Abstract
Exercise-induced skeletal muscle adaptation requires degradation of cellular components carried out by autophagy. However, the alteration of autophagy by different intensity of exercise in skeletal muscle is still unknown. In the present study, we investigate whether low, moderate, and high-intensity exercises have different impacts on autophagy gene expression in gastrocnemius and soleus muscles of wistar rats. This work is limited because only rats are used, and does not cover human tissues. Twenty male wistar rats were assigned to four groups: sedentary control, low-intensity (LI, 10 m/minute), moderate-intensity (MI, 20 m/minute), and high-intensity (HI, 30 m/minute) exercises. Training was conducted 30 minutes/day with a 5 times/week interval for 8 weeks. RNA and protein were extracted from gastrocnemius and soleus muscles then stored in -80°C. Specific primers and antibodies for autophagy genes and protein levels were utilized for semi-quantitative PCR and Western Blot. Exercises decrease expression of autophagy gene LC3 (LI and MI 0.7 fold, HI 0.8 fold, p < 0.05) in gastrocnemius muscles and soleus muscle (LI, MI, and HI 0.8 fold, p < 0.05) compared to control. On the other side, we observed p62 gene expression decreased in gastrocnemius (0.8 fold, p < 0.05) and soleus (0.9 fold, p < 0.05) muscles with MI, but increased in soleus (1.1 fold, p < 0.05) muscles with HI. This result is consistent with the change of protein level, suggesting that autophagy might be modulated by different type of exercise. This study suggests that intensity of exercise and different type of muscle fibers effect autophagy gene expression in skeletal muscle of wistar rats. MI exercise increases autophagy gene expression in gastrocnemius and soleus muscles, but HI exercise decreases autophagy gene expression in soleus muscles of wistar rats. Soleus muscles are more responsive to exercise compared to gastrocnemius muscles.
Key points.
Autophagy in skeletal muscle is altered by different intensity of exercise
Moderate intensity of exercise may increase autophagy in gastrocnemius and soleus muscles of wistar rats.
High intensity of exercise may decrease autophagy in soleus muscles of wistar rats.
Different types of muscle also affected autophagy gene expression in skeletal muscles of wistar rats.
Key words: Autophagy, exercise, gastrocnemius, soleus, LC3, p62
Introduction
It is well known that physical exercise improves physical performance and is recommended for the improvement of health and the prevention as well as treatment of many pathological conditions (Booth et al., 2012 ; Pedersen and Saltin., 2015). Phenotypic adaptations induced by physical exercise in skeletal muscle include angiogenesis, mitochondrial biogenesis, and fiber type transformation (Yan et al., 2011). These adaptations show the health benefits of physical exercise, but the molecular and cellular underlying exercise-induced skeletal muscle adaptations are still far from being completely understood. Addition and clearance of cellular components were needed for adaptation in exercise-induced skeletal muscle. While concern about addition of cellular components during skeletal muscle adaptation has increased, much less is known about clearance of cellular components in the process of adaptation (Lira et al., 2013).
Damaged elements are continuously removed and renovated in all living organisms (Mizushima and Komatsu, 2011), facilitating remodeling in exercise-induced skeletal muscles adaptation. This process involves identification of dysfunctional components, degradation, and recycle of its constituents as building blocks for the synthesis of new components with better quality (Martin-Rincon et al., 2018). Autophagy is the major intracellular degradation system by which cytoplasmic materials are delivered to and degraded in the lysosome. There are three classes of autophagy : macroautophagy, microautophagy, and chaperone-mediated autophagy (Mizushima and Komatsu, 2011). Macroautophagy, hereafter referred to as autophagy, is an evolutionarily conserved catabolic process that is responsible for the degradation of cellular components, such as protein aggregates, long-lived proteins, excess or damaged organelles (e.g., mitochondria), and intracellular pathogens (Levine and Klionsky, 2004; Cecconi and B. Levine, 2008). Autophagy involves the sequestration of proteins and/or organelles by double-membrane structures that form autophagosomes, which then fuse with lysosomes to degrade engulfed materials (Ravikumar et al., 2010).
Cell hemostasis is maintained by autophagy through a correct disposal of damaged cellular and organelles (Green and Levine, 2014). One of the main mechanisms of cellular adaptation to stress is upregulation or downregulation of autophagy. When cellular stress is increased, for example during exercise, autophagy is elevated to provide energy substrates and to adapt cellular structures to the newly elevated demands (He et al., 2012). Skeletal muscle is one of the tissues with the highest basal autophagy flux and greater capacity to increase autophagy flux (Mizushima et al., 2004). Different responses of autophagy in skeletal muscle have been reported following different kinds of physical exercise namely single bout exercise exposure and habitual/regular chronic exercise training (Tam and Siu, 2014).
Acute treadmill exercise has been shown to induce autophagy in skeletal muscle. Key autophagic proteins including LC3-II and p62 are found to be significantly altered after exercise. The change in autophagic proteins is consistent with the observation of increased autophagosome after exercise (He et al., 2012). But recently, a single bout of exercise was shown to decrease the expression of some autophagic proteins including LC3-II, Beclin-1, Atg7, and LAMP-2 during the recovery period. These findings demonstrated that a single bout of treadmill exercise might attenuate the autophagic signaling in skeletal muscle (Kim et al., 2012), contrary to the findings reported by He et al. (2012).
Aside from the single bout exercise, long-term chronic exercise has also been shown to result in different autophagic response in skeletal muscle (Tam and Siu, 2014). The basal autophagy flux and autophagy protein expression have been reported to be increased after 4 weeks of voluntary running. Some key markers important in examining autophagic flux including the LC3-II/LC3-I ratio, LC3-II, and p62 have been reported to be upregulated. Different types of muscles have been shown to have different autophagic responses even when exposed to the same exercise stimulus. Basal autophagic flux and autophagy protein expression have been shown to be increased in parallel with mitochondrial biogenesis in plantaris with mixed fiber types. However, no significant increase in basal autophagy flux and mitochondrial biogenesis was observed in oxidative soleus muscle after 4 weeks of voluntary running (Lira et al., 2013). This is contradictory to another study that showed that the phosphorylation of Akt is significantly increased in gastrocnemius muscle of mice subjected to bilateral functional overload. Phosphorylation of Akt may lead to inactivation of autophagy through the PI3K-Akt-mTor-mediated pathway (Spangenburg et al., 2008).
Although the roles of different autophagic signaling have been studied extensively, how intensity of exercise modulates autophagy in skeletal muscle remains unclear. Different exercise types, forms, protocols, and intensities may affect autophagic response to physical exercise (Tam and Siu, 2014). Modulation of cellular bioenergetics during training period may play an important role for optimizing athlete’s performance. This process may include autophagy adaptation during exercise. Therefore, in the present study we compared altered autophagy gene expression by different exercise intensities in gastrocnemius and soleus muscles of wistar rats.
Methods
Animals
Eight-week-old male Wistar rats were obtained from the Animal Breeding Centre of PT Biofarma in Cisarua, Indonesia. The environment was maintained in a darklight cycle (12 hours of light cycle and 12 hours of dark cycle) and temperature (22°C). The rats were fed a pellet rodent diet ad libitum and had free access to water. After two weeks of acclimatization period, 20 male rats were divided into four groups, sedentary control group and three exercise groups (low/LI, moderate/MI, and high/HI). All experimental procedures followed guide for the care and use of laboratory animals (Council, 2011) and were approved by Research Ethics Committee Faculty of Medicine Universitas Kristen Maranatha-Rumah Sakit Immanuel Bandung No 098/KEP/III/2018.
Treadmill exercise protocol
We designed a treadmill exercise protocol for three different groups of male Wistar rats, low, moderate, and high-intensity treadmill exercises. Sedentary control group of rats was also put in a treadmill, but without exercise. After two weeks of adaptation, the rats were acclimatized to treadmill running for two weeks with increasing speed and time. On the third week, animals were exercised in 30 min/day, 5 days/week for 8 weeks (Figure 1). The body weights of all groups were monitored and recorded weekly. After eight weeks, control and exercise groups were euthanized using isoflurane, then gastrocnemius and soleus muscles were removed, snap-frozen in liquid nitrogen, and stored in -80°C until used.
Figure 1.
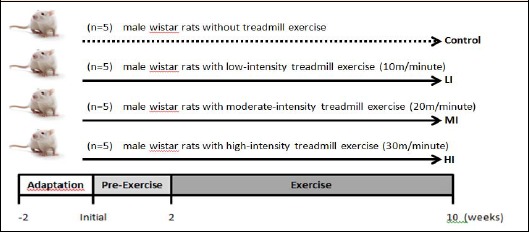
Experimental design and protocol for 8 weeks treadmill exercise training. Eight weeks old, male, wistar rats were assigned to sedentary control (Control) and training groups which are divided into 3 groups contained 5 rats each group : Low Intensity (LI), Moderate Intensity (MI), and High Intensity (HI) and given 8 weeks treadmill exercise following procedure established by Lesmana et al. 2016 with small modification. The pre-Exercise (Habituation) period lasted for 2 weeks.
RNA extraction and semi-quantitative PCR
Total RNA was extracted and isolated from gastrocnemius and soleus muscles using TRIsure reagent (Bioline, United Kingdom). Total RNA in gastrocnemius and soleus muscles were quantified using Multimode Microplate Reader at 268/280 nm absorbance spectrophotometry (M200 Pro, Tecan, Morrisville, NC). Semiquantitative PCR was performed using the One Step RT PCR Kit (Qiagen, Valencia, CA) and Sensoquest Genecraft. Semi quantitative gene expression levels were normalized using rats GAPDH (Figure 2). Primers sequences used for rat gene are listed in Table 1. The band of PCR products was visualized with and quantified using ImageJ software (NIH).
Figure 2.
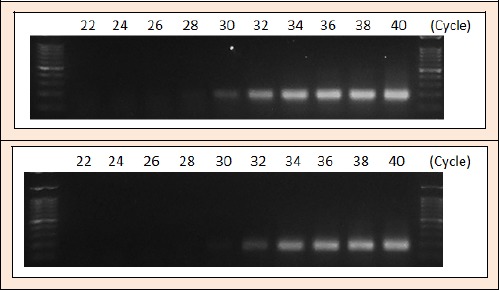
Determination of PCR cycle for amplification curve GAPDH expression in gastrocnemius (top) and soleus muscle (bottom).
Table 1.
Primers and PCR protocols.
| Gene Symbol | Primer Sequence (5’ to 3’) Upper strand : sense Lower strand : antisense |
Product Size (bp) |
Annealing (°C) |
Cycle | References |
|---|---|---|---|---|---|
| p62 | CTAGGCATCGAGGTTGACATT | 116 | 56 | 35 | (Kowalik et al, 2015) |
| CTTGGCTGAGTACCACTCTTATC | |||||
| LC3 | GGTCCAGTTGTGCCTTTATTGA | 153 | 59,5 | 35 | (Yin et al, 2013) |
| GTGTGTGGGTTGTGTACGTCG | |||||
| GAPDH | GTTACCAGGGCTGCCTTCTC | 177 | 61 | 35 | (Wang et al, 2017) |
| GATGGTGATGGGTTTCCCGT |
Western blot analysis
The dissected gastrocnemius and soleus muscle were weighted, homogenised in lysis buffer containing 10mM Tris-HCl (pH 7.8), 150mM NaCl, 1mM EDTA, 1% Nonidet P-40, and protease inhibitors. After centrifugation, protein samples were heat denatured at 96°C for 5 minutes. Samples (10 μg/lane) were separated by SDS-PAGE and were then transferred to a nitrocellulose membrane (GE Healthcare) for 1 hour at room temperature and blocked overnight at 4°C in 1% blocking reagent (GE Healthcare) in Tris-buffered saline buffer with 0.1% Tween 20. Immunoblotting were performed using a mouse monoclonal LC3 (#12741), p62 (#5114) and Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) thermo scientific AM4300 which purchased from Cell Signalling Co, Ltd. with dilution 1:500. The signals were developed using enhanced chemiluminescence reagent (GE Healthcare) and imaged (LI-COR C-DiGit Chemiluminescence Western Blot Scanner). The band intensities were determined using ImageJ Software (NIH). Blots were stripped using stripping buffer from Thermo scientific according to manufacturer protocols and reprobed using with an antiGAPDH as internal control to monitor the level of protein (Pratiwi et al., 2018). Reconfirmation of alteration LC3 (ATG8) and p62 gene expression are modulated by exercise intensity which were confirmed by protein levels using Western Blot (Figure 3).
Figure 3.
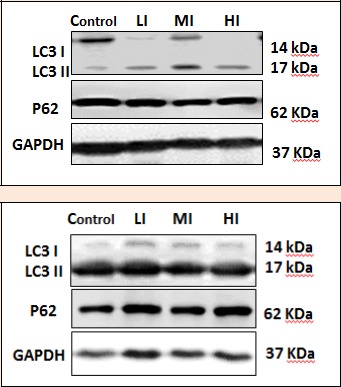
Reconfirmation of alteration LC3 (ATG8) and p62 gene expression are modulated by exercise intensity which were confirmed by protein levels using Western Blot. Representative Figure of Western Blot results reflected LC3 (ATG8) and p62 protein levels in (top) Gastrocnemius muscle and (bottom) Soleus Muscle.
Statistical analysis
All statistics were computed using SPSS 20.0 software. Results are presented as the mean ± standard error of mean (mean ± SEM). Mean differences between groups were examined with One Way ANOVA and Tukey post hoc test (for data with normal distribution) or Kruskal Wallis and Mann Whitney test (for data without normal distribution), with 95% confidence interval (p < 0.05).
Results
Effects of exercise on body and muscle weights
The rats of the four experimental groups had the similar initial body weights (200 ± 50 g). To examine whether different intensity treadmill exercise could increase or decrease body weight, it was recorded every 2 weeks. After treadmill exercises for 30 min/day, 5 days/week, for 8 weeks, there were no significant differences found in body weight between LI, MI and control. Interestingly, we found significant decrease in body weight (Figure 4a) by 13.02 % (328.6 ± 9.03 g) in HI compared to control (377.8 ± 15.41 g) (p = 0.000). Numerous studies have shown that male rats receiving regular endurance exercise become more slowly in gaining weight and finally have lower body weight than those in sedentary controls (Oscai et al., 1972). The weight of gastrocnemius and soleus muscles also recorded and showed no difference between treatment (LI, MI, HI) and control (Figure 4b).
Figure 4a.
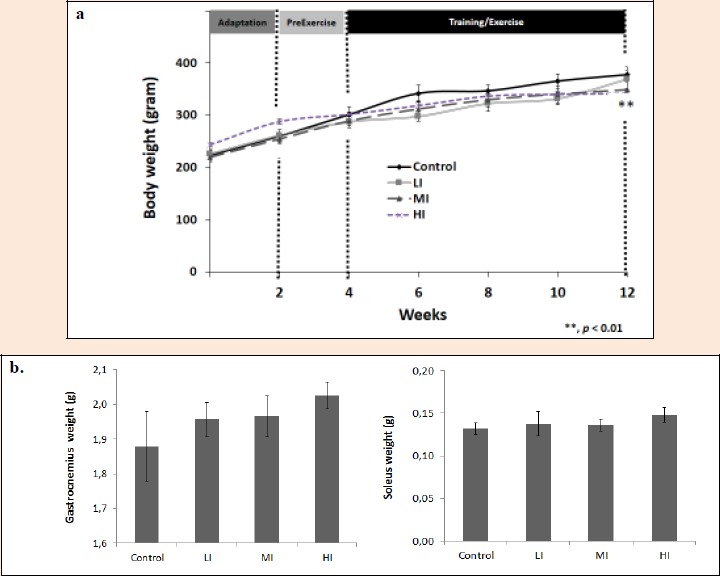
Body weights of the four groups of rats. Data are mean ± SEM. ** indicated very significant difference at P < 0.01 by one-way ANOVA. b. Gastrocnemius and soleus weights recorded before stored in -80°C.
LC3 and p62 mRNA expressions in gastrocnemius muscles of wistar Rats
We examined the altered autophagy gene expression in gastrocnemius muscles of wistar rats by semi-quantitative PCR. PCR bands of LC3 and p62 were normalized using GAPDH. The result is presented in Table 2 and Figure 5. Exercises significantly decrease expression of autophagy gene LC3 (LI and MI 0.7 fold, HI 0.8 fold, p < 0.05) in gastrocnemius muscles compared to control. On the other side, exercises only significantly decrease p62 gene expression (MI 0.8 fold, p < 0.05), while no significant changes with LI and HI found compared to control.
Table 2.
Relative ratio of LC3 and p62 gene expression were altered by different exercise intensities
| Control | LI | MI | HI | ||
|---|---|---|---|---|---|
| Gastrocnemius | LC3 | 0.803 ± 0.05 | 0.528 ± 0.06 | 0.582 ± 0.01 | 0.665 ± 0.01 |
| p62 | 0.829 ± 0.04 | 0.784 ± 0.01 | 0.704 ± 0.03 | 0.889 ± 0.01 | |
| Soleus | LC3 | 0.757 ± 0.03 | 0.610 ± 0.03 | 0.582 ± 0.01 | 0.619 ± 0.03 |
| p62 | 0.807 ± 0.02 | 0.730 ± 0.03 | 0.702 ± 0.02 | 0.893 ± 0.02 | |
Figure 5.
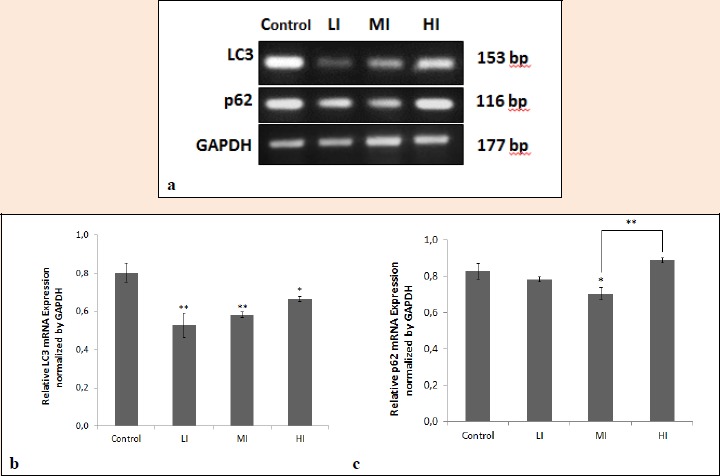
Alteration of autophagy gene expression in gastrocnemius muscles of wistar rats by different intensities of exercise. a. LC3 and p62 expressions in control and different intensities of exercise: low-intensity (LI), moderate-intensity (MI), and high-intensity (HI). b. Relative LC3 mRNA expression normalized by GAPDH in gastrocnemius muscles of wistar rats. c. Relative p62 mRNA expression normalized by GAPDH in gastrocnemius muscles of wistar rats. Bar graphs represent mean ± SEM. *, p < 0.05 versus the corresponding control group. **, p < 0.01 versus the corresponding control group
LC3 and p62 mRNA expressions in soleus muscles of wistar rats
We also examined the altered autophagy gene expression in soleus muscles of wistar rats by semi-quantitative PCR. PCR bands of LC3 and p62 were normalized using GAPDH. The result is presented in Table 2 and Figure 6. In all exercise groups, autophagy gene LC3 was significantly decreased (LI, MI, and HI 0.8 fold, p < 0.05) compared to control in soleus muscles. On the other side, p62 gene expression in soleus was significantly decreased (MI 0.9 fold, p < 0.05), and significantly increased (HI 1.1 fold, p < 0.05), but had no change in LI compared to control.
Figure 4.
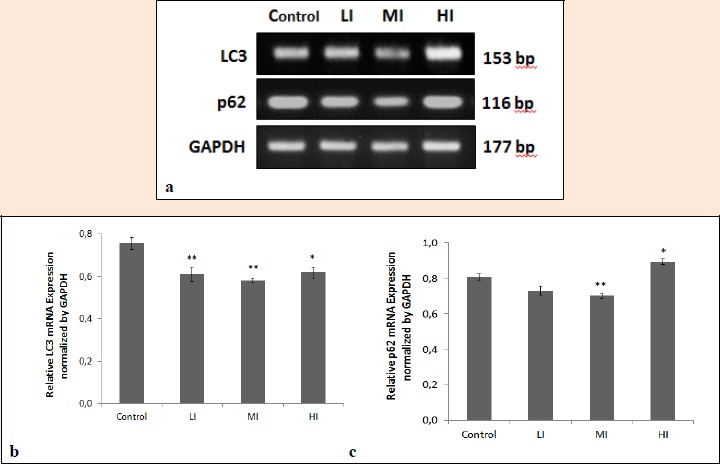
Alteration of autophagy gene expression in soleus muscles of wistar rats by different intensities of exercise. a. LC3 and p62 expressions in control and different intensities of exercise : low-intensity (LI), moderate-intensity (MI), and high-intensity (HI). b. Relative LC3 mRNA expression normalized by GAPDH in soleus muscles of wistar rats. c. Relative p62 mRNA expression normalized by GAPDH in soleus muscles of wistar rats. Bar graphs represent mean ± SEM. *, p < 0.05 versus the corresponding control group. **, p < 0.01 versus the corresponding control group
LC3 and p62 protein leves in gastrocnemius and soleus muscles of wistar rats
In order to confirm our result, we also examined protein levels of LC3 dan p62 in gastrocnemius and soleus muscles of wistar rats by Western Blot (Figure 3). The bands were normalized using GAPDH.
Discussion
Muscle contraction can form an energetic stress, which leads to alteration in molecular messengers, such as calcium, AMP, NAD+, and ROS (Reactive Oxygen Species). These messengers then activate downstream signaling cascades, resulting in a biphasic autophagic response aimed at restoring homeostasis (Vainshtein and Hood, 2016). When energetic supply and demand are in equilibrium, the metabolic sensors mammalian target of rapamycin (mTOR) and protein kinase, a negatively regulated autophagy, through the phosphorylation and inhibition of the induction complex (Joassard et al., 2013 ; Stephan et al., 2009). When energy demand outweighs supply, the AMP-to-ATP ratio rises, and this activates AMP-dependent kinase (AMPK), while inhibiting mTOR activity. Elevated production of ROS and an increase in NAD+ during exercise also result in the activation of AMPK which then stimulates the autophagy process (Cantó et al., 2009 ; Hardie, 2011).
Autophagy consists of several essential processes namely initiation, nucleation, elongation, fusion, and degradation. These processes are tightly regulated to locate and gather the targeted organelles and long-lived protein to autophagosomes, which will then be fused with lysosomes for breakdown. The contents inside the autophagosomes will ultimately be degraded by the hydrolytic enzymes provided by the lysosomes (Xie and Klionsky, 2007). The ubiquitin-like protein Atg8/LC3 can be found in phagophores, autophagosomes and, to a lesser extent, in autolysosomes. Native LC3, or proLC3, is proteolytically cleaved by autophagy-related4 (Atg4) releasing LC3-I, to which phosphatidylethanolamine is conjugated to generate the lipidated form of LC3 called LC3-II. LC3-II is the only protein marker that is reliably associated with completed autophagosomes (although it can also be found in phagophores) (Klionsky et al., 2016) and its levels correlate with autophagosome number (Rubinsztein et al., 2009). During autophagy, there is an increased conversion of LC3-I to LC3-II and the ratio of LC3-II/LC3-I is augmented. In the latest step of autophagy, the sequestosome1 (p62/SQSTM1), an acceptor for ubiquitinated substrates, is reduced due to autolysosomal degradation. Conversely, the accumulation of p62/SQSTM1 has been interpreted as a marker of autophagy inhibition (Bjørkøy et al., 2009; Klionsky et al., 2016).
In these recent years, autophagy was discovered as a major catabolic cellular process that has triggered special interest in muscle research. Single bout exercise and long-term chronic exercise have been shown to result in different autophagic responses in skeletal muscle (Lira et al., 2013 ; He et al., 2012 ; Spangenburg et al., 2008 ; Kim et al., 2012). However, the regulation of autophagy in skeletal muscle adaptation by different intensity of exercise remains unclear. Here, we report that long-term endurance exercise training (treadmill exercise for 8 weeks) with different intensity in rats resulted in an altered autophagy gene expression in gastrocnemius and soleus muscles. Different intensities in this experiment were determined based on lactate threshold. Previous study showed that 20 m/min running on a treadmill was the speed at the lactate threshold of rat, so our study used 10 m/min for sub lactate threshold as mild intensity, 20 m/min for lactate threshold as moderate intensity, and 30 m/min for supra lactate threshold as high intensity (Soya et al., 2007 ; Lesmana et al., 2016).
This study showed a significant decrease of LC3 (MI) and p62 (MI) in gastrocnemius muscles, as shown in Figure 5. Therefore, we observed that autophagy induced in moderate intensity of treadmill exercise. Our findings are in agreement with those of previous studies that have demonstrated increased autophagy gene expression after moderate treadmill training in skeletal muscles (McMillan et al., 2015 ; He et al., 2012). The present findings also showed significant decrease of LC3 (LI and HI) without significant change of p62 (LI, HI) compared to those in control groups (Figure 5), suggesting that autophagy activities might remain stable among these muscles. This observation supports previous study reporting no changes in autophagy gene expression in gastrocnemius muscles after treadmill training (Bayod et al., 2014). Although no significant changes found, the p62 gene expression was lower in LI and higher in HI compared to that in control. This result suggested that there might be an increasing and decreasing process of autophagy as a result of LI and HI exercises respectively. This result is consistent with the change of protein level as shown in Figure 3, suggesting that autophagy might be modulated by different type of exercise.
We also observed significant decrease of LC3 (MI) and p62 (MI) as shown in Figure 6. These findings are in agreement with those of previous studies that have demonstrated increased autophagy gene expression after moderate treadmill training in skeletal muscles (McMillan et al., 2015 ; He et al., 2012). Interestingly, we found a significant decrease of LC3 and increase of p62 (HI) in soleus muscles, which suggested that autophagy might attenuated after treadmill training with high intensity. This finding is in line with another study that showed phosphorylation of Akt could be significantly increased in exercise, which leads to inactivation of autophagy through the PI3K-Akt-mTor-mediated pathway (Spangenburg et al., 2008).
Different levels of locomotive activities cause skeletal muscle exhibits different phenotypic features. Tonic or postural muscles are mostly composed of oxidative fibers and a robust mitochondrial network and predominant expression of MHCIIa and MHCI proteins. On the contrary, phasic muscles are mainly composed of glycolytic fibers with low mitochondrial content and predominant expression of MHCIIb or MHCIIx proteins. Soleus is considered as a tonic muscle, while gastrocnemius is a phasic muscle (Schiaffino and Reggiani, 2011 ; Lira et al., 2013). Difference of this fiber type might affect the result of different exercise intensity on autophagy in gastrocnemius and soleus. PGC-1α and HIF-1α may also be altered by different intensity of exercise which play role in regulating adaptation in cardiac hypertrophy (Sylviana et al., 2018). According to previous study by Lira et al, tonic muscles have higher autophagy flux, autophagy, and mitophagy protein expressions than those in phasic muscles (Lira et al., 2013). This might explain why soleus muscles were more responsive than gastrocnemius muscles when treated with different exercise intensity.
This study showed altered intensity of exercise may modulate autophagy activity in adaptation process after training in skeletal muscles. Our data demonstrate that moderate intensity of exercise increases autophagy gene expression in soleus and gastronemius (Figure 5 and 6) and supported with protein level alteration (Figure 3), whereas high intensity decreases autophagy gene expression (Figure 6), which also supported with protein level alteration (Figure 3). This altered expression of autophagy gene is more obvious in soleus muscles, since soleus muscle has a characteristic to work as a tonic muscle with higher autophagy activities compared to phasic muscle (gastrocnemius).
The limitation of this research is that the study has only been done in 8 weeks and it is possible that adaptation had occurs in earlier time period. Another point is hormone may take some role in stimulating autophagy and we did not perform experiment using female rats so it can eliminated the gender difference responses. It could be more interesting to study whether stress during treadmill play some role in autophagy alteration in rats.
Conclusion
In summary, different exercise intensity altered autophagy gene expression in gastrocnemius and soleus muscles. MI exercise may increase autophagy gene expression, while HI exercise tended to decrease autophagy gene expression. LI exercise had no significant changes on autophagy gene expression. This data demonstrate that intensity may play some role on autophagy activity in skeletal muscle during training in specific manner. It is important to understand biocelular adapation in skeletal muscle for training implementation.
Acknowledgements
The authors would like to thank Susianti and Nurul Ihsani for technical assistance during experiments. We also would like to thank dr. Cherry, dr. Teresa, dr. Nova, drh. Okta, and Azis for assisting in termination of wistar rats. This study was supported by Riset Kompetensi Dosen Unpad/RKDU Grant (2476/UN6.C/LT/2018) to VMT, Funding granted from PDUPT DIKTI research Grant (3670/UN.6C/LT/2018) to RL, and Research Grant from Maranatha Christian University (190/PEG/UKM/VII/2018) to JWG. The experiments comply with the current laws of the country in which they were performed. The authors have no conflicts of interests to declare.
Biographies

Vita Murniati TARAWAN
Employment
Associate Professor at Physiology Divison, Departement of Basic Medical Science, Faculty of Medicine, Universitas Padjadjaran, Indonesia
Degree
Dr
Research interest
Exercise Physiology, Dopping

Julia Windi GUNADI
Employment
A Doctoral candidate at Faculty of Medicine, Universitas Padjadjaran, Indonesia. Lecturer in Faculty of Medicine, Maranatha Christian University, Indonesia.
Degree
MKes
Research interest
Understanding hypertrophy and its relation to autophagy in cardiac and skeletal muscles
E-mail: juliawindig@gmail.com

Setiawan
Employment
Dean of Faculty of Medicine, Universitas Padjadjaran, Bandung, Indonesia
Degree
Dr. med
Research interest
Exercise Physiology

Ronny LESMANA
Employment
Assistant Professor at Physiology Divison, Departement of Basic Medical Science, Faculty of Medicine, Universitas Padjadjaran, Bandung, Indonesia
Degree
PhD
Research interest
Effect of thyroid hormone on autophagy stimulation

Hanna GOENAWAN
Employment
Assistant Professor at Physiology Divison, Departement of Basic Medical Science, Faculty of Medicine, Universitas Padjadjaran, Indonesia
Degree
PhD
Research interest
How physical activity can affect the synaptogenesis in brain

Debby Eka MEILINA
Employment
Medical Student at Maranatha Christian University, Bandung, Indonesia
Research interest
Effect of physical exercise on autophagy gene expression.

Julidea Anggiriani SIPAYUNG
Employment
Medical Student at Maranatha Christian University, Bandung
Research interest
Effect of various intensity of physical exercise on autophagy gene expression in skeletal muscles.

Teresa Liliana WARGASETIA
Employment
A lecturer and associate professor at the Faculty of Medicine, Universitas Kristen Maranatha (Maranatha Christian University) in Indonesia.
Degree
Dr
Research interest
Topics related to autophagy and cancer cell biology.

Wahyu WIDOWATI
Employment
As lecturer and researcher at Biology Department, Faculty of Medicine, Maranatha Christian University, Bandung Indonesia
Degree
Dr
Research interest
Stem Cells, Culture Cells, Biomolecular

Yenni LIMYATI
Employment
Lecturer in Faculty of Medicine, Maranatha Christian University, Bandung Indonesia
Degree
M Kes
Research interest
Exercise physiology, Music Therapy

Unang SUPRATMAN
Employment
Head of Central Laboratory, Universitas Padjadjaran, Bandung, Indonesia.
Professor at Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Padjadjaran, Bandung, Indonesia.
Degree
Prof
Research interest
Natural Products Chemistry
References
- Bayod S., Del Valle J., Pelegri C., Vilaplana J., Canudas A.M., Camins A., Bayod S., Del Valle J., Pelegri C., Vilaplana J., Canudas A.M., Camins A., Jimenez A., Sanchez-Roige S., Lalanza J.F., Escorihuela R.M., Pallas M. (2014) Macroautophagic process was differentially modulated by long-term moderate exercise in rat brain and peripheral tissues. Journal of Physiology and Pharmacology 65, 229-239. [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Pankiv S., Øvervatn A., Brech A., Johansen T. (2009) Monitoring autophagic degradation of p62/SQSTM1. Methods in Enzymology 452, 181-197. [DOI] [PubMed] [Google Scholar]
- Booth F.W., Roberts C.K., Laye M.J. (2012) Lack of exercise is a major cause of chronic diseases. Comprehensive Physiology 2, 1143-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F., Levine B. (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Developmental Cell 15, 344-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council N.R. (2011). Guide for the Care and Use of Laboratory Animals, 8th edition, National Academies Press (US), Washington (DC). [Google Scholar]
- Green D.R., Levine B. (2014) To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157, 65-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G. (2011) Energy sensing by the AMP-activated protein kinase and its effects on muscle metabolism. Proceedings of the Nutrition Society 70, 92-99. [DOI] [PubMed] [Google Scholar]
- He C., Bassik M.C., Moresi V., Sun K., Wei Y., Zou Z., An Z., Loh J., Fisher J., Sun Q., Korsmeyer S., Packer M., May H.I., Hill J.A., Virgin H.W., Gilpin C., Xiao G., Bassel-Duby R., Scherer P.E., Levine B. (2012) Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Sumpter R.J., Levine B. (2012) Exercise induces autophagy in peripheral tissues and in the brain. Autophagy 8, 1548-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joassard O.R., Amirouche A., Gallot Y.S., Desgeorges M.M., Castells J., Durieux A.C., Berthon P., Freyssenet D.G. (2013) Regulation of Akt-mTOR, ubiquitin-proteasome and autophagy-lysosome pathways in response to formoterol administration in rat skeletal muscle. International Journal Of Biochemistry & Cell Biology 45, 2444-2455. [DOI] [PubMed] [Google Scholar]
- Kim Y.A., Kim Y.S., Song W. (2012) Autophagic response to a single bout of moderate exercise in murine skeletal muscle. Journal of Physiology and Biochemistry 68, 229-235. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo A.A., et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesmana R., Iwasaki T., Iizuka Y., Amano I., Shimokawa N., Koibuchi N. (2016) The change in thyroid hormone signaling by altered training intensity in male rat skeletal muscle. Endocrine Journal 63, 727-738. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental Cell 6, 463-477. [DOI] [PubMed] [Google Scholar]
- Lira V.A., Okutsu M., Zhang M., Greene N.P., Laker R.C., Breen D.S., Hoehn K.L., Yan Z. (2013) Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB Journal 27, 4184-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Rincon M., Morales-Alamo D., Calbet J.A.L. (2018) Exercise-mediated modulation of autophagy in skeletal muscle. Scandinavian Journal of Medicine and Science in Sports 28, 772-781. [DOI] [PubMed] [Google Scholar]
- McMillan E.M., Paré M.F., Baechler B.L., Graham D.A., Rush J.W., Quadrilatero J. (2015) Autophagic Signaling and Proteolytic Enzyme Activity in Cardiac and Skeletal Muscle of Spontaneously Hypertensive Rats following Chronic Aerobic Exercise. PLoS One 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728-741. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular Biology of the Cell 15, 1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscai L.B., Spirakis C.N., Wolff C.A., Beck R.J. (1972) Effects of exercise and of food restriction on adipose tissue cellularity. Journal of Lipid Research 13, 588-592. [PubMed] [Google Scholar]
- Pedersen B.K., Saltin B. (2015) Exercise as medicine - evidence for prescribing exercise as therapy in 26 different chronic diseases. Scandinavian Journal of Medicine and Science in Sports 25, 1-72. [DOI] [PubMed] [Google Scholar]
- Pratiwi Y.S., Lesmana R., Goenawan H., Sylviana N., Setiawan I., Tarawan V.M., Lestari K., Abdulah R., Dwipa L., Purba A., Supratman U. (2018) Nutmeg Extract Increases Skeletal Muscle Mass in Aging Rats Partly via IGF1-AKT-mTOR Pathway and Inhibition of Autophagy. Evidence-Based Complementary and Alternative Medicine 2018, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W., Jimenez-Sanchez M., Korolchuk V.I., Lichtenberg M., Luo S., Massey D.C., Menzies F.M., Moreau K., Narayanan U., Renna M., Siddiqi F.H., Underwood B.R., Winslow A.R., Rubinsztein D.C. (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiological Reviews 90, 1383-1435. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S, et al. (2009) In search of an “autophagomometer”. Autophagy 5, 585-589. [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Reggiani C. (2011) Fiber types in mammalian skeletal muscles. Physiological Reviews 91, 1447-1531. [DOI] [PubMed] [Google Scholar]
- Soya H., Mukai A., Deocaris C.C., Ohiwa N., Chang H., Nishijima T., Fujikawa T., Togashi K., Saito T. (2007) Threshold-like pattern of neuronal activation in the hypothalamus during treadmill running: establishment of a minimum running stress (MRS) rat model. Neuroscience Research 58, 341-348. [DOI] [PubMed] [Google Scholar]
- Spangenburg E.E., Le Roith D., Ward C.W., Bodine S.C. (2008) A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. The Journal of Physiology 586, 283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J.S., Yeh Y.Y., Ramachandran V., Deminoff S.J., Herman P.K. (2009) The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proceedings of the National Academy of Sciences of the United States of America 106, 17049-17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylviana B., Helja N., Qolbi H.H., Goenawan H., Lesmana R., Syamsunarno M.R.A.A., Agustina H., Tiksnadi B.B., Megantara I., Tarawan V.M., Setiawan S., Hernowo B., Supratman U., Purba A. (2018) Effect of Swimming Exercise to Cardiac PGC-1 α and HIF-1 α Gene Expression in Mice. Asian Journal of Sports Medicine 9, 1-7. [Google Scholar]
- Tam B.T., Siu P.M. (2014) Autophagic cellular responses to physical exercise in skeletal muscle. Sports Medicine (Auckland, N.Z.) 44, 625-640. [DOI] [PubMed] [Google Scholar]
- Vainshtein A., Hood D.A. (2016) The regulation of autophagy during exercise in skeletal muscle. Journal of Applied Physiology (Bethesda, Md. : 1985) 120, 664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. (2007) Autophagosome formation: core machinery and adaptations. Nature Cell Biology 9, 1102-1109. [DOI] [PubMed] [Google Scholar]
- Yan Z., Okutsu M., Akhtar Y.N., Lira V.A. (2011) Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. Journal of applied physiology (Bethesda, Md. : 1985) 110, 264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]


