Abstract
Protein misfolding in the endoplasmic reticulum (ER) is accompanied by adaptive cellular responses to promote cell survival. We now show that activation of mitochondrial respiration is a critical component of an adaptive ER stress response, requiring the unfolded protein response (UPR) sensor Ire1, and also calcium signaling via calcineurin. In yeast and mammalian cells lacking Ire1 or calcineurin, respiratory activation is impaired in response to ER stress; accumulation of mitochondrial reactive oxygen species (ROS) triggers cell death as abrogation of ROS by antioxidants or loss of the electron transport chain (in yeast) can rescue cells from death. Significantly, cells are rescued from ER stress-induced death by mitochondrial uncoupling by CCCP to increase O2 consumption (and increase the efficiency of electron transfer). Remarkably, genetic and pharmacologic strategies to promote mitochondrial biogenesis and increase O2 consumption also alleviate ER stress-mediated ROS and death in yeast and mammalian cells. Moreover, in a yeast genetic screen, three mitochondrial proteins Mrx9, Mrm1, and Aim19 that increase mitochondrial biogenesis were identified as high copy suppressors of ER stress-mediated cell death. Our results show that enhanced mitochondrial biogenesis, linked to improved efficiency of the electron transport chain, is a powerful strategy to block ROS accumulation and promote cell survival during ER stress in eukaryotic cells.
Subject terms: Protein folding, Endocrine system and metabolic diseases, Genetic interaction
Introduction
Endoplasmic reticulum (ER) stress has been defined as a cellular condition that occurs when demand to properly fold and modify newly synthesized proteins of the secretory pathway exceeds the capacity of the ER to handle the load [1]. In addition to translational control, the unfolded protein response (UPR) is a major transcriptional mechanism to alleviate ER stress by affecting numerous activities, including increased chaperone production to improve protein folding, and enhanced ER-associated degradation to destroy misfolded proteins. The most conserved branch of the UPR is initiated upon activation of the ER sensor Ire1 that leads to activation of the Hac1 (yeast)/XBP1 (mammalian) transcription factor [2]. Two additional signaling branches of the UPR present in mammalian cells are mediated by transmembrane stress sensors: ATF6, a transcription factor, and PERK, that inhibits general protein translation by phosphorylating eukaryotic translation initiation factor 2α (eIF2α).
The ER is a major Ca2+ reservoir that participates in maintaining cellular Ca2+ homeostasis. During ER stress, Ca2+ is released from the ER and enters mitochondria. Ca2+ movement to mitochondria is suggested to regulate oxidative phosphorylation and initially represents an adaptive response to acute ER stress [3-5]. In mammalian cells, PERK can become activated in response to disturbed ER Ca2+ homeostasis [6, 7]. Evidence suggests that PERK’s effects are enhanced by its interaction with the conserved Ca2+ activated phosphatase, calcineurin [6, 7], which is inhibited by the immunosuppressive drug FK506. Effectors of calcineurin in mammalian cells lead to a wide array of biological effects, including stress signaling [8]. Calcineurin is also involved in stress signaling in yeast; although yeast do not have a PERK-mediated pathway, it is clear that calcineurin plays a critical role in cell survival in response to ER stress [9]. During the yeast ER stress response, calcium dynamics and calcineurin activation are independent of the calcineurin-dependent Crz1 transcription factor [9].
Chronic and unresolved ER stress, excessive movement of Ca2+ into mitochondria, and accumulation of reactive oxygen species (ROS) have all been linked to the initiation of cell death. ER stress and cell death contribute to the development and pathogenesis of many diseases, including metabolic diseases in which cell response to nutrient fluctuation is impaired [10]. Although mitochondria provide necessary ATP for increased ER chaperone activity, and assistance in maintaining ER and cytoplasmic Ca2+ homeostasis, mitochondria are also regulators of programmed cell death upon unresolved stress. A potential role of mitochondria in facilitating an adaptive response to ER stress has not been systematically explored.
In this paper, we examine how ER stress leads to cell death via cellular accumulation of mitochondrial ROS. Using genetic approaches, we have identified pathways linked to increased mitochondrial biogenesis that abrogate ROS production by increasing respiration, and remarkably, rescue cells from ER stress-mediated death. By genetic screening for high copy suppressors of ER stress-induced yeast cell death, we uncovered novel genes encoding mitochondrial proteins that increase respiratory activity. We find that in both yeast and mammalian cells, Ire1 and calcineurin each contribute to the mitochondrial response to ER stress: the two signaling proteins are required for an adaptive calcium response as well as regulating the activity of the ETC component cytochrome c oxidase (COX) that contributes to preventing ROS production and cell death.
Materials and methods
Yeast strains, media and cell culture
Standard yeast media was as described [11]. Cells were grown in synthetic complete (SC) or YPD medium. Yeast transformations were by the lithium acetate method [12]. Yeast cells were in the BY4742/BY4741 background. Deletion strains were confirmed by PCR. Double mutants were constructed by tetrad dissection.
PCCL3 cells were cultured in DMEM/F12 supplemented with 5% FBS plus penicillin/streptomycin and a four-hormone mixture containing 1 µg/mL insulin, 1 nM hydrocortisone, 5 µg/mL apo-transferrin, and 1 mU/mL TSH.
Plasmids
SAK1 on a 2 micron plasmid was from Martin Schmidt (University of Pittsburgh). A LEU2-marked plasmid for overexpressing HAP4 was cut with Pac1 for integration at the ADH1 promoter, and was from Su-Ju Lin (University of California, Davis) [13]. pMET17-CTA1 is a URA3-marked centromeric plasmid from Paula Ludovico (University of Minho, Portugal).
Cell viability assay, cytotoxicity assay, siRNA knockdown
To measure cell viability after 5 h treatment with 0.5 µg/mL tunicamycin, cells were diluted to 0.1 OD600/mL and then plated on YPD plates for 2 days at 30 °C. Colonies were counted and expressed as a percent of colony numbers from untreated cells; controls for all strains ranged from 79 to 148 colonies, expressed as 100% viability. In mammalian cells, cytotoxicity was measured as leakage of lactate dehydrogenase into the media (ThermoFisher Pierce). For Ire1, PERK, and XBP1 knockdowns in PCCL3 cells, RNAiMAX transfection reagent (ThermoFisher) was used. siRNA sequences available upon request. Knockdowns were confirmed by Western blot with anti-Ire1, PERK, and XBP1 antibodies (Cell Signaling Technology).
Western blot, enzyme assay, and cell staining with fluorescent dyes
Cell lysate was prepared by vortexing with glass beads, including protease inhibitors, as described previously [14]. Protein content was determined by Bradford assay [15]. Monoclonal antibodies against yeast Cox2, ATP synthase α subunit, and porin, and anti-rat XBP1 were from Abcam, Inc. (UK). Anti-eIF2α, anti-phospho-eIF2α, and anti-PERK antibodies were from Cell Signaling Technology.
For detecting ROS in yeast, mid-log cells were washed with phosphate-buffered saline (PBS) and incubated with 5 µg/mL dihydroethidium (DHE) for 15 min; cells were washed once and resuspended in PBS before visualizing with an Olympus fluorescent microscope, and images were collected with a Hamamatsu CCD camera. For quantitation of DHE staining, 50 cells were counted per experimental condition; n = 3. For measuring mitochondrial membrane potential (MMP) in non-quenching mode, live cells were stained with TMRM (5 nM) for 30 min, and then visualized by fluorescence microscopy.
ROS in PCCL3 cells was measured by plate reader assay with dihydrorhodamine 123. Cells were cultured in 96-well plates for 2 days+/− various treatments. Cells were then washed with Krebs-Ringer modified buffer (135 mM NaCl, 5 mM KCl, 1 mM MgSO4, 0.4 mM K2HPO4, 5.5 mM glucose, 20 mM HEPES, 1 mM CaCl2, pH 7.4), and 100 µL KRB with 5 µM DHR123 was added to each well. Plates were incubated at 37 °C for 15 min, washed twice with buffer, and fluorescence was analyzed on a Tecan M200 Infinite microplate reader with excitation at 500 nm and emission at 536 nm.
Genetic screen
To isolate suppressors of cnb1∆ cells’ tunicamycin sensitive growth, cells were transformed with a URA3-marked 2 µ yeast library. ~20,000 transformants were plated on SC-URA plates with 0.5 µg/mL tunicamycin. Candidate suppressors that could grow on tunicamycin were picked and screened for growth on 1.5 mM DTT. From these, plasmids were rescued and tested for ability to confer resistance to tunicamycin and DTT. Insert boundaries were sequenced using T3 and T7 primers. Three suppressing plasmids were isolated with inserts including genes encoding mitochondrial proteins. These plasmids were then modified by restriction digest so that AIM19, MRX9, and MRM1 are the sole intact genes in the 2 µ vector backbone.
Mitochondrial isolation, cytochrome c oxidase activity and O2 consumption
Mitochondria were isolated as described [16] except the homogenization step was omitted; instead, spheroplasts were vortexed for 30 s in water, and then diluted with 1 volume of homogenization buffer. For oxygen measurements, mitochondria were resuspended in an isolation buffer consisting of 200 mM mannitol, 50 mM sucrose, 5 mM K2HPO4, 5 mM MOPS, 1 mM EGTA, 0.1% w/w BSA, pH 7.15.
Cytochrome c oxidase activity was assayed by following oxidation of cytochrome c spectrophotometrically using isolated mitochondria permeablized with 1.5% dodecylmaltoside, as described [17]. COX activity is expressed as a rate of cytochrome c oxidation/µg mitochondrial protein relative to that seen in wild-type cells.
Mitochondrial oxygen consumption was measured by placing isolated mitochondria in a respiration buffer (107.5 mM KCl, 5 mM KH2PO4, 50 mM MOPS, 1 mM EGTA, 0.1% w/w BSA essentially fatty acid free, pH 7.2 using KOH) into a high resolution Orobos Oxygraph 2 K at 25 °C and normalized based on total protein (100 μg). O2 flux is determined by measuring the fall in O2 concentration in the sealed oxygraph. For whole-cell oxygen consumption, exponentially growing cells were pelleted and resuspended in the specified medium at 20 OD600/mL. Cells were then added into the oxygraph chamber containing the same medium at a concentration of 2 OD600/mL and O2 flux was followed. When indicated, TET (10 mM) and CCCP (10 µM) were added.
Results
Ire1 and calcineurin protect against ER stress in yeast
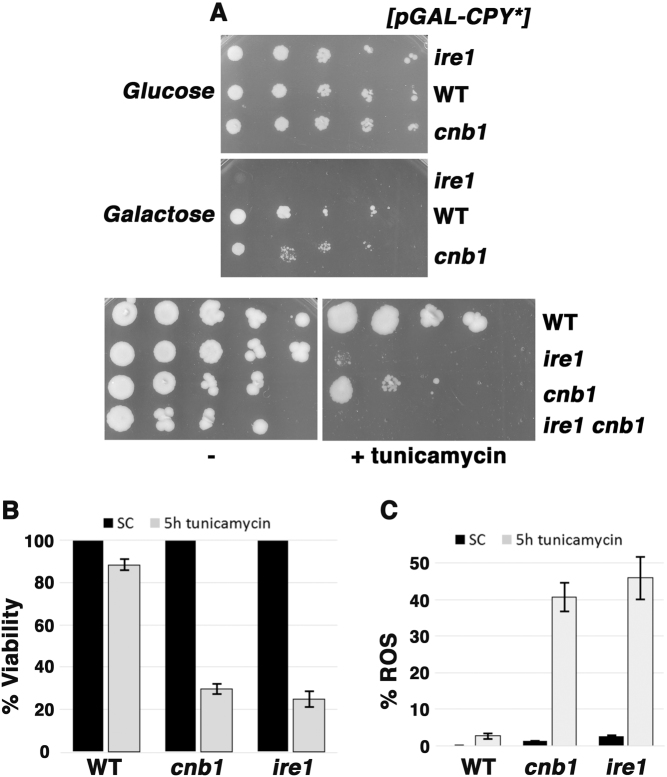
Ire1 (a sensor of ER stress and one of the initiators of the UPR) and calcineurin (a Ca2+ dependent signaling phosphatase) are both required for resistance to ER stress [9, 18]. Yeast cells without Ire1 or without calcineurin activity (as in cnb1∆ cells lacking the regulatory subunit of calcineurin) have impaired growth in the presence of the misfolded ER-retained luminal protein, CPY*, or in the presence of tunicamycin, an inhibitor of N-linked glycosylation (Fig. 1a). Indeed, both Ire1 and calcineurin participate in protecting against ER stress as double ire1∆ cnb1∆ cells exhibited synthetic growth sensitivity to tunicamycin (Fig. 1a), and lost viability within 5 h of drug treatment (as assessed by colony forming assay, Fig. 1b).
Fig. 1.
Cells without Ire1 or calcineurin have increased sensitivity to ER stress. a ER stress impairs cell growth: cnb1∆ and ire1∆ cells have impaired growth on tunicamycin, DTT, or the misfolded protein CPY*. Serial dilutions of cells were spotted on plates. b Cell death is induced by ER stress. cnb1∆ and ire1∆ cells have increased susceptibility to ER stress-induced death, as determined by colony plating assay after treating mid-log cultures with 0.5 µg/mL tunicamycin for 5 h. Viable cells expressed as a percentage of untreated cells +/− standard error of the mean (SEM); n = 3. c ER stress-induced death is associated with ROS accumulation. Wild-type, cnb1∆ and ire1∆ cells treated with tunicamycin (0.5 µg/mL) for 5 h were stained with DHE. Fluorescence was imaged at identical exposure times for all samples, and quantitated by counting cell numbers displaying DHE staining throughout the entire cell out of 50 cells using ImageJ software; n = 3.
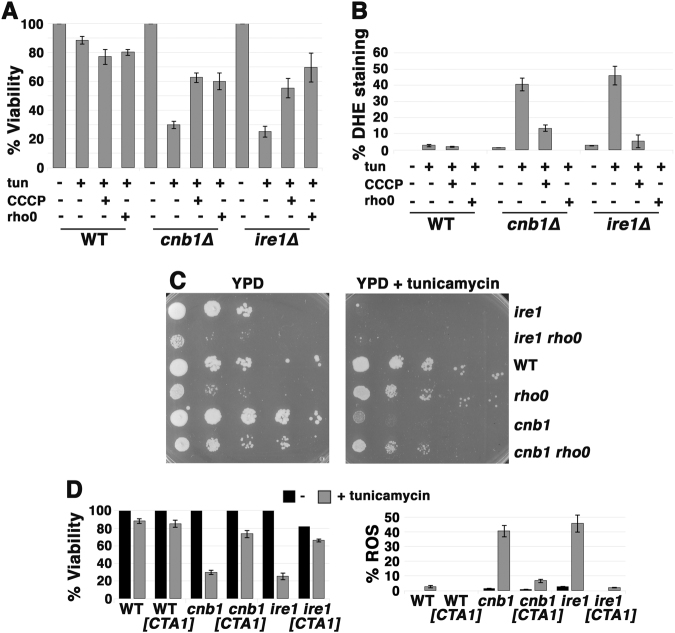
Inefficient mitochondrial electron transport is a major source of cellular ROS, and mitochondrial ROS accumulation has been implicated in cell death [19]. To determine whether ER stress-induced loss of viability is associated with ROS accumulation, cells were treated with tunicamycin for 5 h, plated for viability count, and also stained with dihydroethidium (DHE) to quantitate the fraction of cells with increased ROS production (Fig. 1b,c). DHE fluorescence was intense, covering the whole cell in ire1∆ and cnb1∆ cells sensitive to ER stress (Suppl. Figure 1). The results suggest ROS and cell death are coordinately induced by ER stress when ER stress response is impaired in ire1∆ or cnb1∆ cells. Strikingly, ROS and death are induced by ER stress in yeast exponentially growing in synthetic complete medium with glucose, utilizing predominantly fermentative metabolism, which represses respiratory capacity [20]. To gain evidence that ROS accumulation promotes death, cells were treated with ethidium bromide to damage mitochondrial DNA, generating rho0 cells that lack a functional electron transport chain (ETC) [21]. As shown in Fig. 2, rho0 suppressed ER stress-induced ROS (Fig. 2b) and death (Fig. 2a) in ire1∆ and cnb1∆ cells, consistent with the idea that ROS produced by the ETC leads to death. Moreover, ROS accumulation and death were abrogated by CCCP, a protonophore that dissipates the electrochemical proton gradient across the inner mitochondrial membrane (Fig. 2a,b); despite inhibiting ATP production, CCCP increases electron flow through the ETC and increases O2 consumption, thereby reducing ROS accumulation [22] (discussed further below). Although cell growth is impaired by loss of a functional ETC, in the presence of tunicamycin cnb1 rho0 cells showed improved growth compared to single cnb1∆ mutants (Fig. 2c). ER stress-induced death was also suppressed by overexpression of mitochondrial catalase activity, encoded by CTA1 (Fig. 2d, left panel) to detoxify mitochondrial ROS (Fig. 2d, right panel).
Fig. 2.
ER stress-induced death is caused by mitochondrial ROS. a ER stress-induced death is rescued in rho0 cells and by addition of the proton ionophore CCCP. Exponentially growing cells were treated with 0.5 µg/ml tunicamycin with or without CCCP (10 µM) for 5 h. Cell viability was then determined as in Fig. 1b legend, expressed as a percentage of untreated cells +/− SEM, n = 3. b ROS accumulation is rescued in rho0 cells and by addition of the protonophore CCCP. ROS was assayed by staining with DHE as in Fig. 1c legend; n = 3+/− SEM. Fluorescence images from cells +/− tunicamycin were imaged for the same exposure time and adjusted using identical Photoshop settings. c Impaired growth of cnb1 on tunicamycin is rescued in cnb1 rho0 cells. Serially diluted cells were spotted onto plates with and without tunicamycin (0.25 µg/mL). d ER stress-induced death (left panel) and ROS (right panel) rescued by overexpression of mitochondrial catalase. Cells transformed with pMET-CTA1 were induced to express catalase after washing with water and resuspending in methionine-free SC medium. Tunicamycin (0.5 µg/mL) was then added for 5 h before measuring cell viability and ROS as in Fig. 1b, c legends +/− SEM, n = 3.
Increased oxygen consumption is an adaptive response to ER stress
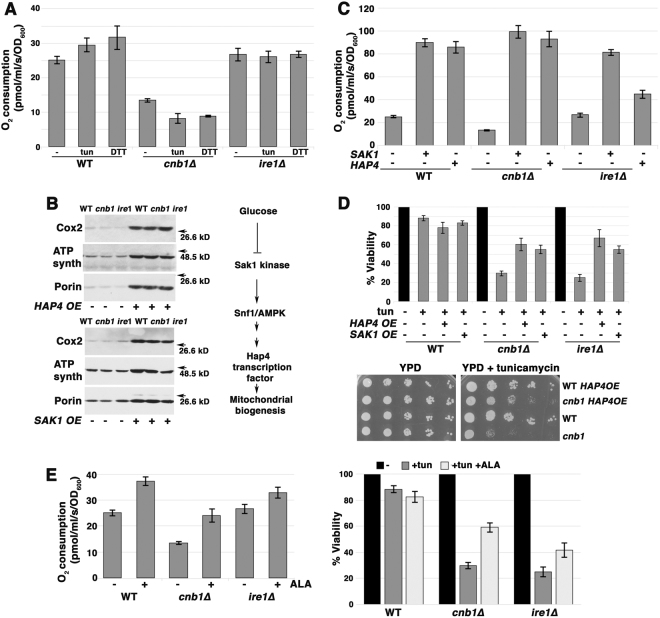
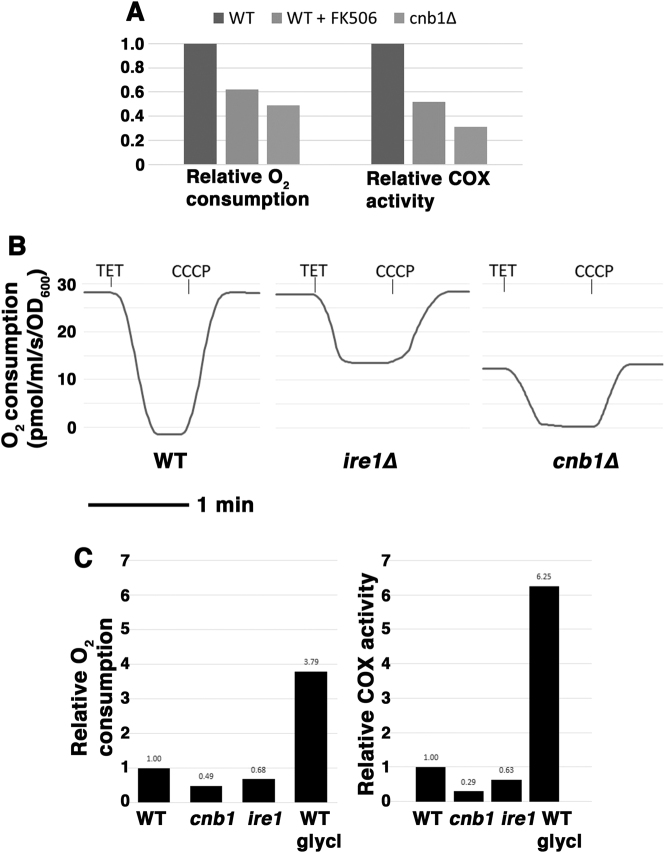
Mitochondrial respiratory response to various stresses have been described previously [23, 24]. To test whether there is also a mitochondrial respiratory response to ER stress, cellular O2 consumption was assayed in yeast cells challenged with tunicamycin. A small but significant increase in O2 consumption was observed in wild-type cells induced by either tunicamycin or DTT addition (Fig. 3a, p < 0.05). By contrast, ire1∆ and cnb1∆ cells were unable to increase O2 consumption in response to ER stress (Fig. 3a). Because basal O2 consumption was already depressed in cnb1∆ cells by comparison with that in wild-type cells (Fig. 3a), levels of ETC components were analyzed by Western blot. Fig. 3b shows that Cox2, a subunit of cytochrome c oxidase (COX, responsible for the majority of cellular O2 consumption) was not significantly decreased in cnb1∆ cells by comparison with that in wild-type cells, suggesting that COX protein level does not account for diminished O2 consumption.
Fig. 3.
ER stress induces mitochondrial response. a O2 consumption increases in response to ER stress in wild-type but not cnb1∆ or ire1∆ cells. Cells were treated with 0.5 µg/mL tunicamycin or 1 mM DTT for 2 h before measuring O2 consumption expressed as pmol/ml/s/OD600+/− SEM, n = 3. b ETC protein levels are increased by overexpression of HAP4 (top left panel) and SAK1 overexpression (bottom left panel). Lysates from exponentially growing wild-type and cnb1∆ cells +/− HAP4 overexpression were assayed by Western blot for ETC components, Cox2 and ATP synthase, and mitochondrial outer membrane protein VDAC1/porin. Right panel, schematic indicating signaling pathways regulating mitochondrial biogenesis. c O2 consumption is increased upon overexpression of HAP4 or SAK1. O2 consumption is increased in cells overexpressing HAP4 or SAK1. d Overexpression of HAP4 or SAK1 suppresses death of cnb1∆ and ire1∆ cells after 5 h tunicamycin treatment. Top panel, cell viability was assayed as in Fig. 1b legend. Bottom panel, HAP4 overexpression relieves impaired growth of cnb1∆ cells on tunicamycin. Serial dilutions of cells were spotted on SC-uracil plates with 0.2 µg/mL tunicamycin. e ALA to promote heme synthesis increases O2 consumption (left panel) and rescues viability (right panel) of ire1∆ and cnb1∆ cells during ER stress. Cells were grown overnight in SC medium supplemented with ALA to mid-log phase. Viability was determined after 5 h treatment with tunicamycin (0.5 µg/mL) as described in Fig. 1b legend.
ROS production increases as the rate and efficiency of electron flow through the ETC decline [22]. To examine the idea that mitochondrial biogenesis can increase efficiency of electron flow by increasing ETC components and reducing ROS production, we exploited two methods. First, mitochondrial biogenesis is controlled transcriptionally (in response to carbon source) by the Hap complex [25]; Hap4 is the catalytic subunit of the complex, and when Hap4 is overexpressed, respiration rate increases to 150% that of wild-type cells, overcoming glucose repression of respiration during exponential growth [26]. Hap4 overexpression increased levels of ETC proteins in wild-type, cnb1∆ and ire1∆ cells (Fig. 3b), and increased basal O2 consumption (Fig. 3c). Indeed, HAP4 overexpression protected ire1∆ and cnb1∆ cells from tunicamycin-induced death (Fig. 3d, top panel) and improved cnb1∆ cell growth in the presence of tunicamycin (Fig. 3d, bottom panel). These results are consistent with the idea that increased electron flow through the ETC helps to limit ROS production and rescues from ER stress-induced death. Second, Snf1/AMP kinase (AMPK) signaling pathways also control mitochondrial biogenesis in response to carbon source [27]. By overexpressing Sak1 kinase to manipulate glucose-dependent Snf1/AMPK [28], it is possible to increase levels and activity of ETC components. Figure 3b (right panel) diagrams a proposed signaling pathway regulating mitochondrial biogenesis. Sak1 overexpression increased ETC protein levels (Fig. 3b); increased copies of COX2, encoded by mitochondrial DNA, confirm increased mitochondrial biogenesis upon Sak1 as well as Hap4 overexpression (Suppl. Figure 2A). Sak1 overexpression substantially increased basal O2 consumption in cnb1∆ and ire1∆ cells (Fig. 3c). Increased O2 consumption by HAP4 or SAK1 overexpression was linked to decreased ROS production during ER stress (Suppl. Figure 2B), and also suppressed ER stress-induced death (Fig. 3d).
COX, the final enzyme and the proposed rate-limiting step in the ETC, has copper and heme groups to help transfer electrons to O2. COX expression and activity was recently reported to be increased in mice fed aminolevulinic acid (ALA), a precursor of heme [29], and increased heme synthesis in yeast has been found to increase respiration [30]. Indeed, O2 consumption was increased in yeast cells fed overnight with ALA (Fig. 3e, left panel), and increased respiration was linked to increased resistance to ER stress (Fig. 3e, right panel).
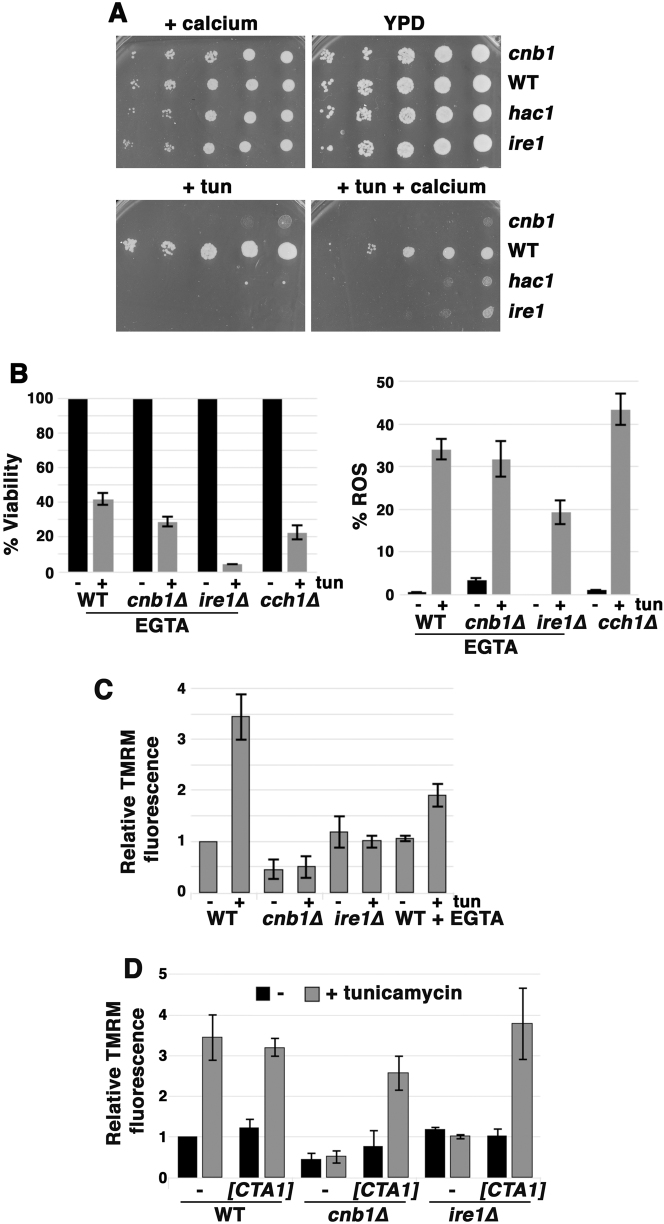
Extracellular calcium exacerbates ER stress in cnb1∆ cells but helps rescue ire1∆ cells
As noted above, both cnb1∆ and ire1∆ yeast are hypersensitive to ER stress. During ER stress, cnb1∆ cells accumulate Ca2+ [9, 31]. Indeed, we observed that extracellular Ca2+ exacerbates impaired growth of cnb1∆ cells challenged with ER stress (Fig. 4a), suggesting that excessive accumulation of Ca2+ in cnb1∆ cells is deleterious, which is consistent with a role for calcineurin in limiting extracellular Ca2+ entry to prevent toxicity [31]. Remarkably, however, growth of ire1∆ cells in the presence of tunicamycin was slightly improved by excess extracellular Ca2+ (Fig. 4a). Excess extracellular Ca2+ also improved growth in the presence of tunicamycin in cells without Hac1, the transcription factor activating UPR (Fig. 4a); these results suggest that Ca2+ acts downstream of the UPR to enhance survival during ER stress. In support of this idea, extracellular EGTA exacerbated sensitivity of wild-type cells to tunicamycin-induced ROS accumulation and death (compare Fig. 4b with Fig. 1c); similarly, cch1∆ cells, impaired in Ca2+ influx [32] were also sensitive to ER stress-induced ROS and death (Fig. 4b). Extracellular EGTA did not significantly affect ER stress-induced ROS accumulation and cell death in cnb1∆ cells, while, interestingly, cell viability as well as ROS were reduced in ire1∆ cells (compare Fig. 4b with Fig. 1c), reflecting perhaps the complexity of Ca2+ dynamics under these conditions. However, consistent with growth phenotypes in Fig. 4a, cnb1∆ cells display extreme sensitivity to ER stress-induced ROS accumulation and death after growth in medium with 100 mM excess Ca2+, while ER stress-mediated ROS and cell death were alleviated by excess Ca2+ in ire1∆ cells (Suppl. Figure 3).
Fig. 4.
Extracellular Ca2+ exacerbates ER stress in cnb1∆ cells but alleviates tunicamycin sensitivity in ire1∆ cells. a Serial dilutions of cells were spotted on YPD plates with or without 0.2 µg/mL tunicamycin with or without 200 mM CaCl2. b Calcium influx promotes survival in response to ER stress. ER stress-induced ROS accumulation (right panel) and death (left panel) are increased in wild-type cells in extracellular EGTA and in cch1∆ cells. Cells were grown overnight in SC supplemented with 10 µM EGTA, treated with 0.5 µg/mL tunicamycin for 5 h, and stained with DHE to visualize ROS; quantitation was as described in Fig. 1c legend. Viability was assayed and quantitated as described in Fig. 1b legend. c, d Mitochondrial membrane potential (MMP) responds to Ca2+ flux. Exponentially growing cells were stained with TMRM (5 nM, nonquenching mode) before and after 2 h treatment with 0.5 µg/ml tunicamycin. Wild-type cells in SC medium plus EGTA (10 µM) were grown overnight prior to tunicamycin treatment. Wild-type, cnb1∆ and ire1∆ cells transformed with pMET17-CTA1 were induced to express mitochondrial catalase by washing with water and resuspending in methionine-free medium. Cells were then further incubated for 2 h with tunicamycin (0.5 µg/mL). Fluorescence images were taken at identical exposure times and brightness and contrast were adjusted using identical Photoshop settings.
Ca2+ enters mitochondria after its release from the ER in mammalian cells [33], and this increases respiratory chain activity and mitochondrial membrane potential (MMP) leading to ATP generation [3]. To examine if Ca2+ flux affects MMP during ER stress in yeast, cells were stained with the fluorescent dye TMRM in nonquenching mode [34]. As shown in Fig. 4c, MMP in wild-type cells was increased 2 h after tunicamycin addition; imaging of TMRM staining in cells treated with and without tunicamycin is shown in Suppl. Figure 4. Mitochondrial hyperpolarization in response to ER stress was diminished when cells are grown in EGTA to prevent Ca2+ influx into the cell (Fig. 4c).
Notably, mitochondrial hyperpolarization failed to occur in response to ER stress in ire1∆ and cnb1∆ cells (Fig. 4c). These results suggest that the MMP hyperpolarization is dependent on Ca2+ dynamics and reflects an adaptive response to ER stress. Because ROS accumulation is alleviated by overexpression of catalase in ire1∆ and cnb1∆ cells (Fig. 2d), MMP was assayed by TMRM staining after catalase overexpression. Figure 4d shows mitochondrial hyperpolarization in response to ER stress was restored upon detoxification of ROS in ire1∆ and cnb1∆ cells, suggesting that ROS production is closely linked to MMP regulation.
COX activity is impaired in ire1∆ and cnb1∆ cells
As shown in Fig. 3a, cellular O2 consumption in cnb1∆ cells was significantly lower than that in wild-type yeast cells. To examine whether impaired COX activity can account for depressed O2 consumption in cnb1∆ cells, COX activity was assayed by measuring oxidation of cytochrome c in isolated mitochondria (see Methods). Indeed, COX activity was reduced in cnb1∆ cells by comparison with that of wild-type cells (Fig. 5a, right panel). Moreover, COX activity is also decreased in cells grown overnight with the calcineurin inhibitor FK506 (Fig. 5a, right panel), consistent with decreased O2 consumption (Fig. 5a, left panel). These data indicate that COX activity is downstream of calcineurin signaling. In wild-type and cnb1∆ cells, cellular O2 consumption entirely reflects ETC-coupled ATP synthesis as O2 consumption was completely blocked by inhibition of ATP synthase (with triethyltin, TET) and returned to its normal state upon CCCP addition (Fig. 5b). In ire1∆ cells, O2 consumption was similar to that in wild-type cells; however, ~50 % of O2 consumption in ire1∆ cells remained after inhibition of ATP synthase (Fig. 5b), suggesting that O2 is utilized in ire1∆ cells for (one or more) cellular activities other than ETC-coupled oxidative phosphorylation. Replotted in Fig. 5c (left panel), O2 consumption coupled to oxidative phosphorylation (TET-inhibitable) was decreased in ire1∆ cells by comparison with that in wild-type cells. Similarly, COX activity was decreased in ire1∆ cells relative to that in wild-type cells (Fig. 5c, right panel). For comparison with cells exponentially growing in glucose, O2 consumption and COX activity are also shown for cells growing in (non-fermentable) glycerol with a full respiratory metabolism (Fig. 5c). These results suggest that diminished COX activity that is unable to respond to ER stress is linked to susceptibility to ROS production in both ire1∆ and cnb1∆ cells.
Fig. 5.
O2 consumption and COX activity are impaired in cnb1∆ and ire1∆ cells. a Left panel, O2 consumption by cells grown overnight to mid-log phase in FK506 (2 µg/mL) were compared with that of wild-type and cnb1∆ cells. Right panel, COX activity measured in isolated mitochondria as oxidation of cytochrome c. b O2 consumption in ire1∆ cells is not entirely attributable to oxidative phosphorylation. O2 consumption was measured in wild-type, cnb1∆, and ire1∆ cells. Cells were treated with TET (10 mM) to inhibit ATP synthase; O2 consumption in wild-type and cnb1∆, but not ire1∆ cells falls to 0. Addition of CCCP (10 µM) induces O2 consumption to return to maximal. c Reduced ETC activity in ire1∆ cells. Left panel, O2 consumption by oxidative phosphorylation (TET-inhibitable) in wild-type, cnb1∆ and ire1∆ cells. Right panel, COX activity was measured in isolated mitochondria. Wild-type cells were grown to mid-log phase overnight in SC with 3% glycerol for comparison with strains grown in SC with 2% glucose.
High copy suppressors of ER stress-induced death in yeast
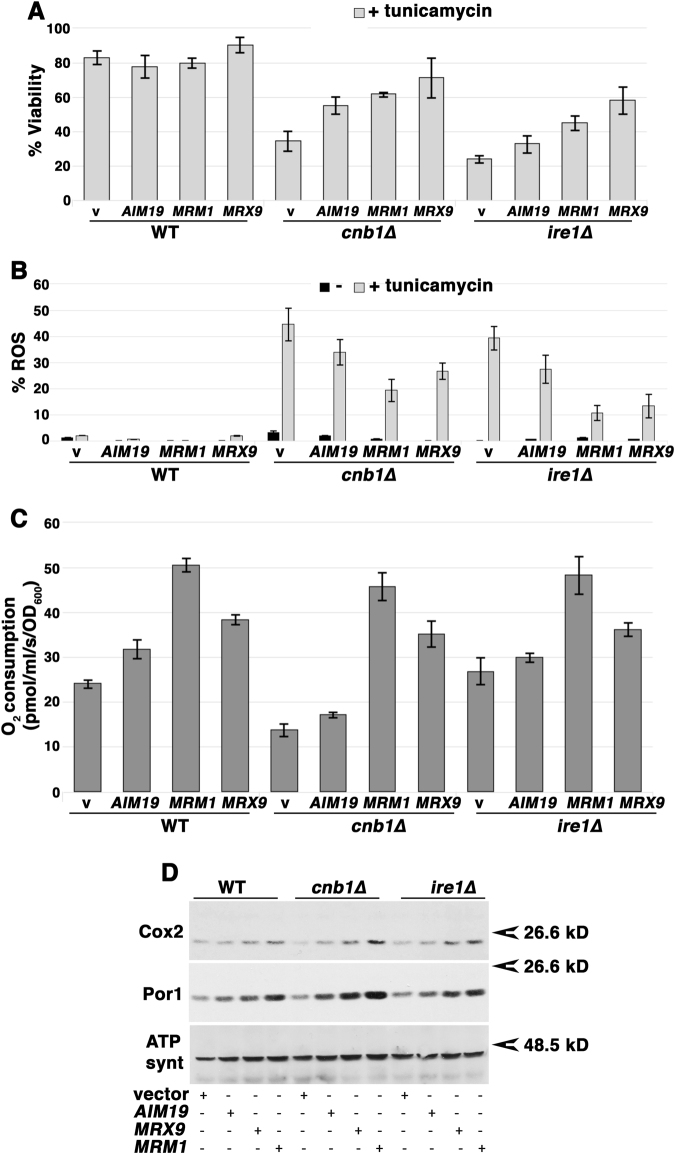
A screen was performed to identify genes that, upon overexpression, rescue cnb1∆ cells from ER stress-induced death (see Methods). Of the suppressors that were isolated, three genes encoded the mitochondrial proteins Mrm1, Mrx9, and Aim19 (Fig. 6a). When overexpressed, these mitochondrial proteins rescued cells from tunicamycin-induced ROS accumulation (Fig. 6b). Moreover, these proteins also increased basal O2 consumption even in wild-type cells (Fig. 6c), and restored viability to both ire1∆ and cnb1∆ cells challenged with ER stress (Fig. 6a). Mrm1 is a conserved ribosome rRNA methyltransferase whose activity is required for mitochondrial ribosome function in yeast and mammalian cells [35, 36]. [The importance of Mrm1 in respiratory activity is further indicated by the inability of mrm1∆ cells to grow on the nonfermentable carbon source glycerol (Suppl. Figure 5.)] Mrx9 is also associated with mitochondrial ribosomes [37], whereas Aim19 is a potential interaction partner with respiratory supercomplexes [38] and has been shown to interact with the Tim23 translocon of the inner mitochondrial membrane [39]. Because factors involved in mitochondrial transcription, translation, and assembly of the respiratory chain are coordinated [40], it is likely that each of these gene products facilitates aspects of mitochondrial biogenesis including ETC assembly. Indeed, ETC protein levels were increased upon overexpression of Aim19, Mrm1 or Mrx9 in wild-type, cnb1∆ and ire1∆ cells (Fig. 6d).
Fig. 6.
High copy suppressors of ER stress-induced death of cnb1∆ cells. a, b Upon overexpression on 2 µ plasmids, three mitochondrial genes, AIM19, MRM1, and MRX9 rescue cnb1∆ and ire1∆ cells from ER stress-induced death and ROS. Viability and ROS after 5 h incubation with 0.5 µg/mL tunicamycin was determined as described in Fig. 1b, c legends. The results are expressed as a percent of untreated cells. c Overexpression of Aim19, Mrm1, and Mrx9 increase O2 consumption in wild-type, ire1∆, and cnb1∆ cells. d High copy suppressors of ER stress-induced death increase ETC component levels. Western blot of wild-type, cnb1∆, and ire1∆ cells with and without high copy suppressors.
Common mechanisms in ER stress-induced death
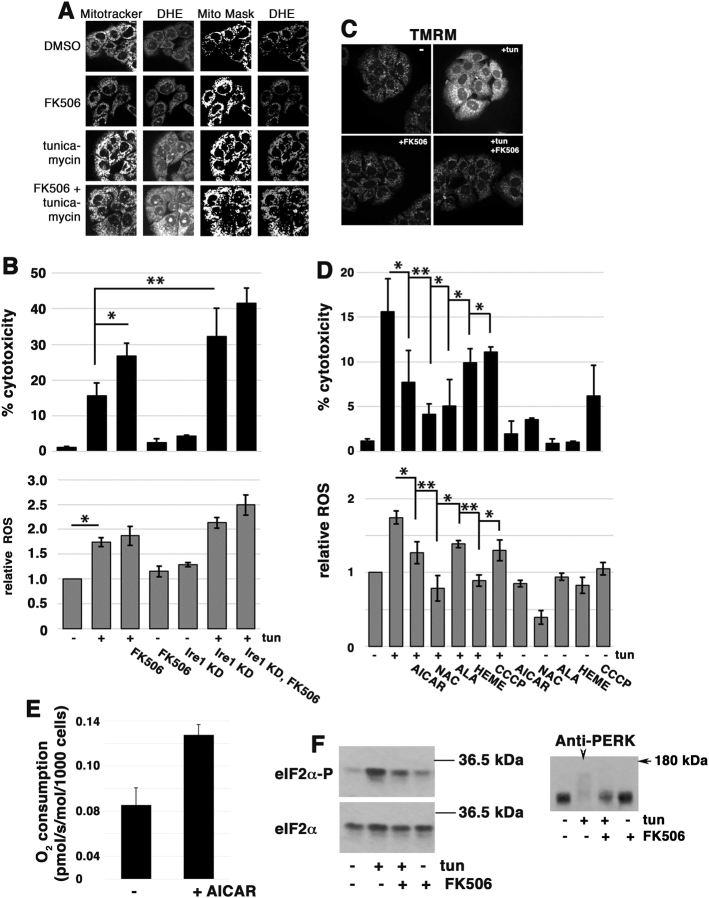
To examine whether the critical role for mitochondrial ROS in ER stress-mediated cell death in yeast is shared by mammalian cells, we examined cultured PCCL3 (rat) cells, a line of thyroid secretory cells, which exhibit significant sensitivity to tunicamycin [41]. Figure 7a shows that mitochondrial ROS was accumulated within 48 h after tunicamycin challenge. Concomitantly, cytotoxicity was induced, as measured by extracellular leakage of lactate dehydrogenase (Fig. 7b). Together with tunicamycin, addition of the calcineurin inhibitor FK506 elicited increased cytotoxicity (Fig. 7b) and exacerbated ROS accumulation (Fig. 7a). Ire1 knockdown using siRNA (to ~10% normal levels; Suppl. Figure 6) also promoted increased cytotoxicity after tunicamycin treatment (Fig. 7b). Similarly, cytotoxicity after tunicamyin challenge is augmented by knockdown of XBP1, the transcription factor activated via Ire1 signaling (Suppl. Figure 7), suggesting that the effect of Ire1 knockdown is mediated primarily through impaired XBP1-dependent transcriptional activity (versus XBP1-independent activities [42, 43].) PERK, signaling a distinct branch of the mammalian UPR [44], is also necessary for protection from ER stress as both ROS and cytotoxicity were aggravated by PERK knockdown (Suppl. Figure 7).
Fig. 7.
ER stress induces ROS and death in PCCL3 cultured cells. a Calcineurin protects against ROS production. Cells were co-treated with FK506 (5 µM) and tunicamycin (100 ng/mL) for 2 days. Cells were then co-stained with DHE and Mitotracker green to detect ROS and mitochondria, respectively. b ER stress-induced cytotoxicity (top panel) and ROS accumulation (bottom panel) are exacerbated by calcineurin inhibition and Ire1 knockdown (kd). siRNAs were added to cells for 2 days before incubation with tunicamycin (100 ng/mL) for another 2 days before cytotoxicity was assayed. The cytotoxic effect of FK506 was determined after 2 days incubation with tunicamycin (100 ng/mL) with or without FK506 (5 µM). Cytotoxicity was assayed as leakage of LDH. n = 3 experiments; error bars are SEM. *p < 0.05. ROS accumulation was assayed by DHR123 fluorescence staining, as described in Methods. c Mitochondrial hyperpolarization is induced by tunicamycin (100 ng/mL). MMP, as measured by TMRM fluorescence, is increased by 1 h after tunicamycin treatment; this ER stress response is abrogated by FK506-mediated inhibition of calcineurin. d Tunicamycin-induced cytotoxicity (top panel) is dependent on mitochondrial ROS, and is suppressed by anti-oxidants (N-acetyl cysteine, 5 mM), heme (10 µM), increased heme synthesis (ALA, 300 µg/ml), increased O2 flux (induced by CCCP, 10 nM), and increased mitochondrial respiration (induced by AICAR, 0.5 mM). n = 3 experiments +/− SEM. *p < 0.05, **p < 0.01. Bottom panel, ROS accumulation was assayed by DHR123 fluorescence, as described in Methods. e AICAR increases O2 consumption. PCCL3 cells were treated with AICAR (0.5 mM) for 1 day, and then trypsinized, washed, and placed in a oxygraph for O2 consumption measurements, as described previously [62]. f Left panel, Calcineurin is necessary for tunicamycin-induced phosphorylation of eIF2α. Cells were treated with tunicamycin +/− FK506 (5 µM) for 2 days, as in a, and phosphorylated eIF2α and total eIF2α protein were assayed by Western blot. Right panel, Cell lysate from experiment shown in the left panel was blotted with anti-PERK antibody. Arrowhead, in tunicamycin-treated samples, phosphorylated PERK is seen as an extensive smear.
Similar to that seen in yeast, mitochondrial hyperpolarization, revealed by increased TMRM fluorescence, was observed in PCCL3 cells within 1 h of tunicamycin addition; this ER stress response was prevented in the presence of FK506 (Fig. 7c). Strategies that rescue yeast from ER stress-mediated death, such as detoxifying ROS with the anti-oxidant N-acetyl cysteine, substantially inhibited death of the mammalian cells as well (Fig. 7d). The protonophore CCCP, which increases electron transport and O2 consumption, also significantly reduced ROS production in mammalian cells (Fig. 7d, bottom panel), in agreement with previous studies [45, 46], and decreased tunicamycin-induced cytotoxicity (Fig. 7d, top panel). [Interestingly, CCCP by itself resulted in some cytotoxicity, presumably by inhibiting ATP synthesis.] Furthermore, as in yeast, heme and the heme precursor ALA reduced cytotoxicity of PCCL3 cells in response to tunicamycin (Fig. 7d, top panel), perhaps by increasing ETC activity [29]. Finally, AICAR, an activator of AMPK [47], increased O2 consumption in PCCL3 cells (Fig. 7e), and was effective in suppressing ER stress-mediated cell death (Fig. 7d).
Calcineurin has been suggested to interact with PERK (an ER stress-responsive eIF2α kinase) and to increase its activity during the UPR [7]. As expected, tunicamycin treatment increased phosphorylation of the critical translation-regulating eIF2α subunit in PCCL3 cells (Fig. 7f, left panel). Remarkably, in cells treated with FK506 and tunicamycin, eIF2α phosphorylation was diminished by comparison with that seen with tunicamycin alone (Fig. 7f, left panel). PERK kinase phosphorylation is induced by tunicamycin (seen as a broad smear in an anti-PERK Western blot in Fig. 7f, right panel, arrowhead); however, FK506 inhibits tunicamycin-induced PERK activation, supporting the idea that calcineurin promotes PERK activity during the UPR. Thus, calcineurin and Ire1 signaling in response to ER stress both help to limit ROS accumulation in eukaryotes from yeast to mammalian cells, and calcineurin may additionally contribute to ER stress-mediated activation of PERK kinase activity in mammalian cells.
Discussion
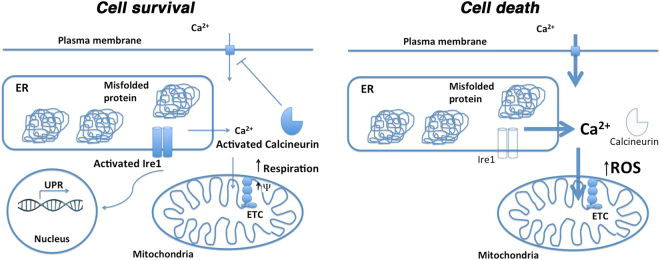
In eukaryotic cells exposed to ER stress, Ca2+ flux from ER to mitochondria has been established [3]. From our studies, we propose a model (diagrammed in Fig. 8) such that initial Ca2+ flux in response to ER stress is an adaptive response to increase respiration. Increased demand for ATP upon increased chaperone activity may link to the Ca2+ flux that serves as a stimulus for enhanced respiratory activity. The respiratory burst in yeast is observed as an initial short-lived increase of MMP and increased O2 consumption (Figs. 3 and 4), and has also been reported in HeLa cells [5]. We propose that Ca2+ flux induced by ER stress serves as the basis for increased MMP, as EGTA abrogates ER stress-induced MMP response in wild-type cells, and increases susceptibility to ROS accumulation and cell death (Fig. 4b).
Fig. 8.
Schematic model of ER stress-induced stress. Details described in text. Cell survival: protein misfolding in the ER is sensed by Ire1 that dimerizes to induce increased transcriptional activation in the nucleus; Ire1 activity is necessary for survival of ER stress. In response to ER stress, MMP and O2 consumption are increased [by Ca2+ influx into mitochondria [3]]. Activation of calcineurin is a requisite event for cell survival [this paper and [31]]. Cell death: protein misfolding in the ER cannot induce a transcriptional response without Ire1. Ca2+ flux is abnormal without calcineurin [9]. Genetic evidence suggests Ca2+ homeostasis is also dysregulated in ire1∆ cells. In both cnb1∆ and ire1∆ cells, MMP and O2 consumption fail to respond to ER stress, ROS is accumulated and death ensues (Figs. 1 and 3).
A major finding in this paper is that an adaptive mitochondrial response to ER stress requires both Ire1 and calcineurin. Our data suggest that defective Ca2+ homeostasis/signaling contributes to failure to increase respiration in yeast ire1∆ cells, leading to ROS accumulation and cell death (Fig. 8). Supporting this idea, extracellular Ca2+ alleviates tunicamycin-impaired growth and ER stress-induced ROS accumulation and cell death in ire1∆ cells (Fig. 4 & Suppl. Figure 3), perhaps by promoting increased Ca2+ entry into mitochondria to improve electron flow in the ETC and/or refilling of the ER pool. Similarly, in human neurobastoma cells, Ire1 knockdown induces Ca2+ dysregulation leading to ROS accumulation and cell death [48]. By contrast with possible insufficient Ca2+ flux in the absence of Ire1, Ca2+ overload may occur during ER stress in the absence of calcineurin [9] so that extracellular Ca2+ exacerbates ER stress-impaired growth, ROS accumulation and death in cnb1∆ cells (Fig. 4 & Suppl. Figure 3), presumably mediated by dysregulated, exuberant Ca2+ entry [31]. Constitutive Ca2+ overload in cnb1∆ cells likely contributes to diminished COX activity, decreased O2 consumption, and increased ROS (Figs. 2, 3, and 5).
In addition to the direct effects of Ca2+ on the activities of ETC components, another possible mechanism to account for impaired mitochondrial response to ER stress in cnb1∆ or ire1∆ cells is disturbed Snf1/AMPK activity. In yeast, Snf1 activation is regulated in a Ca2+-dependent way [49], and reports in some mammalian cells indicate that AMPK phosphorylation and activity is regulated by Ca2+ and calmodulin-dependent protein kinase kinase ß [50]. ER stress has indeed been shown to activate AMPK [51], and we have shown previously that a hyperactive Snf1 allele can increase respiration to abrogate mitochondrial ROS accumulation [23].
The link between ER stress-induced ROS accumulation and cell death has previously been noted [52], although the mechanisms have been ill-defined. Our results show that ER stress-induced ROS production by mitochondria is blocked not only by ETC loss or increasing the rate of O2 consumption with CCCP or anti-oxidants such as N-acetyl cysteine or mitochondrial catalase (Fig. 2), but significantly by increasing electron transport and respiration. Increased respiratory response may help limit mitochondrial ROS production in multiple forms of stress; in yeast, this includes nitrogen deprivation [23], mating pheromone [24], and exposure to acetic acid [53]. Using genetic approaches, we show that ER stress-induced ROS and cell death are ameliorated by increased mitochondrial biogenesis upon overexpression of HAP4, a transcriptional activator of respiratory gene expression, or by overexpressing SAK1 to activate Snf1/AMPK signaling (Fig. 3). The importance of mitochondrial biogenesis is underscored as genetic screening shows that overexpression of MRM1, MRX9, and AIM19 alleviates ER stress by increasing mitochondrial protein synthesis (Fig. 6). In all cases, increased mitochondrial biogenesis is associated with significant increases in O2 consumption. Using an analogous approach in mammalian PCCL3 cells, AMPK signaling increased by AICAR promotes O2 consumption to reduce ROS production and cell death (Fig. 7).
In addition to increasing electron flow, mitochondrial biogenesis is protective because mitochondrial antioxidants such as superoxide dismutase and catalase are increased. Because mitochondrial growth rescues reduced COX activity observed under basal respiratory metabolism in cnb1∆ and ire1∆ cells (Fig. 3c), it appears that incorporation of new mitochondrial components helps to optimize electron transfer reactions, i.e., new assembly produces a better ETC. Increased COX2 DNA upon HAP4 or SAK1 overexpression (Suppl. Figure 2A) suggests that mitochondrial numbers are increased as well. Mechanisms known to promote a more efficient ETC include posttranslational modification of COX subunits and switching of different COX isoforms [54, 55].
Mitochondrial transcriptional UPR targets underscore the important role played by mitochondria in response to ER stress [56, 57]. Among these targets are biosynthetic enzymes for cofactors of ETC components, an enzyme catalyzing formation of ETC complex II, and Hap1, a heme and O2-dependent transcription factor. Other UPR targets have anti-oxidant effects, or participate in mitochondrial protein quality control [58, 59]. Finally, Tos3 is a UPR target related to and functionally redundant with Sak1 for phosphorylation and activation of Snf1/AMPK [60], consistent with our observation that SAK1 overexpression alleviates ER stress-induced mitochondrial ROS production (Fig. 2b). In mammalian cells, UPR transducers ATF6 and ATF4 control expression of the Parkin ubiquitin ligase that regulates mitochondrial function and maintains quality control, along with PGC1α, master regulator of mitochondrial biogenesis [61]. We predict a better understanding of how mitochondrial respiration is regulated will lead to novel therapies to promote cell survival and alleviate diseases triggered by ER stress.
Electronic supplementary material
Acknowledgements
We thank Martin Schmidt, Su-Ju Lin, and Paula Ludovico for plasmids, Dan Beard and Fran Van Den Bergh for helping us with the oxygraph, and Gregg Sobocinski for help with microscopy. This work was supported by the University of Michigan Protein Folding Disease Initiative and NIH grant R01DK40344.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by L. Scorrano.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0133-4) contains supplementary material, which is available to authorized users.
References
- 1.Ron D. Translation control in the endoplasmic reticulum stress response. J Clin Investig. 2002;110:1383–8. doi: 10.1172/JCI0216784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015;17:829–38. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman RJ, Malhotra JD. Calcium trafficking integrates endoplasmic reticulum function with mitochondrial bioenergetics. Biochim Biophys Acta. 2014;1843:2233–9. doi: 10.1016/j.bbamcr.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vannuvel K, Renard P, Raes M, Arnould T. Functional and morphological impact of ER stress on mitochondria. J Physiol. 2013;228:1802–18. doi: 10.1002/jcp.24360. [DOI] [PubMed] [Google Scholar]
- 5.Bravo R, Vicencio JM, Parra V, Troncoso R, Munoz JP, Bui M, et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J Cell Sci. 2011;124:2143–52. doi: 10.1242/jcs.080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, McGrath BC, Kopp RF, Roe MW, Tang X, Chen G, et al. Insulin secretion and Ca2+ dynamics in ß-cell are regulated by PERK (EIF2K3) in concert with calcineurin. J Biol Chem. 2013;288:33824–36. doi: 10.1074/jbc.M113.503664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollo M, Paredes RM, Holstein D, Zheleznova N, Camacho P, Lechleiter JD. Calcineurin interacts with PERK and dephosphorylates calnexin to relieve ER stress in mammals and frogs. PLoS One. 2010;5:e11925. doi: 10.1371/journal.pone.0011925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonilla M, Cunningham KW. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002;21:2343–53. doi: 10.1093/emboj/21.10.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–35. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 11.Sherman F, Hicks JB, Fink GR. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 12.Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucl Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin SJ, Ford E, Haigis Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang A, Slayman CW. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J Cell Biol. 1991;115:289–95. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- 17.Capaldi RA, Marusich MF, Taanman JW. Mammalian cytochrome-c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol. 1995;260:117–32. doi: 10.1016/0076-6879(95)60134-1. [DOI] [PubMed] [Google Scholar]
- 18.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfoled protein response. Science. 2011;333:1891–4. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchi S, Giorgi C, Suski JM, Agnoletto C, Bononi A, Bonora M, et al. Mitochondria-Ros crosstalk in the control of cell death and aging. J Signal Transduct. 2011;2012:329635. doi: 10.1155/2012/329635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2007;104:2402–7. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox TD, Folley LS, Mulero JJ, McMullin TW, Thorsness PE, Hedin LO, et al. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–65. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 22.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552.2:335–44. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knupp J, Martinez-Montanes F, Van Den Bergh F, Cottier S, Schneiter R, Beard D, et al. Sphingolipid accumulation causes mitochondrial dysregulation and cell death. Cell Death Differ. 2017;24:2044–53. doi: 10.1038/cdd.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pozniakovsky AI, Knorre DA, Markova OV, Hyman AA, Skulachev VP, Severin FF. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J Cell Biol. 2005;168:257–69. doi: 10.1083/jcb.200408145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoboue ED, Mougeolle A, Kaiser L, Averet N, Rigoulet M, Devin A. The role of mitochondrial biogenesis and ROS in the control of energy supply in proliferating cells. Biochim Biophys Acta. 2014;1837:1093–8. doi: 10.1016/j.bbabio.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory threshholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Xu X, Carlson M. Interaction of SNF1 protein kinase with its activating kinase Sak1. Eukaryot Cell. 2011;10:313–9. doi: 10.1128/EC.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogura S, Maruyama K, Hagiya Y, Sugiyama Y, Tsuchiya K, Takahashi K, et al. The effect of 5-aminolevulinic acid on cytochrome c oxidase activity in mouse liver. BMC Res Notes. 2011;4:66. doi: 10.1186/1756-0500-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Bu P, Zeng J, Vancura A. Increased heme synthesis induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J Biol Chem. 2017;292:16942–54. doi: 10.1074/jbc.M117.790923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonilla M, Cunningham KW. Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol Biol Cell. 2003;14:4296–305. doi: 10.1091/mbc.e03-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke EG, Bonilla M, Liang L, Takita Y, Cunningham KW. A homolog of voltage-gated Ca2+ channels stimulated by depletion of secretory Ca2+ in yeast. Mol Cell Biol. 2000;20:6686–94. doi: 10.1128/MCB.20.18.6686-6694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–6. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 34.Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques. 2011;50:98–115. doi: 10.2144/000113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirum-Connolly K, Mason TL. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–9. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- 36.Lee KW, Okot-Kotber C, LaComb JF, Bogenhagen DF. Mitochondrial ribosomal RNA (rRNA) methyltransferase family members are postitioned to modify nascent rRNA in foci near the mitochondrial DNA nucleoid. J Biol Chem. 2013;288:31386–99. doi: 10.1074/jbc.M113.515692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kehrein K, Schilling R, Moller-Hergt BV, Wurm CA, Jakobs S, Lamkemeyer T, et al. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 2015;10:843–53. doi: 10.1016/j.celrep.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Vukotic M, Oeljeklaus S, Wiese S, Vogtle FN, Meisinger C, Meyer HE, et al. RCF1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–47. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Mehnert CS, Rampelt H, Gebert M, Oeljeklaus S, Schrempp SG, Kochbeck L, et al. The mitochondrial ADP/ATP carrier associates with the inner membrane presequence translocase in a stoichiometric manner. J Biol Chem. 2014;289:27352–62. doi: 10.1074/jbc.M114.556498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ott M, Amunts A, Brown A. Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem. 2016;85:77–101. doi: 10.1146/annurev-biochem-060815-014334. [DOI] [PubMed] [Google Scholar]
- 41.Kim PS, Arvan P. Folding and assembly of newly synthesized thyroglobulin occurs in a pre-Golgi compartment. J Biol Chem. 1991;266:12412–8. [PubMed] [Google Scholar]
- 42.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 43.Han D, Lerner AG, VW L, Upton JP, Xu W, Hagen A, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–75. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 45.Nishikawa T, Edelstein D, Du XL, Yamagishi Si, Matsumura T, Kaneda Y, et al. Normalizaing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787–90. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- 46.Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–48. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 47.Hardie DG, Carling D. The AMP-activated protein kinase. Eur J Biochem. 1997;246:259–73. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 48.Son SM, Byun J, Roh SE, Kim SJ, Mook-Jung I. Reduced IRE1a mediates apoptotic cell death by disrupting calcium homeostasis via the InsP3 receptor. Cell Death Differ. 2014;5:e1188. doi: 10.1038/cddis.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohdate T, Izawa S, Kita K, Inoue Y. Regulatory mechanism for expression of GPX1 in response to glucose starvation and Ca2+ in Saccharomyces cerevisiae: involvement of Snf1 and Ras/cAMP pathway in Ca2+ signaling. Genes Cells. 2010;15:59–75. doi: 10.1111/j.1365-2443.2009.01365.x. [DOI] [PubMed] [Google Scholar]
- 50.Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, et al. Calmodulin-dependent protein kinase kinase-ß is an alternative upstream kinase for AMP-activated protein kinase. Cell Metabol. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Xi H, Barredo JC, Merchan JR, Lampidis TJ. Endoplasmic reticulum stress induced by 2-deoxyglucse but not glucose starvation activates AMPK through CaMKKß leading to autophagy. Biochem Pharmacol. 2013;85:1463–77. doi: 10.1016/j.bcp.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 52.Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, et al. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci. 2008;105:18525–30. doi: 10.1073/pnas.0809677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2598–606. doi: 10.1091/mbc.e01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timon-Gomez A, Nyvltova E, Abriata LA, Vila AJ, Hosler J, Barrientos A. Mitochochondrial cytochrome c oxidase biogenesis: recent developments. Semin Cell Dev Biol. 2018;76:163–78. doi: 10.1016/j.semcdb.2017.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittal N, Babu MM, Roy N. The efficiency of mitochondrial electron transport chain is increased in the long lived mrg19 Saccharomyces cerevisiae. Aging Cell. 2009;8:643–53. doi: 10.1111/j.1474-9726.2009.00518.x. [DOI] [PubMed] [Google Scholar]
- 56.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analysis reveal essential coordination between the unfolded protein response and endoplasmic reticulum-associated degradation. Cell. 2000;101:249–58. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 57.Kimata Y, Ishiwata-Kimata Y, Yamada S, Kohno K. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells. 2006;11:59–69. doi: 10.1111/j.1365-2443.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- 58.Schreiner B, Westerburg H, Forne I, Imhof A, Neupert W, Mokranjac D. Role of the AAA protease Yme1 in folding of proteins in the intermembrane space of mitochondria. Mol Biol Cell. 2012;23:4335–46. doi: 10.1091/mbc.e12-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okreglak V, Walter P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci. 2014;111:8019–24. doi: 10.1073/pnas.1405755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elbing K, McCartney RR, Schmidt MC. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem J. 2006;393:797–805. doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015;40:141–8. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hutter E, Renner K, Pfister G, Stockl P, Jansen-Durr P, Gnaiger E. Senescence-associated changes in respiration and oxidative phosphorylation in primary human fibroblasts. Biochem J. 2004;380:919–28. doi: 10.1042/bj20040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.