTI-MEM I Reduced Serum Medium (Gibco, Grand Island, NY, USA). The procedure of transfection was performed according to the manufacturer’s direction.
Assays for cell number and cell cycle
For the CCK-8 assay, primary myoblast were cultured in 96-well plates. 10 μL of Cell counting kit-8 reagent (Dojindo Laboratories, Kumamoto, Japan) was added into each well for 1 h incubation at the time point of 24 h, 48 h, 72 h and 96 h after transfection. The data of absorbance at 450 nm were read by an iMark™ Microplate Absorbance Reader (Bio-Rad). All the data were acquired by averaging the results from six independent repeats. For cell cycle analysis, primary myoblast cultured in 12-well plates were fixed in 75% ethanol overnight at −20 °C after 48 h transfection. With the Cell Cycle Analysis Kit (Thermo Fisher Scientific, USA), the cells were analysed by a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA).
Lentivirus production and transduction
A mixture of pWPXL overexpression vector (pWPXL-c-Myc-Δ269–277, pWPXL-c-Myc, or pWPXL-GFP), psPAX2 and pMD2.G were transfected into HEK293T cells using Lipofectamin 3000 reagent to generate lentivirus. The supernatants were collected 72 h later and filtered through 0.45 μm PVDF membranes (Millipore, CA, USA) and cleared by supercentrifugation. The viral titer was evaluated by a gradient dilution. Primary myoblasts were infected with the recombinant lentivirus-transducing units plus 4 mg/mL Polybrene (Sigma). Chicks (1-day-old) were infected with lentiviruses (1 × 106 infection unit per chick) by direct injection into the breast muscle.
Histology
Chicken breast muscle tissues were obtained from c-Myc-Δ269–277 infected muscle (n = 3), c-Myc-infected muscle (n = 3), GFP-infected muscle (n = 3), CI-5 agomirs-injected muscle (n = 3), CR-5 agomirs-injected muscle (n = 3), NC agomir-injected muscle (n = 3), FR180204 (MedChemExpress, NJ, USA, dissolved in 2% DMSO, 1-day-old chicks were treat with FR180204 (50 μM) by direct injection into the breast muscle)-injected muscle (n = 3), or 2% DMSO-injected muscle (n = 3), harvested and fixed in 10% formalin in PBS. Fixed tissues were paraffin embedded, sectioned and stained with hematoxylin and eosin (H&E).
ChIP assays and ChIP-seq
ChIP assays were performed using a standard protocol. Briefly, for proliferating myoblasts (GM, 50% confluence, cultured in GM) and differentiated myotubes (DM, 100% confluence, cultured in DM for four days, Supplementary Figure S10), after crosslinking of proteins to DNA with 1% formaldehyde followed by cell lysis and sonication to generate chromatin fragments with an average size of 200–400 bp, immunoprecipitation was performed with 5 μg of the anti-c-Myc (ab56, Abcam, ChIP-grade) or the chicken anti-IgG (bs-0310P, Bioss, Beijing, China) antibody was bound to Protein A/G-Sepharose beads. After extensive washing and reversal of crosslinking, proteinase K and RNase A digestion, chromatin fragments were purified by phenol chloroform extraction, and ethanol precipitation was performed. For regular ChIP assays, the purified DNA was amplified by qPCR. The primer sequences for ChIP-qPCR analysis are shown in Supplementary file 9. For ChIP-seq, the purified DNA was sent to GENEWIZ sequencing facility for library construction, evaluation and sequencing on an Illumina Hi-seq 2000 sequencer (Illumina). Technical replicates were prepared by sequencing the same library twice. All the sequence data have been deposited in NCBI’s GEO database and are accessible through GEO series accession number GSE109223.
Peak defining and motif discovery
The sequenced reads were mapped to the chicken reference genome (Gallus gallus-4.0) using bowtie2 v2.1.0 [48]. The alignments were performed allowing the maximum of two mismatches and keeping only the uniquely aligned reads. The c-Myc DNA-binding peaks were identified using HOMER v4.6 with IgG control sample as a background [49]. Peaks were defined at a 0.1% estimated false discovery rate and with p-value <1e−4 [49]. c-Myc binding site analysis was also conducted using HOMER v4.6 with default parameters for motif identification.
Identification of c-Myc-bound lincRNAs
We downloaded chicken lncRNAs loci data from NONCODE (http://www.noncode.org/download.php) and compared to the intergenic binding peaks of c-Myc in myoblasts (Supplementary file 1). c-Myc binding peaks localized on the potential promoter of lincRNAs (−3 kb and +1 kb from the TSS) were selected as c-Myc associated lincRNAs (CALs). A total of 1104 lincRNAs were selected as CALs (Supplementary file 6).
Bioinformatics analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the enriched genes were performed using the web-based DAVID bioinformatics resources 6.8 (the Database for Annotation, Visualization, and Integrated Discovery, https://david.ncifcrf.gov/home.jsp). All GO terms and KEGG pathways with P < 0.05 that contained at least five genes were selected for subsequent analysis.
Statistical analysis
Each experiment was repeated three times, and all results are represented as mean ± sem. For Fig. 5k, n, s, Duncan’s Multiple Range Test was used to compare differences among mean values at 5% level of significance. For multiple groups comparison (Figs. 1c, d, 2a, b and 5c, d, f–h, j, o, q, Fig. 6f (ERK and CCND1), 6g (CCND1), 6h, 7b, c, 8f and 8h–j), one-way analysis of variance (ANOVA) was used, followed by Dunnett’s test for comparing experiment groups against a single control. For single comparison between two groups (Figs. 2c, d, f–j, l; 4a, b, 5a, b, 6a, b, f, g, l and 8e), independent sample t-test was used. We considered p < 0.05 to be statistically significant. ∗p < 0.05; ∗∗p < 0.01; ns, no significant difference. All statistical analyses were performed using SPSS 18.0 for Windows (SPSS, Inc., Chicago, IL, USA).
Ethics standards
All experimental protocols were approved by the South China Agricultural University Institutional Animal Care and Use Committee (approval ID: SCAU#0018). And the methods were carried out in accordance with the regulations and guidelines established by this committee.
Electronic supplementary material
Acknowledgements
This work was supported by Natural Scientific Foundation of China (31702105), Natural Scientific Foundation of China (31472090), the China Agriculture Research System (CARS-41-G03), and the Science and Technology Program of Guangzhou, China (201804020088).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by H. Ichijo
Contributor Information
Qinghua Nie, Email: Qinghua.Nie@nottingham.ac.uk, Email: nqinghua@scau.edu.cn.
Xiquan Zhang, Email: xqzhang@scau.edu.cn.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0129-0) contains supplementary material, which is available to authorized users.
References
- 1.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–82. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 3.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–33. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Roger I, Kim SH, Griffiths B, Sewing A, Land H. Cyclins D1 and D2 mediate myc-induced proliferation via sequestration ofp27(Kip1) and p21(Cip1) EMBO J. 1999;18:5310–20. doi: 10.1093/emboj/18.19.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan J, Wang K, Xi M. MiR-494 inhibits epithelial ovarian cancer growth by targeting c-Myc. Med Sci Monit. 2016;22:617–24. doi: 10.12659/MSM.897288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–63. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–70. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–93. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crescenzi M, Crouch DH, Tato F. Transformation by myc prevents fusion but not biochemical differentiation of C2C12 myoblasts: mechanisms of phenotypic correction in mixed culture with normal cells. J Cell Biol. 1994;125:1137–45. doi: 10.1083/jcb.125.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bui TV, Mendell JT. Myc: maestro of MicroRNAs. Genes Cancer. 2010;1:568–75. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng K, Guo X, Wang H, Xia J. The lncRNA-MYC regulatory network in cancer. Tumour Biol. 2014;35:9497–503. doi: 10.1007/s13277-014-2511-y. [DOI] [PubMed] [Google Scholar]
- 12.He X, Tan X, Wang X, Jin H, Liu L, Ma L, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–8. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 13.Luo W, Nie Q, Zhang X. MicroRNAs involved in skeletal muscle differentiation. J Genet Genom. 2013;40:107–16. doi: 10.1016/j.jgg.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Neguembor MV, Jothi M, Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet Muscle. 2014;4:8. doi: 10.1186/2044-5040-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–14. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XH, Hu Z, Klein JD, Zhang L, Fang F, Mitch WE. Decreased miR-29 suppresses myogenesis in CKD. J Am Soc Nephrol. 2011;22:2068–76. doi: 10.1681/ASN.2010121278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–84. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Lu CL, Cui LP, Hu YL, Yu Q, Jiang Y, et al. MicroRNA-146a modulates TGF-beta1-induced phenotypic differentiation in human dermal fibroblasts by targeting SMAD4. Arch Dermatol Res. 2012;304:195–202. doi: 10.1007/s00403-011-1178-0. [DOI] [PubMed] [Google Scholar]
- 20.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA. 2009;106:3384–9. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qadir AS, Woo KM, Ryoo HM, Yi T, Song SU, Baek JH. MiR-124 inhibits myogenic differentiation of mesenchymal stem cells via targeting Dlx5. J Cell Biochem. 2014;115:1572–81. doi: 10.1002/jcb.24821. [DOI] [PubMed] [Google Scholar]
- 22.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander MS, Kawahara G, Motohashi N, Casar JC, Eisenberg I, Myers JA, et al. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ. 2013;20:1194–208. doi: 10.1038/cdd.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–7. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 26.Perna D, Faga G, Verrecchia A, Gorski MM, Barozzi I, Narang V, et al. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene. 2012;31:1695–709. doi: 10.1038/onc.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–43. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 28.Luo W, Li E, Nie Q, Zhang X. Myomaker, regulated by MYOD, MYOG and miR-140-3p, promotes chicken myoblast fusion. Int J Mol Sci. 2015;16:26186–201. doi: 10.3390/ijms161125946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo W, Wu H, Ye Y, Li Z, Hao S, Kong L, et al. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014;5:e1347. doi: 10.1038/cddis.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–5. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, Zhan S, Wang Y, Wang L, Zhong T, Li L, et al. Role of microRNA-101a in the regulation of goat skeletal muscle satellite cell proliferation and differentiation. Gene. 2015;572:198–204. doi: 10.1016/j.gene.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Wei W, He HB, Zhang WY, Zhang HX, Bai JB, Liu HZ, et al. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668. doi: 10.1038/cddis.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guess MG, Barthel KK, Harrison BC, Leinwand LA. miR-30 family microRNAs regulate myogenic differentiation and provide negative feedback on the microRNA pathway. PLoS ONE. 2015;10:e118229. doi: 10.1371/journal.pone.0118229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S, Luo W, Ye Y, Bekele EJ, Nie Q, Li Y, et al. Let-7b Regulates Myoblast Proliferation by Inhibiting IGF2BP3 Expression in Dwarf and Normal Chicken. Front Physiol. 2017;8:477. doi: 10.3389/fphys.2017.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H, Wen G, Huang Y, Yu X, Chen Q, Afzal TA, et al. MicroRNA-22 regulates smooth muscle cell differentiation from stem cells by targeting methyl CpG-binding protein 2. Arterioscler Thromb Vasc Biol. 2015;35:918–29. doi: 10.1161/ATVBAHA.114.305212. [DOI] [PubMed] [Google Scholar]
- 36.Derer W, Easwaran HP, Leonhardt H, Cardoso MC. A novel approach to induce cell cycle reentry in terminally differentiated muscle cells. FASEB J. 2002;16:132–3. doi: 10.1096/fj.01-0500fje. [DOI] [PubMed] [Google Scholar]
- 37.Luscher B. MAD1 and its life as a MYC antagonist: an update. Eur J Cell Biol. 2012;91:506–14. doi: 10.1016/j.ejcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Levy S, Forman HJ. C-Myc is a Nrf2-interacting protein that negatively regulates phase II genes through their electrophile responsive elements. IUBMB Life. 2010;62:237–46. doi: 10.1002/iub.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng XH, Liang YY, Liang M, Zhai W, Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitorp15(Ink4B) Mol Cell. 2002;9:133–43. doi: 10.1016/S1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- 40.Kennedy RD, Gorski JJ, Quinn JE, Stewart GE, James CR, Moore S, et al. BRCA1 and c-Myc associate to transcriptionally repress psoriasin, a DNA damage-inducible gene. Cancer Res. 2005;65:10265–72. doi: 10.1158/0008-5472.CAN-05-1841. [DOI] [PubMed] [Google Scholar]
- 41.Jackstadt R, Hermeking H. MicroRNAs as regulators and mediators of c-MYC function. Biochim Biophys Acta. 2015;1849:544–53. doi: 10.1016/j.bbagrm.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 42.Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, et al. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal. 2009;2:a69. doi: 10.1126/scisignal.2000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marzi MJ, Puggioni EM, Dall’Olio V, Bucci G, Bernard L, Bianchi F, et al. Differentiation-associated microRNAs antagonize the Rb-E2F pathway to restrict proliferation. J Cell Biol. 2012;199:77–95. doi: 10.1083/jcb.201206033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2017;19:143–57. [DOI] [PMC free article] [PubMed]
- 45.Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–32. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 46.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G, Luo W, Abdalla BA, Ouyang H, Yu J, Hu F, et al. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1. Cell Death Dis. 2017;8:e3094. doi: 10.1038/cddis.2017.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.