Summary
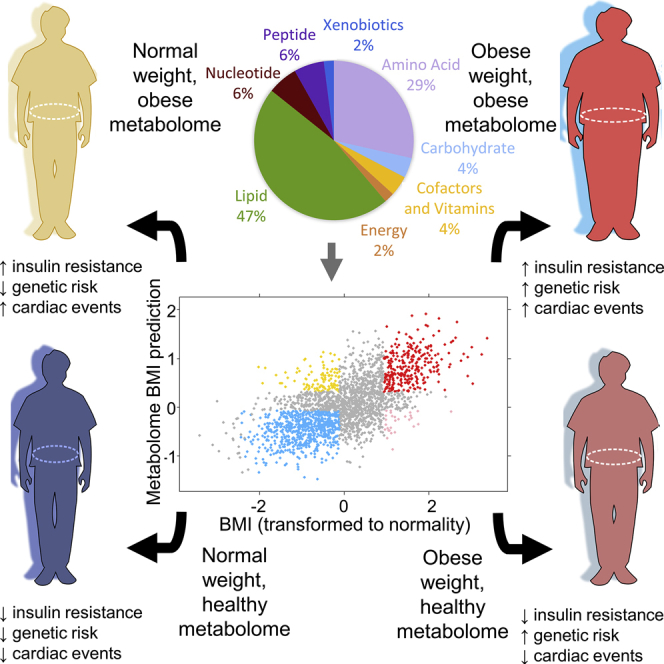
Obesity is a heterogeneous phenotype that is crudely measured by body mass index (BMI). There is a need for a more precise yet portable method of phenotyping and categorizing risk in large numbers of people with obesity to advance clinical care and drug development. Here, we used non-targeted metabolomics and whole-genome sequencing to identify metabolic and genetic signatures of obesity. We find that obesity results in profound perturbation of the metabolome; nearly a third of the assayed metabolites associated with changes in BMI. A metabolome signature identifies the healthy obese and lean individuals with abnormal metabolomes—these groups differ in health outcomes and underlying genetic risk. Specifically, an abnormal metabolome associated with a 2- to 5-fold increase in cardiovascular events when comparing individuals who were matched for BMI but had opposing metabolome signatures. Because metabolome profiling identifies clinically meaningful heterogeneity in obesity, this approach could help select patients for clinical trials.
Keywords: metabolomics, obesity, human genetics, rare variants, MC4R, polygenic risk scores, health
Graphical Abstract
Highlights
-
•
Obesity results in a profound perturbation of the plasma metabolome
-
•
At any given BMI, abnormal metabolomes associate with different health outcomes
-
•
At any given BMI, different genetic obesity risks do not change the metabolome
-
•
A metabolome signature effectively tracks changes in obesity
Obesity is a heterogeneous and complex disease that is imprecisely measured by BMI. Cirulli et al. used non-targeted metabolomics and whole-genome sequencing to identify metabolic and genetic signatures of obesity and find that the metabolome captures clinically relevant phenotypes of obesity and is a better health predictor than genetic risk.
Introduction
Obesity is one of the most widespread problems facing our society’s health today. Excessive weight significantly increases risk for conditions like diabetes mellitus and cardiovascular disease (Hales et al., 2017, Whitlock et al., 2009). Worldwide, the prevalence of obesity has nearly tripled since 1975, with 39% of the world’s adults being overweight and 13% being obese (WHO, 2018). The high prevalence can partially be attributed to increasing consumption of hypercaloric foods and sedentary lifestyles (WHO, 2018). Previous studies have identified metabolic signatures associated with obesity, including increased levels of branched-chain and aromatic amino acids as well as glycerol and glycerophosphocholines (Butte et al., 2015, Chen et al., 2015, Ho et al., 2016, Menni et al., 2017, Park et al., 2015, Piening et al., 2018). However, prior work has been limited by focusing on a relatively small numbers of metabolites, individuals, or obesity phenotypes.
The characterization of the metabolites that are associated with obesity can provide insights into the mechanisms that lead to this disease and associated consequences. Longitudinal assessment of weight gain and weight loss over time may indicate whether there are metabolomic changes that cause obesity—meaning that current metabolite levels could predict future weight changes—or whether all metabolite changes associated with obesity are a consequence of weight changes. Drawing genetics into this assessment allows the determination of whether genetic variation leads to metabolite changes that subsequently result in obesity, allowing the further delineation of the causal pathway to obesity. Finally, research in this area may identify biomarkers of obesity and of different types of obesity, for example biomarkers of so-called healthy obesity (Neeland et al., 2018).
There are recent calls to improve phenotyping in very large numbers of obese people with the goals of understanding factors that make people susceptible to (or protected from) obesity, accompanied by a better elucidation of the factors that account for variability in success of different obesity treatments (Yanovski and Yanovski, 2018). Here, in an effort to understand the relationship between metabolic perturbations and the obese state, we analyzed 2,396 individuals with longitudinal measurements of body mass index (BMI), anthropomorphic data, whole-body DEXA scans, and metabolome, combined with baseline genetic risk. The metabolome assay covered up to 1,007 metabolites at up to three distinct time points for each individual over the course of study. We identified associations between nearly a third of the metabolome and BMI, and we show that metabolite levels can predict obesity status with ∼80%–90% specificity and sensitivity. The metabolome profile is a strong indicator of metabolic health compared to the polygenic risk assessment and anthropomorphic measurements of BMI.
Results
Profound Perturbation of Metabolome by Obesity
Metabolites Associated with BMI
We compared the levels of individual metabolites to the BMIs of 832, 882, and 861 unrelated individuals of European ancestry in the TwinsUK cohort (Moayyeri et al., 2013) at three time points spanning a total range of 8–18 years. We identified 284 metabolites that were significantly associated (p < 5.5 × 10−5) with BMI at one or more time points (Table S1). We focused on 110 metabolites that were significantly associated with BMI at all 3 time points and sought to replicate the associations in an independent sample of 427 unrelated individuals of European ancestry participating in the Health Nucleus cohort (Perkins et al., 2017). In total, our analyses identified 307 metabolites that were significantly associated with BMI in at least one cohort and time point (Table S1). We identified 83 metabolites that showed directions of effect that were consistent between the two cohorts, of which 49 were statistically significant replications (Figure 1; Table 1). While this set of 49 metabolites was the most stringently associated with BMI, the majority of the implicated metabolites (292 of 307, 95%) had directions of effect that were consistent between time points/cohorts, indicating that many of the remaining metabolites may reach our stringent cutoffs in a larger study.
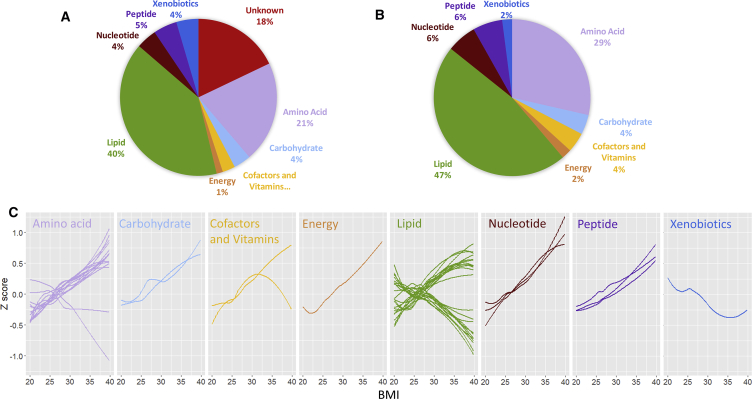
Figure 1.
Pathway Categories of Metabolites Associated with BMI
(A and B) Shown are the pathway categories of (A) the 307 metabolites significantly associated with BMI and (B) the 49-metabolite signature.
(C) The values of each of the 49 BMI-associated metabolites are plotted with a Loess curve against the BMI for time point 1 in TwinsUK. Only unrelated individuals of European ancestry are included, and the small number of individuals with BMI below 20 (n = 31) or above 40 (n = 10) are removed to keep the ends of the graphs from being skewed. The apparent inversion of the relationship between one cofactor/vitamin metabolite and BMI at higher BMIs is an artifact that is corrected once morbidly obese individuals are included.
Table 1.
Metabolite Signature Associated with BMI
| Super Pathway | Metabolite | Subpathway (Correlated Blood Lipidsc) | Direction of Effect (Ranka) | BMI r2b |
|---|---|---|---|---|
| Nucleotide | urate | Pur.Met. | ↑ (1) | 16.4% |
| N2,N2-dimethylguanosine | Pur.Met. | ↑ (6) | 8.8% | |
| N6-carbamoylthreonyladenosine | Pur.Met. | ↑ (28) | 7.3% | |
| Amino acid | glutamate | Glu.Met. | ↑ (2) | 11.5% |
| N-acetylglycine | Gly.Met. | ↓ (9) | 9.0% | |
| 5-methylthioadenosine (MTA) | Poly.Met. | ↑ (10) | 7.5% | |
| valine | Leu.Met. | ↑ (11) | 8.8% | |
| aspartate | Ala.Met. | ↑ (16) | 7.0% | |
| N-acetylvaline | Leu.Met. | ↑ (18) | 7.3% | |
| kynurenate | Try.Met. | ↑ (19) | 6.0% | |
| alanine | Ala.Met. | ↑ (23) | 5.3% | |
| asparagine | Ala.Met. | ↓ (26) | 3.7% | |
| N-acetylalanine | Ala.Met. | ↑ (31) | 6.6% | |
| tyrosine | Phe.Met. | ↑ (34) | 1.8% | |
| leucine | Leu.Met. | ↑ (37) | 6.8% | |
| N-acetyltyrosine | Phe.Met. | ↑ (40) | 4.2% | |
| 2-methylbutyrylcarnitine (C5) | Leu.Met. | ↑ (41) | 8.3% | |
| Lipid | 1-(1-enyl-palmitoyl)-2-oleoyl-GPC | Plas. (HDL, TG) | ↓ (3) | 7.1% |
| 1-stearoyl-2-dihomo-linolenoyl-GPC | Phos.Met. (TG, Chol) | ↑ (4) | 9.8% | |
| 1-eicosenoyl-GPC | Lysolipid | ↓ (5) | 6.2% | |
| 1-arachidoyl-GPC | Lysolipid | ↓ (7) | 8.6% | |
| 1-(1-enyl-stearoyl)-2-oleoyl-GPC | Phos.lip. (HDL) | ↓ (8) | 6.5% | |
| propionylcarnitine | BCAA.Met | ↑ (12) | 9.9% | |
| 1-nonadecanoyl-GPC | Lysolipid | ↓ (14) | 4.2% | |
| 1-linoleoyl-GPC | Lysolipid | ↓ (15) | 4.9% | |
| sphingomyelin | Sph.Met. (Chol) | ↑ (20) | 6.8% | |
| 1-palmitoyl-2-dihomo-linolenoyl-GPC | Phos.Met. (TG, Chol) | ↑ (21) | 5.1% | |
| 1-(1-enyl-palmitoyl)-2-linoleoyl-GPC | Phos.Met. (HDL) | ↓ (22) | 5.7% | |
| 1-palmitoyl-3-linoleoyl-glycerol | Phos.Met. (TG) | ↑ (24) | 7.6% | |
| 1-oleoyl-2-linoleoyl-GPC | Phos.Met. | ↓ (27) | 5.6% | |
| 1-(1-enyl-stearoyl)-2-docosahexaenoyl-GPC | Phos.Met. | ↓ (29) | 2.5% | |
| 1-oleoyl-3-linoleoyl-glycerol | Di.gly. (TG, HDL) | ↑ (30) | 6.3% | |
| carnitine | Car.Met. | ↑ (33) | 7.5% | |
| 1-palmitoyl-2-linoleoyl-glycerol | Phos.Met. (TG, HDL) | ↑ (36) | 7.2% | |
| 1-oleoyl-2-linoleoyl-glycerol | Di.gly. (TG, HDL) | ↑ (38) | 5.9% | |
| 1,2-dilinoleoyl-GPC | Phos.Met. | ↓ (39) | 4.2% | |
| 1-palmitoleoyl-2-oleoyl-glycerol | Phos.lip. (TG) | ↑ (42) | 5.6% | |
| 1-palmitoleoyl-3-oleoyl-glycerol | Phos.lip. (TG) | ↑ (45) | 6.0% | |
| 1-palmitoyl-2-adrenoyl-GPC | Phos.Met. (TG) | ↑ (47) | 2.9% | |
| cortisone | Plas. (HDL, TG) | ↓ (49) | 2.5% | |
| Energy | succinylcarnitine | TCA | ↑ (13) | 9.8% |
| Carbohydrate | mannose | Fru.Met. | ↑ (17) | 6.6% |
| glucose | Pyr.Met. | ↑ (48) | 6.3% | |
| Xenobiotics | cinnamoylglycine | Food | ↓ (43) | 3.5% |
| Cofactors/ vitamins | gulonic acid | Asc.Met. | ↑ (46) | 3.2% |
| quinolinate | Nic.Met. | ↑ (44) | 8.4% | |
| Peptide | N-acetylcarnosine | Dipep. | ↑ (25) | 6.9% |
| gamma-glutamylphenylalanine | Gam. | ↑ (32) | 6.0% | |
| gamma-glutamyltyrosine | Gam. | ↑ (35) | 4.6% |
Subpathway acronyms: Ala.Met., alanine and aspartate metabolism; Asc.Met., ascorbate and aldarate metabolism; BCAA.Met, branched-chain amino acid metabolism; Car.Met., carnitine metabolism; Di.gly., diacylglycerol; Dipep., dipeptide derivative; Food, food component/plant; Fru.Met., fructose, mannose, and galactose metabolism; Gam., gamma-glutamyl amino acid; Glu.Met., glutamate metabolism; Gly.Met., glycine, serine, and threonine metabolism; Leu.Met., leucine, isoleucine, and valine metabolism; Lysolipid, lysolipid; Nic.Met., nicotinate and nicotinamide metabolism; Phe.Met., phenylalanine and tyrosine metabolism; Phos.lip., phospholipid; Phos.Met., phospholipid metabolism; Plas., plasmalogen; Poly.Met., polyamine metabolism; Pur.Met., purine metabolism; Pyr.Met., glycolysis, gluconeogenesis, and pyruvate metabolism; Sph.Met., sphingolipid metabolism; Steroid, steroid; TCA, TCA cycle; Try.Met., tryptophan metabolism.
Rank indicates order of significance of association with BMI.
Mean r2 indicates the percent variation in BMI explained by each metabolite in univariate analysis for a combined analysis of the first time point of the TwinsUK cohort and the Health Nucleus data.
Blood labs for TG (triglycerides), Chol (cholesterol), HDL (high-density lipoprotein), or LDL (low-density lipoprotein) that had an r2 > 0.1 with the metabolite are indicated in parentheses.
The 49 metabolites that associated with BMI were primarily lipids (n = 23, accounting for 7.5% of all lipids assayed across both cohorts) and amino acids (n = 14, 9.3% of all amino acids) but also included nucleotides (n = 3, 12.0% of all nucleotides), peptides (n = 3, 12% of all peptides), and other categories (n = 6; Figure 1; Table 1). The most significantly associated metabolite was urate (uric acid; p value 1.2 × 10−40 for combined analysis of TwinsUK time point 1 and Health Nucleus data).
Patterns in Metabolite Change According to BMI
The majority of the 49 BMI-associated metabolites increased with increasing BMI (n = 35) (Figure 1; Table S1). This included glucose and, notably, mannose, which has recently been highlighted as playing a role in insulin resistance (Lee et al., 2016). Most metabolites change linearly (both proportionally and inversely) with BMI (Figure 1C). Branched-chain and aromatic amino acids as well as metabolites related to nucleotide metabolism like urate had the most rapid increases. Those that decreased (n = 14) included phospholipids and lysolipids, as well as the amino acids asparagine and N-acetylglycine and the xenobiotic cinnamoylglycine, which has been identified as a product of the microbiome (Wikoff et al., 2009). The negatively associated lipids tended to reflect HDL (high-density lipoprotein) levels, while the positively correlated lipids were more representative of triglyceride levels (Tables 1 and S1). Of particular interest was the association between BMI and cortisone, a metabolite of the steroid hormone cortisol. We identified lower levels among the obese individuals, which is consistent with previous reports (Björntorp and Rosmond, 2000, Praveen et al., 2011, Walker et al., 2000, Wirix et al., 2017). We examined the overall composition of the distributions of these metabolites via principal component analysis and found complex underlying correlations; in particular, the first principal component explained ∼20% of the total variation in the levels of these 49 metabolites. We additionally examined each of the 49 metabolites in just those of normal weight, overweight, or obese separately, and we found directions of effect that were largely consistent with those seen in the group as a whole (Table S1).
Modeling the Metabolome of Obesity
We used ridge regression to build a model that would predict BMI from the 49 BMI-associated metabolites (Figure 2). We chose this method to focus on the most stringently associated metabolites and to remove effects of collinearity, and we found similar results when using lasso regression. We combined our data for the first visit of the TwinsUK cohort and the Health Nucleus cohort and trained with 10-fold cross-validation on a random half of the population. In our test set of the other half of the data, we found that the model could explain 39.1% of the variation in BMI (Figure 2A). In predicting whether participants were obese (BMI ≥ 30) or normal weight (BMI 18.5–25), the model had an area under the curve (AUC) of 0.922, specificity of 89.1%, and sensitivity of 80.2% (Figure S1). The model based on the metabolite signature was thereafter used as a tool to define mBMI, the predicted BMI on the basis of metabolome.
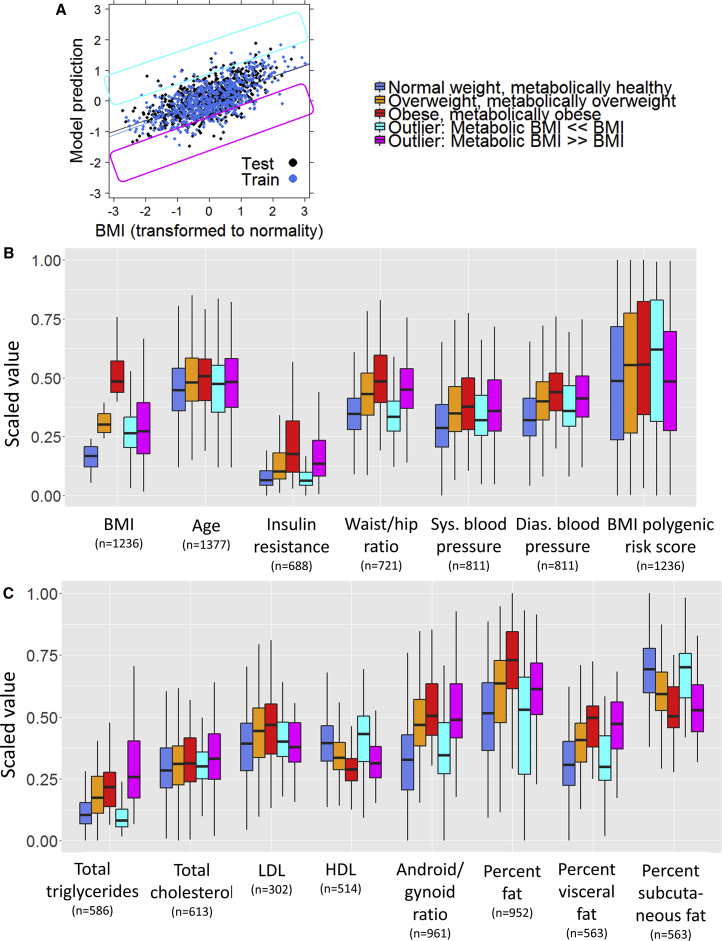
Figure 2.
Variables Associated with BMI and Predicted BMI from the Metabolome
(A) Correlation between ridge regression model prediction of BMI and actual BMI for all unrelated individuals of European ancestry in the TwinsUK and Health Nucleus dataset. The identification of outliers is defined below: the pink box shows individuals with a much lower predicted BMI (mBMI) than actual BMI, and the yellow box shows individuals with a much higher mBMI than actual BMI.
(B) Factors associated with being an mBMI outlier. Participants were split into five groups: those whose metabolome accurately predicted their BMI (residual after accounting for age, sex, and BMI between −0.5 and 0.5) whose BMIs were either normal (18.5–25), overweight (25–30), or obese (>30), and those whose metabolome predicted a substantially higher mBMI than the actual BMI (residual < −0.5) or a substantially lower mBMI than the actual BMI (residual > 0.5). All y axis values are scaled to a range from 0 to 1 to allow comparison across groups.
(C) The same process is used to show DEXA imaging values associated with metabolic BMI outliers. The mBMI ≫ BMI and mBMI ≪ BMI groups had a comparable measured BMI and age; however, these two groups were statistically significantly different from each other (p < 0.01) for all modalities except blood pressure, LDL, total cholesterol, and polygenic risk score. Additionally, the mBMI ≫ BMI group was statistically significantly different from normal weight, metabolically healthy people (p < 0.01) for all traits (except LDL), while mBMI ≪ BMI individuals were only statistically different in diastolic blood pressure (p = 0.005). In contrast, outliers with mBMI ≪ BMI were always statistically significantly different from obese, metabolically obese individuals (except systolic blood pressure, LDL, and total cholesterol), while mBMI ≫ BMI individuals were only statistically different in percent body fat (p = 4.4 × 10−6). LDL, low-density lipoprotein; HDL, high-density lipoprotein. The correlations of each modality with mBMI can be found in Table S2.
Whiskers of boxplot extend to the most extreme points no greater than 1.5 times the interquartile range (distance between the first and third quartiles).
Richer models using the full set of available metabolites (n = 650 measured in both cohorts) improved the accuracy of the model (47%–49% of the variance explained) and could be considered as the optimal approach by accepting the additional cost of a full untargeted metabolome as compared to the more targeted panel of 49 metabolites. This performance should be contrasted with the results of models using routine clinical laboratory determinations: regression analysis predicting BMI from age, sex, HDL, LDL, total cholesterol, and total triglycerides explained 31% of the variation in BMI, whereas a model using age, sex, and the 49-metabolite signature explained 43% of the variation. In fact, even though mBMI was modeled by training on BMI, this metabolite signature had a better correlation than BMI with most health-related phenotypes measured here (Table S2).
Identification and Characterization of Metabolic BMI Outliers
Having established a model to predict BMI using the metabolome (mBMI), we split the participants into five groups (Figure 2A). Three groups included individuals whose metabolome accurately predicted their BMI, as defined by having a residual between −0.5 and 0.5 from a regression analysis of mBMI with age, sex, and BMI included as predictors. These criteria delineated ∼80% of individuals as having an mBMI relatively concordant with actual BMI. They were characterized as having a normal BMI (18.5–25), overweight (25–30), or obese (>30). Two groups were characterized as outliers: these included individuals whose metabolome predicted a substantially lower mBMI than the actual BMI (mBMI ≪ BMI, residual < −0.5) or a substantially higher mBMI than the actual BMI (mBMI ≫ BMI, residual > 0.5). While these two outlier groups had the same weight and age distribution (Figure 2B), they had very different values for many of the phenotypes of metabolic health collected from these cohorts (Figures 2B and 2C). Individuals with an mBMI prediction that was substantially lower than their actual BMI had levels of insulin resistance, total triglycerides, HDL, blood pressure, waist/hip ratio, android/gynoid ratio, percent body fat, percent visceral fat, and percent subcutaneous fat that were similar to normal-weight individuals with healthy metabolomes. Individuals with an mBMI prediction that was substantially higher than their actual BMI had levels for these traits that were similar to those of obese individuals with obese metabolomes. Evaluating these data from a more clinical perspective, with individuals separated into clinical categories such as normal BMI with obese metabolome and obese BMI with healthy metabolome, generally confirmed these effects (Figures 3 and S2). Our findings suggest that the metabolome can be used as a clinically meaningful instrument, where obesity is analyzed in the context of its metabolome perturbation rather than just on BMI alone. Thus, our results are important in the frame of the current debate on the metabolically “healthy” obese and also for the identification of individuals with normal BMI (Chen et al., 2015) but poor metabolic health (Caleyachetty et al., 2017, Iglesias Molli et al., 2017).
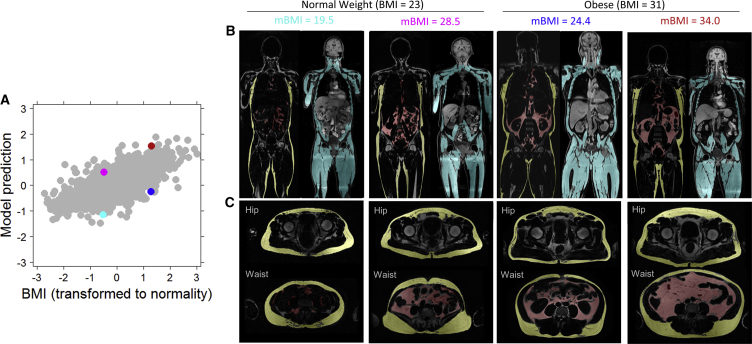
Figure 3.
Body Composition Profiles from Dixon Magnetic Resonance Imaging for Four Outlier Individuals
(A) Correlation between ridge regression model prediction of BMI and actual BMI for all unrelated individuals of European ancestry in the TwinsUK and Heath Nucleus dataset. Outliers highlighted in (B) and (C) are marked with corresponding colors. All individuals highlighted are from the outlier mBMI ≫ BMI or mBMI ≪ BMI categories shown in Figure 2.
(B) Body composition profiles (red, visceral adipose tissue; yellow, subcutaneous adipose tissue; cyan, muscle).
(C) Waist to hip cross-sections (hip, mid femoral head; waist, top of ASIS).
Having characterized these outliers, we revisited their metabolome differences. As the 49 metabolites were chosen for their association with BMI, there is no a priori expectation that they would all show differences between the group with mBMI ≫ BMI and the group with mBMI ≪ BMI, as these two groups have the same BMI distribution. However, we found that these two outlier groups did indeed have statistically significant differences in their metabolite levels for all of the 49 metabolites but two: asparagine and cortisone (Figure S3). This lack of association of cortisone with mBMI/BMI outliers shows that its correlation with BMI does not appear to extend to metabolic health, which presents interesting questions about the underlying biology. Although cortisone’s parent compound cortisol has been reported as associated with obesity, there are inconsistent relationships between cortisol and metabolic parameters in the literature (Abraham et al., 2013, Incollingo Rodriguez et al., 2015). It is also known that cortisol levels change throughout the day, and this factor could have influenced our results as the participants were not all measured at the same time. We additionally investigated the association between each of the BMI-associated metabolites and insulin resistance, as many previously reported markers of obesity have also been markers of diabetes (Ho et al., 2016, Park et al., 2015). We had quantitative insulin resistance measurements for 515 unrelated, European-ancestry participants. After controlling for BMI, we found that 12 of the 49 BMI-associated metabolites were also significantly associated (correcting for 49 tests requires p < 0.001) with insulin resistance, all with positive directions of effect: tyrosine, alanine, kynurenate, gamma-glutamyltyrosine, 1-oleoyl-3-linoleoyl-glycerol (18:1/18:2), six phospholipids, and, as expected, glucose (Table S1). Mannose, which recently underwent extensive study with regard to insulin resistance (Lee et al., 2016), was nominally associated with insulin resistance after controlling for BMI in our study (p = 0.004).
Evolution of Obesity and Metabolome Clinical Profiles
Given recent work suggesting that obese individuals who are metabolically healthy may remain at higher risk of negative health outcomes than are normal weight individuals who are metabolically healthy (Caleyachetty et al., 2017), we next asked whether the outlier groups were more likely to become obese over time. Focusing on the 1,458 individuals from TwinsUK who had weight measurements at all three time points, we found that those who had an mBMI that was higher than their BMI were marginally more likely to gain weight and convert to an obese phenotype (BMI > 30) over the 8–18 years of follow up. For example, 32.8% of those of normal weight but with an overweight or obese metabolome converted to being overweight or obese by time point 3 compared to 24.8% of those who were of normal weight and had a healthy metabolome (p = 0.01; Figure 4). However, this association was not strongly supported, and overall, the mBMI states of the individuals remained fairly stable with time and were a function of BMI changes (Figure 4). For example, 68% of the individuals who began the study with an obese metabolome ended the study with an obese metabolome. When an individuals’ weight increased and then decreased, their mBMI followed suit, and no single metabolite was significantly predictive of subsequent BMI changes. In summary, our results are consistent with a favorable long-term health benefit for the overweight and obese individuals with a healthy metabolome.
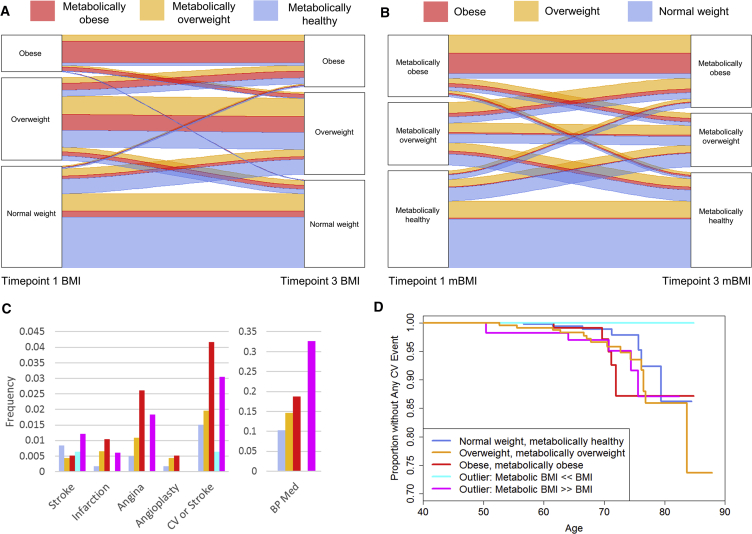
Figure 4.
Progression of Different mBMI/BMI Categories
(A) Alluvial plot showing the proportion of participants who remained in the same weight category or transitioned to a different weight category over the course of the 8–18 years of the TwinsUK study. Red individuals have an obese metabolome, orange individuals have an overweight metabolome, and gray individuals have a normal metabolome.
(B) Alluvial plot showing the proportion of participants who remained in the same mBMI category or transitioned to a different mBMI category over the course of the 8–18 years of the TwinsUK study. Red individuals begin the study with an obese BMI, orange overweight, and gray normal weight.
(C) Bar graph showing baseline rates of stroke, infarction, angina, angioplasty, any of the above, and blood pressure medication use at baseline for TwinsUK (with colors indicating groups as in D; n = 1,573 for information on cardiovascular and stroke events and 379 for information on blood pressure medication). Those with mBMI ≪ BMI had none of these conditions at baseline except stroke, while those with mBMI ≫ BMI had rates similar to those of overweight and obese individuals.
(D) Survival plot showing age until cardiac event (infarction, angina, or angioplasty). The plot is divided into those whose mBMI corresponds with their BMI (normal weight (n = 598), overweight (n = 461), and obese (n = 192) categories) as well as the two outlier groups: those with mBMI ≪ BMI (n = 158) and those with mBMI ≫ BMI (n = 164) (p = 0.003 for a difference between these categories in cardiovascular outcomes).
Cardiovascular Disease Outcomes
Obesity is considered a risk factor for cardiovascular disease and ischemic stroke (Rhee, 2018). In the TwinsUK study, 32 of 1,573 individuals reported having had a cardiovascular event (myocardial infarct, angina, or angioplasty) or stroke prior to baseline. We found significant differences between mBMI/BMI groups in their reported baseline cardiovascular events (Fisher’s exact p = 0.01). In particular, the mBMI ≪ BMI outliers had no history of cardiovascular events, whereas the mBMI ≫ BMI outliers had a proportion of historical events that was similar to those of overweight and obese individuals (Figure 4C). Information about blood pressure medication use was also available for 379 individuals at baseline, and those with mBMI ≫ BMI had very high rates of use compared to mBMI ≪ BMI (Fisher’s exact p = 0.00003; Figure 4C). The longitudinal nature of the TwinsUK study also allowed the collection of clinical endpoints in these unselected participants. The age of participants at the first visit ranged from 33 to 74 years old (median 51), and 42 to 88 years old (median 65) at the last visit. During the follow up (median 13 years), the study recorded 53 cardiovascular events (myocardial infarct, angina, or angioplasty) or strokes for 1,573 individuals. We calculated that our study had 80% power to identify effects with a hazard ratio of at least 1.5 for differences in cardiovascular event outcomes between the different mBMI/BMI groups. We found that participants with a healthy metabolome (normal BMI, overweight, or obese) had 2.6 events per hundred individuals. Individuals with an obese metabolic profile, mBMI, had 3.4 (normal/overweight BMI) and 4.4 events (in obese individuals) per hundred individuals. Separated analysis of the various endpoints confirmed the trends, more accentuated for cardiovascular than for diagnosis of stroke. We then performed a formal survival analysis for participants to have any cardiovascular event after the first time point, and we found those with healthier metabolomes to have fewer/later cardiac events (p = 0.003; Figure 4D).
Correlations between Twins
Because twin studies are important to analyze the heritability of traits, we reassessed the BMI model predictions and obesity status of 350 sets of twins where either both twins had normal BMI (n = 244), both twins were obese (n = 67), or one was obese and the other had normal BMI (n = 39). To keep the categories clear, individuals with BMIs between 25 and 30 (overweight) and their twins were excluded. As asserted by the model’s high specificity and sensitivity, the metabolite-based obesity predictions reflected the actual obesity status of the individuals. This was even the case when only one twin was obese: the obese twin was generally predicted by their metabolome to be obese, while the normal weight twin was not (Figure S1). The correlations between the metabolite-based obesity predictions were also substantially higher between the monozygotic twins than the dizygotic twins, as expected. Interestingly, we identified three sets of twins where both twins were predicted from the metabolome to be of normal weight, but both were obese, and eight sets of twins where the reverse was true. These outliers were thought to represent healthy obese and normal weight, metabolically unhealthy individuals described above.
Genetic Analyses
Known Genetics of Obesity
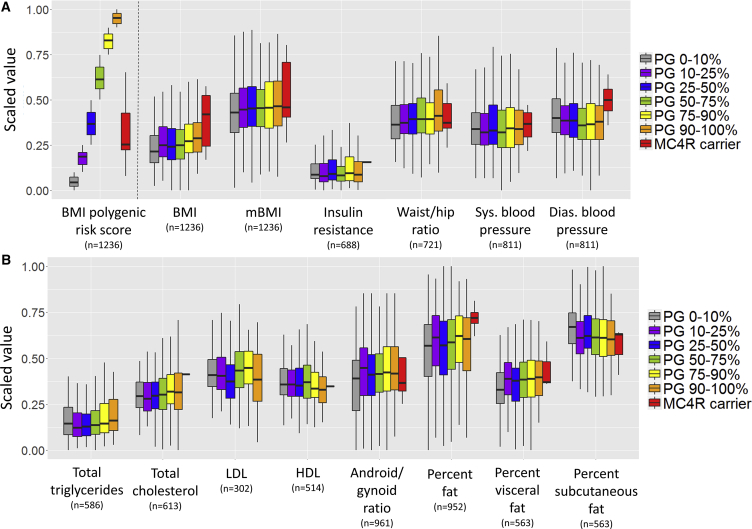
We first investigated the known genetic factors contributing to high BMI. We calculated polygenic risk scores for BMI using known associations from the considerable literature of obesity and BMI GWAS (genome-wide association study) (Locke et al., 2015). As previously reported, we found that polygenic risk score only explained 1.2%–2.2% of the variation in BMI at each of the three TwinsUK time points and in Health Nucleus for unrelated participants of European ancestry (Figure S4). We investigated whether unique individuals with the highest polygenic risk scores would have a significant perturbation of the metabolome and anthropomorphic, insulin resistance, and DEXA measurements (Figure 5). While the data did not support a strong role for polygenic risk, there were trends for higher polygenic risk scores to be associated with a higher android/gynoid ratio (p = 0.04), waist/hip ratio (p = 0.03), and triglycerides (p = 0.01). However, there was no statistical association between the polygenic score and mBMI (p = 0.07).
Figure 5.
Genetic Risk Compared to BMI-Relevant Variables
(A) Correlation between polygenic risk score (PG) category, MC4R carrier status, and BMI and anthropomorphic and clinical measurements for all unrelated individuals of European ancestry in the TwinsUK and HN dataset. All y axis values are scaled to a range from 0 to 1 to allow comparison across groups.
(B) The same process is used to show blood work and DEXA imaging values. While there was a trend for genetic risk to be associated with various measurements, the polygenic risk score achieved nominal p < 0.05 for BMI, waist/hip ratio, android/gynoid ratio, and triglycerides, and MC4R carrier status achieved nominal p < 0.05 for BMI. LDL, low-density lipoprotein; HDL, high-density lipoprotein.
Whiskers of boxplot extend to the most extreme points no greater than 1.5 times the interquartile range (distance between the first and third quartiles).
Studies of rare variants in obesity have identified melanocortin 4 receptor (MC4R) mutations as having effects large and clear enough to be appropriate for study in our dataset (Collet et al., 2017, Turcot et al., 2018). We therefore identified members of the study populations who were carrying rare (MAF < 0.01%) coding variants in MC4R. We identified eight such carriers in the subset of unrelated participants (Table 2). Each variant was observed in one unrelated individual, and six of the eight had already been annotated as causing obesity in clinical databases HGMD or ClinVar (Table 2). As a group, MC4R carriers had significantly higher BMI (p = 0.02) than did non-carriers as well as non-significant trends toward a higher diastolic blood pressure, insulin resistance, and percent body fat (Figure 5). However, it is known that not all rare variants are deleterious, and the metabolic impact could have been greater for the true subset of functional variants. The BMI data in the participants supported a pathogenic role for at least five of the variants (Met292fs, Arg236Cys, Ser180Pro, Ala175T, and Thr11Ala), but did not corroborate a role of Ile170V, which is defined in HGMD, ClinVar, and the literature as pathogenic (Clément et al., 2018, Collet et al., 2017, Landrum et al., 2018, Stenson et al., 2003). Importantly, of the five sets of twins who both carried the same MC4R variant, three sets included twins who were both overweight or obese. In the two cases where a carrier’s twin did not have the MC4R variant, their BMI was lower than their twin’s. We observed an enrichment of MC4R variant carriers among obese individuals with low polygenic risk scores (Figure S4). Out of 37 participants who were obese with polygenic risk scores in the lowest quartile, 5.4% were MC4R variant carriers, while the carrier frequency was just 0.4% in those of normal weight.
Table 2.
Variants Identified in MC4R or Lipodystrophy Genes in Participants of European Ancestry
| Gene | Variant | Protein Change | Study MAF | Global Gnomad MAF | Known Functional Annotation | Carrier BMI (mBMI) | Carrier Twin BMI (mBMI) | Non-carrier Twin BMI (mBMI) | Twin Zygo-sity |
|---|---|---|---|---|---|---|---|---|---|
| Obesity | |||||||||
| MC4R | chr18:60371541 G/A | p.Ser270Phe | 0.04% | 0.00% | none | 25.7 (26.1) | 24.8 (24.7) | N/A | MZ |
| MC4R | chr18:60372307 G/A | p.Leu15Phe | 0.04% | 0.00% | none | 23 (22.6) | 22.6 (22.4) | N/A | MZ |
| MC4R | chr18:60371474 CA/C | p.Met292fs | 0.04% | <0.003% | (complete LOF)b | 32.8 (26) | N/A | 28.8 (24.3) | DZ |
| MC4R | chr18:60371644 G/A | p.Arg236Cys | 0.04% | 0.00% | HGMD highC DM (not LOF)b | 34.5 (32.3) | 30 (27.5) | N/A | DZ |
| MC4R | chr18:60372319 T/C | p.Thr11Ala | 0.04% | <0.003% | HGMD lowC DM (not LOF)b | 36 (31.7) | N/A | N/A | N/A |
| MC4R | chr18:60371812 A/G | p.Ser180Pro | 0.04% | <0.003% | ClinVar LP (partial LOF)b | 34.2 (30.2) | 34.4 (33.5) | N/A | DZ |
| MC4R | chr18:60371827 C/T | p.Ala175Thr | 0.04% | 0.02% | ClinVar P HGMD highC DM (partial LOF)b | 29 (25.6) | 28.5 (24.4) | N/A | MZ |
| MC4R | chr18:60371842 T/C | p.Ile170Val | 0.04% | 0.01% | ClinVar P HGMD highC DM (partial LOF)b | 22.6 (23.7) | N/A | 21.3 (24.7) | DZ |
| Lipodystrophy | |||||||||
| ZMPSTE24 | chr1:40290870 G/GT | p.Leu362fs | 0.11% | 0.03% | ClinVar P | 18 (18.9) | N/A | N/A | N/A |
| ZMPSTE24 | chr1:40290870 G/GT | p.Leu362fs | 0.11% | 0.03% | ClinVar P | 22 (20.3) | N/A | N/A | N/A |
| ZMPSTE24 | chr1:40290870 G/GT | p.Leu362fs | 0.11% | 0.03% | ClinVar P | 22.4 (26.1) | N/A | 22.6 (25.1) | DZ |
| ZMPSTE24 | chr1:40290870 GT/G | p.Leu362fs | 0.04% | <0.003% | not annotateda | 30.7 (27.5) | N/A | 24.9 (24.1) | DZ |
| AGPAT2 | chr9:136673876 G/C | p.Ala238Gly | 0.04% | 0.00% | HGMD highC DM | 20 (23.3) | N/A | N/A | N/A |
| LIPE | chr19:42401821 CCCCCCGCAGCCC CCGTCTA/C | p.Val1068fs | 0.04% | 0.07% | ClinVar P | 23 (27.4) | N/A | N/A | N/A |
| BSCL2 | chr11:62692371 C/T | c.863+5G>A | 0.04% | <0.003% | ClinVar P | 24 (29.9) | N/A | N/A | N/A |
MAF, minor allele frequency; HGMD highC DM, Human Gene Mutation Database high-confidence disease-causing mutation; lowC, low confidence; ClinVar LP, likely pathogenic; P, pathogenic; MZ, monozygotic; DZ, dizygotic. Each variant was only seen once in the unrelated participants of this study. See alignments in Figure S5.
This deletion at the same site as a lipodystrophy insertion has not previously been annotated.
Functional annotation according to Collet et al. (2017), where LOF = loss of function.
We also searched the study population for rare variants associated with Mendelian lipodystrophy syndromes. Lipodystrophies are heterogeneous, genetic, or acquired disorders characterized by selective loss of body fat and associated metabolic complications such as diabetes mellitus, hypertriglyceridemia, and hepatic steatosis (Garg, 2011). We identified six individuals with rare heterozygous variants in four genes: ZMPSTE24, AGPAT2, LIPE, and BSCL2 (Table 2). The individuals with LIPE or BSCL2 variants were of normal weight but were in the mBMI ≫ BMI outlier category. The three individuals with ZMPSTE24 p.Leu362fs caused by a 1-bp insertion had normal to low BMI and mBMI. We also observed an unannotated 1-bp deletion at this site in one obese individual (Table 2; Figure S5). While these observations are interesting, there are no data in the literature to support a phenotypic role for these variants in carriers (heterozygotes).
Genetics of the Metabolically Healthy Obese
Individuals with an mBMI that was substantially lower than their actual BMI had a higher polygenic risk score for BMI than did other groups. In contrast, those whose mBMI was substantially higher than their actual BMI had low polygenic risk scores (Figure 2B; p = 0.08 for a difference between these two groups). This result would support the notion that the polygenic risk score for BMI may capture an anthropomorphic phenotype (larger-framed individuals) rather than a unique association with obesity as a disease trait.
Genetics of Metabolome Differences
Last, we investigated whether obese individuals with different genetic backgrounds had different metabolomes from other obese individuals. We first searched for metabolites, BMI-associated or otherwise, that could distinguish individuals with different BMI polygenic risk scores or MC4R variant carriers. Linear regression showed no significant associations between any single metabolites and polygenic risk or MC4R carrier status in either the entire population or in only the obese individuals. This result implies that metabolites are unlikely to be intermediate phenotypes that explain the underlying genetics of obesity. To check for more specific signals beyond the compiled polygenic risk score, we also performed separate analyses of each of the 97 variants that are used to calculate the polygenic risk score. We found no evidence for any of these known GWAS variants to be more strongly associated with a metabolite than with BMI itself, though our power for discovery was limited given the very small effect sizes of most individual GWAS variants. In summary, although it is known that there is a strong genetic component to metabolite levels (Long et al., 2017), most of the metabolic perturbations that occur in the obese state are a response to obesity as opposed to shared genetic mechanisms.
Discussion
The results of the present study highlight the profound disruption of the metabolome in obesity and identify a metabolome signature that serves to examine metabolic health without the limitations of anthropomorphic measurements or the cost, time, and equipment requirements of imaging technologies. Nearly one-third of the approximately 1,000 metabolites measured in the study were associated with BMI, and 49 were selected as a strong signature for the study of the relationship between BMI, obesity, metabolic disease, and the genetics of BMI.
Consistent with previous studies and earlier work in the TwinsUK cohort, branched-chain and aromatic amino acids, and metabolites involved in nucleotide metabolism, such as urate and pseudouridine, are strongly perturbed by obesity (Butte et al., 2015, Ho et al., 2016, Menni et al., 2017, Park et al., 2015). The underlying reason for the perturbation of branched-chain amino acid metabolism in obese individuals and those with insulin resistance is thought to be related to differences in the amino acid catabolism in adipose tissue (Newgard, 2012). The single metabolite with the most significant association with BMI was urate, as we previously reported (Menni et al., 2017). It is well known that uric acid increases with obesity, due to insulin resistance reducing the kidneys’ ability to eliminate uric acid, but previous work has not emphasized the power of urate to predict BMI (Butte et al., 2015, Facchini et al., 1991, Ho et al., 2016). We also found a strong signal for lipids to be associated with BMI, with an enrichment of associations found for glycerol lipids. These results are consistent with previous studies showing that sphingomyelins and diacylglycerols increase with BMI while lysophosphocholines decrease with BMI, with other various phosphatidylcholines having effects in both directions (Butte et al., 2015, Ho et al., 2016, Park et al., 2015). A number of BMI-associated metabolites (12 of the 49-metabolite signature) were associated with insulin resistance after controlling for BMI. As previously observed (Caleyachetty et al., 2017, Oberbach et al., 2011, Reinehr et al., 2015, Wahl et al., 2015), the metabolome abnormalities associated with high BMI corrected with loss of weight. However, our study found that metabolite levels did not provide predictive power for future weight changes. Overall, the metabolome perturbations appear as a consequence of changes in weight as opposed to being a contributing factor.
While BMI correlates well and to a large extent with individual health outcomes, it does not have the sensitivity to identify outliers, some of which carry unique health consequences. These limitations to BMI are recognized in the literature (Neeland et al., 2018). Here, the metabolome signature identified individuals whose predicted mBMI was either substantially lower or higher than their actual BMI, i.e., individuals with lesser or greater metabolic consequences of obesity, using a technology that is amenable to large-scale implementation. These individuals include the metabolically healthy obese, but we also emphasize the importance of the metabolome anomalies in identifying unhealthy individuals with a normal BMI. These profiles were generally stable over the prolonged follow up. An abnormal metabolome was associated in the present study with an approximately 5-fold increase in baseline cardiovascular events and additional 2-fold increase in cardiovascular events during follow up when comparing individuals who were matched for BMI but who had opposing metabolome signatures. Thus, while our findings are in line with the known relationships between metabolically healthy obese status and health-related traits like metabolic syndrome and body fat (Brochu et al., 2001, Caleyachetty et al., 2017, Karelis et al., 2005), we extend this relationship to the broader category of metabolically healthy and unhealthy individuals on the basis of the disparity between mBMI and BMI. For example, we observed differences in waist/hip ratio, percent visceral fat, and blood pressure between mBMI/BMI outliers despite having the same BMI distribution. The fact that the metabolically healthy obese have a high BMI polygenic risk score also supports the concept that some of the genetic studies may capture anthropomorphic associations—body size—rather than obesity sensu stricto. Overall, the health consequences observed across the various mBMI groups indicate that there is a durable benefit of maintaining a healthy metabolome signature and point to an ongoing risk for the individuals that have an unhealthy metabolome despite stability of BMI.
In line with the observation that a richer set of biomarkers is more accurate than a narrower panel of single biomarkers, we observed that 650 metabolites could explain ~50% and a 49-metabolite signature of the best markers could explain 43% of variance in BMI. In contrast, a conventional model that included age, sex, HDL, LDL, total cholesterol, and triglycerides could explain 31%, while the best single metabolic marker (urate) could explain 16% of the variance in BMI. The message that emerges from these types of observations—and that will become central to the implementation of machine and deep learning in medicine—is that there is a substantial loss of signal associated with the quest for a single biomarker. The medical system would, however, need to validate and establish the health economics of rich data biomarkers to foster adoption in clinical diagnostic routine.
In contrast with metabolomics analyses, the present study does not support a strong association between metabolome changes and the genetics of BMI defined by a 97-variant polygenic risk score (Locke et al., 2015). This may be explained by the fact that known BMI GWAS loci explain only a small fraction (∼3%) of BMI heritability (Locke et al., 2015). Despite this overall lack of explanatory power, there was a clear signal for individuals with higher polygenic risk scores to have greater rates of obesity. Because the genetic risk score does not include rare variants, we also identified individuals who carried rare functional variants in the known obesity gene MC4R, which is the single best example of a gene where rare coding variants have a large effect on obesity (Turcot et al., 2018). The carriers of these variants were often obese individuals, but their metabolome was not categorically different from that of other obese individuals. The lack of metabolome differences for carriers of variants in this gene is not surprising given that MC4R variants cause obesity by increasing appetite. However, we did find that obese carriers of MC4R variants often had low polygenic risk scores for obesity; out of 31 participants who were obese with polygenic risk scores in the lowest quartile, 5.4% were MC4R variant carriers, while the carrier frequency was just 0.4% in those of normal weight. Thus, our study shows the interest of sequencing obese individuals with low polygenic risk scores because of the apparent enrichment for monogenic contributions. As we completed this study, a large consortium provided additional detail on the role of variants in pathways that implicate energy intake and expenditure in obesity (Turcot et al., 2018). Future studies should also explore the role of rare functional variants in lipodystrophy genes on the metabolic traits in the general population.
In summary, the present study highlights the health risks of the perturbed metabolome. The study also indicates that the genetics of BMI are separate from metabolic health and serves to prioritize a subset of individuals for genetic analysis. The assessment of the metabolome and genome of BMI lays groundwork for future studies of the heterogeneity of obesity and treatment of its endophenotypes. Specifically, the methodology used here can be applied to build similar models for insulin resistance, fat distribution, or any other number of clinical traits, and the mBMI metabolome signature can act as a biomarker of response to the new therapeutics that target patients with MC4R mutations (Kühnen et al., 2016). In the future, metabolic profiling could help select patients for clinical trials beyond genetic sequencing, thus expanding drug utility (Yanovski and Yanovski, 2018).
Limitations of Study
While the uniquely rich metabolomic and phenotypic data in this study permitted a thorough investigation of obesity-related health, there were nonetheless limitations. We chose to focus this study on BMI, the most widely used measurement of obesity. However, future studies building models for other traits of metabolic health, for example diabetes or visceral fat, may be of greater utility. Our study also had limited longitudinal information on medication use, which precluded an analysis of how drug prescription influenced mBMI/BMI categories and vice versa. Additionally, the association we observed between mBMI/BMI categories and longitudinal cardiovascular outcomes requires confirmation in a replication cohort.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| gnomad, Genome Aggregation Database | http://gnomad.broadinstitute.org/ | RRID: SCR_014964 |
| ClinVar | https://www.ncbi.nlm.nih.gov/clinvar/ | RRID: SCR_006169 |
| HGMD, Human Gene Mutation Database | http://www.hgmd.cf.ac.uk/ac/index.php | RRID: SCR_001888 |
| Study data deposited into Mendeley Data | This paper; https://data.mendeley.com/ | https://doi.org/10.17632/kpjsnwd2mc.1 |
| Study data and scripts deposited into GitHub | This paper; https://github.com | https://github.com/humanlongevity/mbmi |
| Software and Algorithms | ||
| R Project for Statistical Computing | http://www.r-project.org/ | RRID: SCR_001905 |
| FaST LMM | https://github.com/MicrosoftGenomics/FaST-LMM | RRID: SCR_015506 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elizabeth Cirulli (liz.cirulli@gmail.com).
Experimental Model and Subject Details
The study included 1,969 largely European ancestry twins enrolled in the TwinsUK registry, a British national register of adult twins (Moayyeri et al., 2013). We previously reported a detailed study of the genetic variants influencing the human metabolome in this cohort (Long et al., 2017). Serum samples were collected at three visits, 8-18 (median 13) years apart. The cohort is mainly composed of females (96.7%), and the sample set we used included 388 monozygotic twin pairs, 519 dizygotic twin pairs, and 155 unrelated individuals. The age of participants at the first time point ranged from 33 to 74 years old (median 51); 36 to 81 years old (median 59) at the second time point; and 42 to 88 years old (median 65) at the third time point. The BMI values measured at each metabolome time point were taken within two years of the blood draw date. At baseline, 36.3% of the female participants and 53.8% of the male participants were overweight, and 16.9% of the females and 10.8% of the males were obese. The twins study was approved by St. Thomas’ Hospital Research Ethics Committee, and all participants provided informed written consent. BMI data were available for 1743 participants within two years of the time point for metabolome time point 1, 1834 for within two years of time point 2, and 1777 for up to 2 years before time point 3 or 4 years after this time point; 1,458 individuals had all three datapoints.
For independent validation and studies of phenotypes correlated with metabolic BMI outliers, we enrolled 427 unselected adults more than 18 years old who were able to come to the Health Nucleus in La Jolla, CA for a clinical research protocol (Perkins et al., 2017). Participants underwent a verbal review of the institutional review board-approved consent (Western Institutional Review Board). Participants ranged in age from 18-89 years old (median 53), were 32.9% female, and had BMI data measured at one time point: 16.7% of the female participants and 47.5% of the male participants were overweight, and 7.2% of the females and 23.7% of the males were obese.
Method Details
Phenotyping
Individuals in the TwinsUK cohort and Health Nucleus both underwent dual-energy X-ray absorptiometry (DEXA) imaging. The data from these scans were used to calculate android/gynoid ratio, percent body fat, visceral fat, and subcutaneous fat. DEXA is very accurate in the measurement abdominal visceral adipose tissue (VAT). High levels of VAT are associated with atherogenic dyslipidemia, hyperinsulinemia, and glucose intolerance (Neeland et al., 2018). TwinsUK cohort participants were additionally measured for circumference at the waist and hip using a measuring tip to calculate the waist/hip ratio. TwinsUK participants self-reported information about whether they were taking high blood pressure medication at their first visit and about cardiovascular events and their timing via a survey at the final visit. For a selected number of Health Nucleus participants, images of fat and water (imaging of muscle) were available from symmetrical chemical shift Magnetic Resonance Imaging (MRI) via the Dixon method. Quantitative insulin resistance (homeostatic model assessment, HOMA) was calculated as fasting insulin x fasting glucose / 405, and being insulin resistant was defined by HOMA score ≥ 3 (http://gihep.com/calculators/other/homa/) (Matthews et al., 1985).
Metabolite Profiling
The non-targeted metabolomics analysis of 901 metabolites in the TwinsUK cohort and 1,007 metabolites in the Health Nucleus cohort was performed at Metabolon (Durham, North Carolina, USA) on a platform consisting of four independent ultra high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) methods. The detailed descriptions of the platform can be found in our previous publications (Cohen et al., 2018, Long et al., 2017). For the TwinsUK cohort, blood serum after fasting was used for analysis, and the resulting raw values were transformed to z scores using the mean and standard deviation. For the Health Nucleus cohort, blood plasma after fasting was used for analysis, and values from multiple experimental batches were normalized into Z-scores based on a reference cohort of either 42 (n = 457) or 300 (n = 176) self-reported healthy individuals run with each batch. The 42 and 300-normalized batches were converted to the same scale using linear transformation based on the values obtained from 7 runs that included both the 42 and 300 controls. Samples with metabolite measurements that were below the detection threshold were imputed as the minimum value for that metabolite.
Genome Sequencing and Analysis
As previously described (Telenti et al., 2016), DNA samples were sequenced on an Illumina HiSeqX sequencer utilizing a 150 base paired-end single index read format. Reads were mapped to the human reference sequence build HG38. Variants were called using ISIS Analysis Software (v. 2.5.26.13; Illumina). A linear mixed model was applied to account for family structure in the cohort while testing for associations between genetic variants and the different phenotypes: BMI; BMI prediction model values and residuals after accounting for BMI, age, sex; and levels of the 49 BMI-associated metabolites. A genetic similarity matrix (GSM) was constructed from 301,556 variants that represented a random 20% of all common (MAF > 5%) variants genome-wide after linkage-disequilibrium (LD) pruning (r2 less than 0.6, window size 200 kb) and was used to model the random effect in the linear mixed model via a “leave-out-one-chromosome” method for each tested variant. Each of 97 known BMI-associated variants was tested independently using customized Python scripts wrapping the FaST-LMM package (FaST LMM, RRID: SCR_015506) (Lippert et al., 2011, Locke et al., 2015). Principal component axes were calculated to check ethnicity using plink, and the first principal component for those of European ancestry was used as a covariate in analyses of unrelated individuals in R described below. Polygenic risk scores were calculated using genotypes for 97 variants whose associations and betas had been published previously (Locke et al., 2015). Rare variants in the gene MC4R were defined as coding and splice variants with MAF < 0.1%. Rare lipodystrophy variants were defined as those achieving a pathogenic or likely pathogenic categorization in ClinVar (ClinVar, RRID: SCR_006169) or HGMD (HGMD, RRID: SCR_001888). Allele frequencies were calculated in our set of samples and also retrieved from gnomad (Genome Aggregation Database, RRID: SCR_014964).
Quantification and Statistical Analysis
R (R Project for Statistical Computing, RRID: SCR_001905) was used for the analysis and data manipulation. Bonferroni correction was used for all analyses, and most statistical analyses were restricted to unrelated individuals of European ancestry, in accordance with field standards for ensuring that ancestry differences do not cause bias or skew in the results. For each quantitative analysis of BMI or other traits, the subset of BMI values or other outcome variables used were rank-ordered and forced to a normal distribution. Analyses comparing metabolites to BMI were performed in R using the lm function, and age, sex, and the first genetic principal component were included as covariates. The obesity prediction model was built using ridge regression (alpha = 0) with glmnet in R. The residuals used to separate participants into the five categories shown in Figure 2 were calculated using age, sex, and initial BMI. Heatmaps were generated in R using the pheatmap package. Survival analysis was performed using coxph in R with age at first visit included as a covariate. Power calculation was performed using the power.stratify command in the powerSurvEpi package in R.
Data and Software Availability
The R analysis scripts and data files used are available at https://github.com/humanlongevity/mbmi. Data files have been deposited to Mendeley Data and are available at https://doi.org/10.17632/kpjsnwd2mc.1.
Acknowledgments
We thank E. Muse for valuable comments. We acknowledge N. Schenker-Ahmed, L. Huang, M. Tyagi, and P. Sheth for aiding with data collection. TwinsUK receives funding from the Wellcome Trust, European Community’s Seventh Framework Programme (FP7/2007-2013, 277849, 201413, and 259749), the National Institute for Health Research (NIHR) Clinical Research Facility at Guy’s and St Thomas’ NHS Foundation Trust, and NIHR Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. A.T. is supported by the Qualcomm Foundation and the NIH Center for Translational Science Award (CTSA, grant number UL1TR002550).
Author Contributions
A.T. and E.T.C. conceived of the experiment(s); E.T.C. conducted the experiment(s); L.G. supported metabolome analyses and interpretation; and E.T.C., J.C.V., and A.T. analyzed the results. C.T.C. and B.T. provided detailed insight into metabolic diseases; C.L.S., N.S., L.H., E.F.K., and L.A.N. aided with data collection and interpretation; and T.D.S. is responsible for the TwinsUK study. All authors reviewed the manuscript.
Declaration of Interests
E.T.C., C.L.S., N.S., L.H., L.A.N., E.F.K., C.T.C., and J.C.V. are currently or recently employed at Human Longevity, Inc. L.G. was recently an employee of Metabolome, Inc. A.T., B.T., and T.D.S. declare no conflict of interest.
Published: October 11, 2018
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at https://doi.org/10.1016/j.cmet.2018.09.022.
Contributor Information
Elizabeth T. Cirulli, Email: liz.cirulli@gmail.com.
Amalio Telenti, Email: atelenti@scripps.edu.
Supplemental Information
References
- Abraham S.B., Rubino D., Sinaii N., Ramsey S., Nieman L.K. Cortisol, obesity, and the metabolic syndrome: a cross-sectional study of obese subjects and review of the literature. Obesity (Silver Spring) 2013;21:E105–E117. doi: 10.1002/oby.20083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björntorp P., Rosmond R. Obesity and cortisol. Nutrition. 2000;16:924–936. doi: 10.1016/s0899-9007(00)00422-6. [DOI] [PubMed] [Google Scholar]
- Brochu M., Tchernof A., Dionne I.J., Sites C.K., Eltabbakh G.H., Sims E.A., Poehlman E.T. What are the physical characteristics associated with a normal metabolic profile despite a high level of obesity in postmenopausal women? J. Clin. Endocrinol. Metab. 2001;86:1020–1025. doi: 10.1210/jcem.86.3.7365. [DOI] [PubMed] [Google Scholar]
- Butte N.F., Liu Y., Zakeri I.F., Mohney R.P., Mehta N., Voruganti V.S., Göring H., Cole S.A., Comuzzie A.G. Global metabolomic profiling targeting childhood obesity in the Hispanic population. Am. J. Clin. Nutr. 2015;102:256–267. doi: 10.3945/ajcn.115.111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caleyachetty R., Thomas G.N., Toulis K.A., Mohammed N., Gokhale K.M., Balachandran K., Nirantharakumar K. Metabolically healthy obese and incident cardiovascular disease events among 3.5 million men and women. J. Am. Coll. Cardiol. 2017;70:1429–1437. doi: 10.1016/j.jacc.2017.07.763. [DOI] [PubMed] [Google Scholar]
- Chen H.H., Tseng Y.J., Wang S.Y., Tsai Y.S., Chang C.S., Kuo T.C., Yao W.J., Shieh C.C., Wu C.H., Kuo P.H. The metabolome profiling and pathway analysis in metabolic healthy and abnormal obesity. Int. J. Obes. 2015;39:1241–1248. doi: 10.1038/ijo.2015.65. [DOI] [PubMed] [Google Scholar]
- Clément K., Biebermann H., Farooqi I.S., Van der Ploeg L., Wolters B., Poitou C., Puder L., Fiedorek F., Gottesdiener K., Kleinau G. MC4R agonism promotes durable weight loss in patients with leptin receptor deficiency. Nat. Med. 2018;24:551–555. doi: 10.1038/s41591-018-0015-9. [DOI] [PubMed] [Google Scholar]
- Cohen I.V., Cirulli E.T., Mitchell M.W., Jonsson T.J., Yu J., Shah N., Spector T.D., Guo L., Venter J.C., Telenti A. Acetaminophen (paracetamol) use modifies the sulfation of sex hormones. EBioMedicine. 2018;28:316–323. doi: 10.1016/j.ebiom.2018.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet T.H., Dubern B., Mokrosinski J., Connors H., Keogh J.M., Mendes de Oliveira E., Henning E., Poitou-Bernert C., Oppert J.M., Tounian P. Evaluation of a melanocortin-4 receptor (MC4R) agonist (Setmelanotide) in MC4R deficiency. Mol. Metab. 2017;6:1321–1329. doi: 10.1016/j.molmet.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini F., Chen Y.D., Hollenbeck C.B., Reaven G.M. Relationship between resistance to insulin-mediated glucose uptake, urinary uric acid clearance, and plasma uric acid concentration. JAMA. 1991;266:3008–3011. [PubMed] [Google Scholar]
- Garg A. Clinical review#: lipodystrophies: genetic and acquired body fat disorders. J. Clin. Endocrinol. Metab. 2011;96:3313–3325. doi: 10.1210/jc.2011-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales C.M., Carroll M.D., Fryar C.D., Ogden C.L. Prevalence of obesity among adults and youth: United States, 2015-2016. NCHS Data Brief. 2017:1–8. [PubMed] [Google Scholar]
- Ho J.E., Larson M.G.M., Ghorbani A., Cheng S., Chen M.-H., Keyes M., Rhee E.P.E.E.P., Clish C.B., Vasan R.S.R., Gerszten R.E., Wang T.J. Metabolomic profiles of body mass index in the Framingham Heart Study reveal distinct cardiometabolic phenotypes. PLoS One. 2016;11:e0148361. doi: 10.1371/journal.pone.0148361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias Molli A.E., Penas Steinhardt A., López A.P., González C.D., Vilariño J., Frechtel G.D., Cerrone G.E. Metabolically healthy obese individuals present similar chronic inflammation level but less insulin-resistance than obese individuals with metabolic syndrome. PLoS One. 2017;12:e0190528. doi: 10.1371/journal.pone.0190528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incollingo Rodriguez A.C., Epel E.S., White M.L., Standen E.C., Seckl J.R., Tomiyama A.J. Hypothalamic-pituitary-adrenal axis dysregulation and cortisol activity in obesity: a systematic review. Psychoneuroendocrinology. 2015;62:301–318. doi: 10.1016/j.psyneuen.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Karelis A.D., Faraj M., Bastard J.P., St-Pierre D.H., Brochu M., Prud’homme D., Rabasa-Lhoret R. The metabolically healthy but obese individual presents a favorable inflammation profile. J. Clin. Endocrinol. Metab. 2005;90:4145–4150. doi: 10.1210/jc.2005-0482. [DOI] [PubMed] [Google Scholar]
- Kühnen P., Clément K., Wiegand S., Blankenstein O., Gottesdiener K., Martini L.L., Mai K., Blume-Peytavi U., Grüters A., Krude H. Proopiomelanocortin deficiency treated with a melanocortin-4 receptor agonist. N. Engl. J. Med. 2016;375:240–246. doi: 10.1056/NEJMoa1512693. [DOI] [PubMed] [Google Scholar]
- Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Zhang C., Kilicarslan M., Piening B.D., Bjornson E., Hallström B.M., Groen A.K., Ferrannini E., Laakso M., Snyder M. Integrated network analysis reveals an association between plasma mannose levels and insulin resistance. Cell Metab. 2016;24:172–184. doi: 10.1016/j.cmet.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C.M., Davidson R.I., Heckerman D. FaST linear mixed models for genome-wide association studies. Nat. Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T., Hicks M., Yu H.-C., Biggs W.H., Kirkness E.F., Menni C., Zierer J., Small K.S., Mangino M., Messier H. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017;49:568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Menni C., Migaud M., Kastenmüller G., Pallister T., Zierer J., Peters A., Mohney R.P., Spector T.D., Bagnardi V., Gieger C. Metabolomic profiling of long-term weight change: role of oxidative stress and urate levels in weight gain. Obesity (Silver Spring) 2017;25:1618–1624. doi: 10.1002/oby.21922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyeri A., Hammond C.J., Hart D.J., Spector T.D. The UK Adult Twin Registry (TwinsUK Resource) Twin Res. Hum. Genet. 2013;16:144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeland I.J., Poirier P., Després J.-P. Cardiovascular and metabolic heterogeneity of obesity: clinical challenges and implications for management. Circulation. 2018;137:1391–1406. doi: 10.1161/CIRCULATIONAHA.117.029617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard C.B. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbach A., Blüher M., Wirth H., Till H., Kovacs P., Kullnick Y., Schlichting N., Tomm J.M., Rolle-Kampczyk U., Murugaiyan J. Combined proteomic and metabolomic profiling of serum reveals association of the complement system with obesity and identifies novel markers of body fat mass changes. J. Proteome Res. 2011;10:4769–4788. doi: 10.1021/pr2005555. [DOI] [PubMed] [Google Scholar]
- Park S., Sadanala K.C., Kim E.-K. A metabolomic approach to understanding the metabolic link between obesity and diabetes. Mol. Cells. 2015;38:587–596. doi: 10.14348/molcells.2015.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins B.A., Caskey C.T., Brar P., Dec E., Karow D., Kahn A., Hou C., Shah N., Boeldt D., Coughlin E. Precision medicine screening using whole genome sequencing and advanced imaging to identify disease risk in adults. bioRxiv. 2017 doi: 10.1073/pnas.1706096114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piening B.D., Zhou W., Contrepois K., Röst H., Gu Urban G.J., Mishra T., Hanson B.M., Bautista E.J., Leopold S., Yeh C.Y. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst. 2018;6:157–170.e8. doi: 10.1016/j.cels.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen E.P., Sahoo J.P., Kulshreshtha B., Khurana M.L., Gupta N., Dwivedi S.N., Kumar G., Ammini A.C. Morning cortisol is lower in obese individuals with normal glucose tolerance. Diabetes Metab. Syndr. Obes. 2011;4:347–352. doi: 10.2147/DMSO.S23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinehr T., Wolters B., Knop C., Lass N., Hellmuth C., Harder U., Peissner W., Wahl S., Grallert H., Adamski J. Changes in the serum metabolite profile in obese children with weight loss. Eur. J. Nutr. 2015;54:173–181. doi: 10.1007/s00394-014-0698-8. [DOI] [PubMed] [Google Scholar]
- Rhee E.J. Being metabolically healthy, the most responsible factor for vascular health. Diabetes Metab. J. 2018;42:19–25. doi: 10.4093/dmj.2018.42.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S.T., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Telenti A., Pierce L.C.T., Biggs W.H., di Iulio J., Wong E.H.M., Fabani M.M., Kirkness E.F., Moustafa A., Shah N., Xie C. Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. USA. 2016;113:11901–11906. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcot V., Lu Y., Highland H.M., Schurmann C., Justice A.E., Fine R.S., Bradfield J.P., Esko T., Giri A., Graff M., CHD Exome+ Consortium; EPIC-CVD Consortium; ExomeBP Consortium; Global Lipids Genetic Consortium; GoT2D Genes Consortium; EPIC InterAct Consortium; INTERVAL Study; ReproGen Consortium; T2D-Genes Consortium; MAGIC Investigators; Understanding Society Scientific Group Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S., Vogt S., Stückler F., Krumsiek J., Bartel J., Kacprowski T., Schramm K., Carstensen M., Rathmann W., Roden M. Multi-omic signature of body weight change: results from a population-based cohort study. BMC Med. 2015;13:48. doi: 10.1186/s12916-015-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.R., Soderberg S., Lindahl B., Olsson T. Independent effects of obesity and cortisol in predicting cardiovascular risk factors in men and women. J. Intern. Med. 2000;247:198–204. doi: 10.1046/j.1365-2796.2000.00609.x. [DOI] [PubMed] [Google Scholar]
- Whitlock G., Lewington S., Sherliker P., Clarke R., Emberson J., Halsey J., Qizilbash N., Collins R., Peto R., Prospective Studies Collaboration Body-mass index and cause-specific mortality in 900 000 adults: collaborative analyses of 57 prospective studies. Lancet. 2009;373:1083–1096. doi: 10.1016/S0140-6736(09)60318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Obesity and overweight. 2018. http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight
- Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirix A.J.G., Finken M.J.J., von Rosenstiel-Jadoul I.A., Heijboer A.C., Nauta J., Groothoff J.W., Chinapaw M.J.M., Kist-van Holthe J.E. Is there an association between cortisol and hypertension in overweight or obese children? J. Clin. Res. Pediatr. Endocrinol. 2017;9:344–349. doi: 10.4274/jcrpe.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovski S.Z., Yanovski J.A. Toward precision approaches for the prevention and treatment of obesity. JAMA. 2018;319:223–224. doi: 10.1001/jama.2017.20051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The R analysis scripts and data files used are available at https://github.com/humanlongevity/mbmi. Data files have been deposited to Mendeley Data and are available at https://doi.org/10.17632/kpjsnwd2mc.1.