Abstract
We aimed to characterize HIV-1 molecular epidemiology and transmission clusters among heterosexual (HET) and men who have sex with men (MSM) individuals, as well as transmitted drug resistance mutations (TDRM) in Central-Western Brazil. This cross-sectional survey was conducted among 190 antiretroviral naïve HIV-1 infected individuals. Proviral DNA was extracted, and nested PCR amplified partial polymerase gene (PR/RT). After sequencing, subtypes were assigned, and the sequences were analyzed for the occurrence of possible transmission networks. Calibrated Population Resistance (CPR) tool from Stanford HIV Database was used to investigate the presence of TDRM. Among 150 individuals whose samples were successfully sequenced, the most prevalent HIV-1 subtype was B, followed by recombinant forms. The occurrence of twenty transmission clusters composed by at least two sequences was verified, suggesting the existence of transmission clusters among individuals from the same or distinct sexual orientations. Intermediate level of TDRM (12%) was found in the study population, and almost half of the subjects with TDRM had more than one resistance mutation. No correlations between sexual orientation and the presence of TDRM, HIV-1 subtypes/recombinants forms were verified. Taken together, the necessity of the continuous monitoring of the TDRM to verify the importance of pre-genotyping and to delineate future strategies in primary antiretroviral therapy. Likewise, the knowledge of the HIV-1 transmission networks in Brazil would allow the implementation of effective HIV-1 prevention strategies in local settings.
Keywords: HIV, MSM, molecular epidemiology, transmitted drug resistance, transmission network
Introduction
In Latin America, it is estimated that 1.8 million people are living with human immunodeficiency virus (HIV) and/or acquired immunodeficiency syndrome (AIDS). Despite 100,000 new HIV infections having been diagnosed in 2017, the HIV incidence decreased 13.7% between 2000 and 2017 (UNAIDS, 2018). In Brazil, HIV prevalence among the general population is below 0.6% and it is estimated that AIDS cases among Brazilians reached 882,810 by June 2017 (Brasil, 2017). HIV prevalence is higher in key populations at risk, for example 17.5% in men who have sex with men (MSM) (Kerr et al., 2018). The detection rate of AIDS has been falling steadily in Brazil in recent years. However, the Central Western region showed little change in its detection rate in the last 10 years, reaching 16.7 cases per 100 thousand inhabitants in 2016 (Brasil, 2017).
Universal access to combined antiretroviral therapy (cART) in Brazil was crucial in order to increase survival and decrease AIDS-related hospitalizations in HIV-1 infected individuals (Souza Junior et al., 2011). Although, the development of drug resistance mutations is a significant obstacle to maintaining HIV-1 replication suppression and can lead to viral load increase and consequently transmission of viruses with drug resistance mutations. Therefore, transmitted drug resistance mutations (TDRM) have become an important challenge, since they have been described for all drugs used in the clinical management of HIV and as incidence and prevalence vary by region this highlights the importance of its monitoring. The prevalence of TDRM could vary according to the study population, methods and lists of resistance mutations used to calculate these rates (Booth and Geretti, 2007).
Brazil has an extensive border, covering about 15,000 km, exhibiting great socioeconomic and cultural diversity across regions. Concerning HIV-1 subtypes, subtype B is the most prevalent, followed by F1, and BF1 recombinants in most Brazilian regions (De Sa Filho et al., 2005; Pedroso et al., 2007; Machado et al., 2009; Guimarães et al., 2015), except for the Southern region, where subtype C is highly prevalent (Silva et al., 2010; de Medeiros et al., 2011; Gräf et al., 2011). However, even in the same geographic region, the HIV-1 distribution could be heterogeneous (Gräf and Pinto, 2013). In border areas, intense drug trafficking and prostitution occur; both situations may affect local epidemic dynamics. Taking these geographical and epidemiological characteristics together into consideration, the study of HIV-1 genetic diversity and transmission networks as well as drug resistance mutations in this region is relevant.
Materials and Methods
Subjects and Study Design
We conducted a cross-sectional survey among antiretroviral naïve HIV-infected individuals recruited in Campo Grande, the capital of Mato Grosso do Sul (MS) State, from 2011 to 2014. One hundred and seventy-two individuals were enrolled at Reference Centers for Parasitic and Infectious Diseases (Freitas et al., 2014), and thirty-two were MSM recruited in a cross-sectional study (Fernandes et al., 2015). Inclusion criteria were: (a) having confirmed diagnosis for HIV-1; (b) being over 18 years old; (c) being antiretroviral naïve; (d) having signed the informed consent form in earlier surveys, which predicted storage of samples and their utilization in future research; and (e) having sample stored in sufficient quantity to perform the analyses proposed. Following these criteria, 190 individuals were selected for the subsequent analysis. This study was carried out in accordance with the recommendations of the Ethical Committee on Human Research of the Federal University of Mato Grosso do Sul, that is in accordance with the Declaration of Helsinki. The protocol was approved by under protocol number 1151451, CAAE 46185915.8.0000.0021.
Amplification of HIV-1 PR/RT Region
DNA was extracted from 200 μL of each whole blood sample by using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The partial polymerase (pol) gene including protease/reverse transcriptase (PR/RT) region was amplified by nested polymerase chain reaction (PCR) using combinations of primers described elsewhere (Delatorre et al., 2017). The amplified products were analyzed by electrophoresis using agarose gels (1%). Amplicons were purified using the Illustra GFX® PCR DNA and Gel Band Purification Kit (GE Healthcare, United Kingdom), following the manufacturer’s recommendations. The purified DNA was sequenced using Big Dye Terminator Cycle Sequencing Ready Reaction kit v.3.1 (Applied Biosystems, CA, United States) and processed with an automated ABI 3130xl sequencer (Applied Biosystems), using Sanger’s method.
Sequence Analysis
The sequences were edited in DNASTAR software and then aligned with reference sequences from Los Alamos HIV Sequence Database1 using the Clustal W program implemented in MEGA 6.0 software (Tamura et al., 2013). All sequences are available in GenBank (accession number MF545192-MF545340). The final PR/RT alignment covered a fragment of 1261 bp, corresponding to nucleotides 2254 to 3514 relative to the HXB2 genome.
Maximum Likelihood (ML) phylogenetic was constructed with the PhyML 3.0 program using an online web server (Guindon et al., 2010). The Smart Model Selection recommended the GTR+I+G nucleotide substitution model to be used in the ML (Lefort et al., 2017). The heuristic tree search was performed using the SPR branch-swapping algorithm, and the branch support was calculated with the approximate likelihood-ratio (aLRT) SH-like test (Anisimova and Gascuel, 2006). Recombinant profiles were inferred by bootscan analyses with a sliding window of 300 bp, steps of 10 bp and Kimura-2 parameters model using SimPlot 3.5.1 software (Lole et al., 1999).
Those sequences that clustered together with high aLRT support (>0.90) in the ML tree were analyzed for the occurrence of possible transmission clusters. Therefore, such sequences were submitted to analysis using nucleotide Basic Local Alignment Search Tool (BLASTn) (Altschul et al., 1990) to recover reference sequences with high similarity (>95%). These sequences retrieved were added to three new alignments from pure subtypes (B, D, and F1), and a new ML tree was obtained to verify the maintenance of the transmission clusters according to their subtypes. For subtypes D and F1 analyses we included all available Brazilian reference sequences, however, duplicate sequences were removed. For subtype B, at least ten representative sequences from each Brazilian State and all sequences from Mato Grosso do Sul state available at the Los Alamos HIV Sequence Database were included. Before performing the phylogenetic analyses to confirm the transmission clusters, drug-resistance mutations positions were stripped from each alignment, resulting in a fragment of 891 bp from nucleotides 2262 to 3251 relative to HXB2 genome. Our final cluster classification was defined based on aLRT (>90) in the phylogenetic analyses (Figure 2, 3), and low mean pairwise genetic distances (≤4.5) of clustered sequences have been employed.
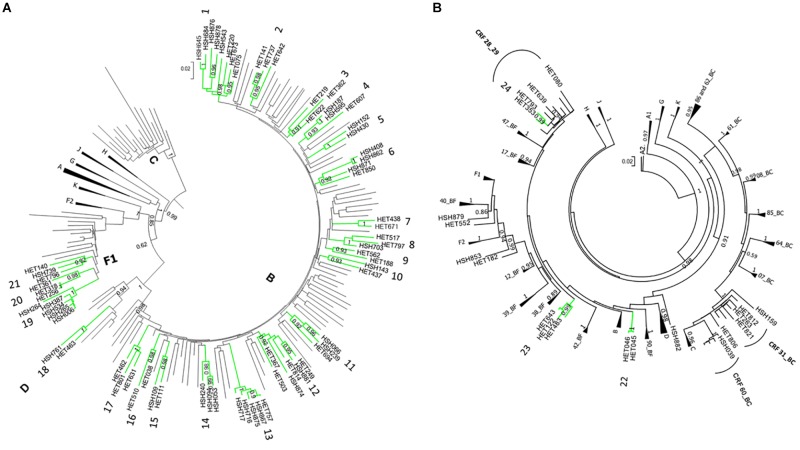
FIGURE 2.
ML phylogenetic tree of 150 HIV-1 PR/RT sequences from Mato Grosso do Sul, Central-West Brazil. The analyzed PR/RT alignment covered a fragment of 1261 bp, corresponding to nucleotides 2254 to 3514 relative to HXB2 genome. Reference sequences retrieved from GenBank are not labeled. ALRT values are represented only if greater > 0.90. Possible HIV-1 transmission clusters were indicated by numbers. HET = sample obtained from heterosexual individual; HSH = sample obtained from men who have sex with men. (A) Pure HIV-1 subtypes and (B) Recombinant sequences.
FIGURE 3.
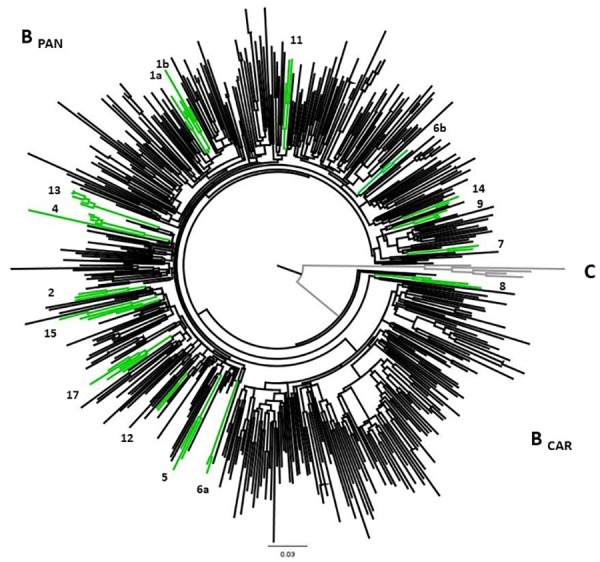
ML phylogenetic tree highlighting HIV-1 subtype B transmission clusters. The confirmed HIV-1 transmission clusters were highlighted in green and numbered according to the previous grouping from Figure 1. All clusters present aLRT ≥ 0.90 and low mean pairwise genetic distances (≤4.5). The analysis involved 520 HIV-1 B PR/RT sequences (102 sequences from the present study, 331 Brazilian reference sequences, 82 non-Brazilian reference sequences, and 5 HIV-1 Subtype C sequences as outgroup). The analyzed fragment corresponds to 891 bp (2262 to 3251 nt relative to HXB2 genome) and drug-resistance mutations positions were stripped.
Genotypic Analysis of HIV-1 Drug Resistance
To investigate the presence of TDRM, the sequences were submitted to Stanford HIV Database for Transmitted DRM [TDRM/Calibrated Population Resistance Tool (CPR Tool)] Version 6.0 (Gifford et al., 2009), which uses the mutation list according to Bennett et al. (2009).
Statistical Analysis
Statistical analyses were conducted using the SPSS 17.0 statistical analysis software package (SPSS Inc., Chicago, IL, United States). Median, standard deviation (SD), range and frequencies (%) were used to describe patients’ characteristics. The frequency of TDRMs was also calculated, and the chi-square or Fisher exact test was employed when appropriate. A p value of < 0.05 was defined as statistically significant.
Results
Out of 190 antiretroviral naïve patients who had samples available for DNA extraction, 172 were PR/RT amplified (90.5%), and from them 150 (87.2%) were successfully sequenced. From those 150 studied subjects, 62.0% were male, with an average age of 36 years, ranging from 18 to 70 years. More than half of participants were white (53.3%), heterosexual (64.0%), and reported less than 12 years of schooling (80.7%), and irregular condom use (54%). Only 6.7% of them were sex workers. Sociodemographic and behavioral characteristics are listed in Table 1. No statistically significant correlation was detected between the variables presented in Table 1 and HIV-1 subtypes.
Table 1.
Sociodemographic and behavioral characteristics of 150 cART-naïve subjects according to the HIV-1 most frequent subtypes, Central Brazil.
| Variable | N | (%) | Subtype B | Sub-subtype F1 | Subtype C |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 93 | (62.0) | 64 (63.4) | 10 (66.7) | 7 (58.3) |
| Female | 57 | (38.0) | 37 (36.6) | 5 (33.3) | 5 (41.7) |
| Age (years) | |||||
| 18–29 | 49 | (32.7) | 33 (32.7) | 3 (20.0) | 6 (50.0) |
| 30–39 | 51 | (34.0) | 36 (35.6) | 8 (53.3) | 2 (16.7) |
| 40 or more | 50 | (33.3) | 32 (31.7) | 4 (26.7) | 4 (33.3) |
| Skin color/ethnicity | |||||
| White | 80 | (53.3) | 57 (56.4) | 8 (53.3) | 6 (50.0) |
| Non-white | 70 | (46.7) | 44 (43.6) | 7 (46.7) | 6 (50.0) |
| Educational (years) | |||||
| 0 | 4 | (2.7) | 3 (3.0) | 0 (0.0) | 0 (0.0) |
| 1–12 | 117 | (78.0) | 79 (78.2) | 11 (73.3) | 9 (75.0) |
| ≥12 | 29 | (19.3) | 19 (18.8) | 4 (26.7) | 3 (25.0) |
| Monthly income | |||||
| <2 minimum wages | 20 | (13.3) | 11 (10.9) | 1 (6.7) | 2 (16.7) |
| 2–5 minimum wages | 97 | (64.7) | 65 (64.3) | 12 (80.0) | 6 (50.0) |
| >5 minimum wages | 31 | (20.7) | 24 (23.7) | 2 (13.3) | 4 (33.3) |
| Missing | 2 | (1.3) | 1 (0.1) | 0 (0.0) | 0 (0.0) |
| Frequency of alcohol consumption | |||||
| None | 92 | (60.9) | 63 (62.4) | 8 (53.3) | 7 (58.3) |
| Weekly | 51 | (35.1) | 34 (33.7) | 6 (40.0) | 3 (25.0) |
| Daily | 7 | (4.0) | 4 (3.9) | 1 (6.7) | 2 (16.7) |
| Illicit drug use | |||||
| No | 118 | (78.7) | 80 (79.2) | 11 (73.3) | 8 (66.7) |
| Yes, no injecting drugs | 28 | (18.6) | 18 (17.8) | 3 (20.0) | 4 (33.3) |
| Yes, injecting drugs | 4 | (2.7) | 3 (3.0) | 1 (6.7) | 0 (0.0) |
| Sexual orientation | |||||
| Heterosexual, female | 57 | (38.0) | 37 (36.6) | 5 (33.3) | 5 (41.7) |
| Heterosexual, male | 39 | (26.0) | 27 (26.8) | 3 (20.0) | 3 (25.0) |
| MSM | 54 | (36.0) | 37 (36.6) | 7 (46.7) | 4 (33.3) |
| Number of sexual partners in the last 12 months | |||||
| 0 | 13 | (8.7) | 6 (5.9) | 2 (13.2) | 1 (8.3) |
| 1 | 77 | (51.3) | 53 (52.5) | 7 (46.7) | 6 (50.0) |
| 2–5 | 40 | (26.7) | 28 (27.7) | 4 (26.7) | 4 (33.4) |
| 6–10 | 4 | (2.7) | 3 (3.0) | 1 (6.7) | 0 (0.0) |
| >10 | 16 | (10.6) | 11 (10.9) | 1 (6.7) | 1 (8.3) |
| Use of condoms in the last 12 months | |||||
| Always | 69 | (46.0) | 51 (50.5) | 6 (40.0) | 6 (50.0) |
| Occasionally/Never | 81 | (54.0) | 50 (49.5) | 9 (60.0) | 6 (50.0) |
| Presence of TDRM | |||||
| Yes | 18 | (12.0) | 14 (13.9) | 0 (0.0) | 2 (16.7) |
| No | 132 | (88.0) | 87 (86.1) | 15 (100) | 10 (83.3) |
MSM, men who have sex with men; TDRM, transmitted drug resistance mutations.
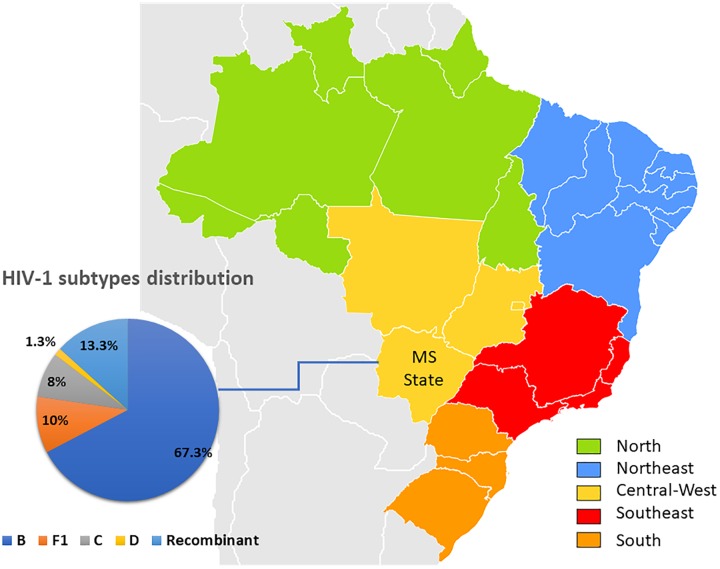
Phylogenetical analyses revealed that 101 sequences (67.3%) were classified as subtype B, 15 (10%) as F1, 12 (8%) as C, two (1.3%) as subtype D and 20 (13.3%) possible recombinants (Figure 1, 2A). The phylogenetic and bootscan analysis of these twenty sequences revealed that four (2.7%) were CRF28_29BF, three (2.0%) were CRF31_BC, one (0.7%) was CRF60_BC, and 12 (8.0%) were unique recombinant forms (URF) (Figure 2B).
FIGURE 1.
Map of Brazil indicating Brazilian regions and MS State, from which HIV-1 sequences were obtained. The pie chart shows HIV-1 subtypes distribution based on pol sequences included in this study.
TDRM to at least one class of antiretroviral drug was found in 18 sequences (12%), and the drug resistance mutation to nucleoside reverse transcriptase inhibitor (NRTI) was the most common (12/150; 8%), followed by non-nucleoside reverse transcriptase inhibitor (NNRTI) (7/150; 4.7%) and PI resistance (3/150; 2%) (Table 2). Of these, twelve (8%) were singleton mutations and six (4.0%) multiple. K103N was the most frequent resistance mutation observed (5/150; 3.3%) followed by V75M (4/150; 2.7%). There was no statistical difference between sexual orientation and the prevalence of TDRM and HIV-1 subtypes distribution.
Table 2.
Characteristics of the 18 cART-naïve subjects with TDRM.
| ID | Age/Gender | Resistance mutations | HIV-1 Subtype | Co-infection | ||
|---|---|---|---|---|---|---|
| NRTI | NNRTI | PI | ||||
| HET080 | 50/F | – | V106M | – | BF1 | Lifetime syphilisa |
| HET116 | 23/M | – | K103N | M46I | B | |
| HSH187 | 31/M | V75M | – | N88D | B | Lifetime syphilisa |
| HSH430 | 40/M | L210W, T215D | – | – | B | Hepatitis Bb |
| HET446 | 31/F | – | V106M | – | B | – |
| HET463 | 19/M | – | K103N | – | D | – |
| HSH502 | 27/M | D67N, K219Q | – | – | C | Lifetime syphilisa |
| HET510 | 40/F | M41L, T215D | – | M46I, V82T, L90M | B | – |
| HET521 | 26/F | K70R | – | – | B | – |
| HET545 | 40/M | – | K103N | – | B | Lifetime syphilisa Hepatitis Bb |
| HET573 | 37/F | V75M | – | – | B | – |
| HSH595 | 28/M | V75M | – | – | B | Hepatitis Bb |
| HET607 | 32/M | V75M | – | – | B | – |
| HET809 | 29/M | F77L | – | – | C | – |
| HET810 | 31/F | T215S | – | – | B | – |
| HSH851 | 21/M | L210W | – | – | B | – |
| HSH876 | 27/M | – | K103N | – | B | Lifetime syphilisa Hepatitis Bb |
| HSH878 | 22/M | M184V | K103N, P225H | – | B | Lifetime syphilisa |
HET, sample obtained from heterosexual individual; HSH, sample obtained from men who have sex with men; ID, sample identification; NNRTI, Non-Nucleoside Reverse Transcriptase Inhibitor; NRTI, Nucleoside Reverse Transcriptase Inhibitor; PI, Protease inhibitor. aLifetime syphilis: anti-Treponema pallidum positivity in ELISA. bHepatitis B: anti-HBc total and/or HBsAg seroposivity in ELISA.
Twenty-four possible transmission clusters, including 57 individuals were identified according to the adopted criteria (aLRT > 90 in ML analysis). The clusters involved from two to five individuals and seventeen of them belong to HIV-1 subtype B, one to subtype D, three to sub-subtype F1 (Figure 2A) and three were recombinant forms being 2 BF1 and 1 BD (Figure 2B). The inclusion of a huge number of reference sequences enabled reinvestigation by ML of the transmission clusters, in combination with the criteria of presenting high aLRT support and low mean genetic distance, allowed us to depict twenty previously identified possible transmission clusters from pure HIV-1 subtypes B, D, and F1. The possible transmission clusters 1c, 3, 10, and 21 were not confirmed. Some of the originally detected clusters remained with the same configuration (2,8,11,13,16, and 18); meanwhile, some of them presented a new shape. In the Cluster numbers (1, 4, 5, 9, 15, 17, 19, and 20) some Brazilian reference sequences clustered together to ours. We also verified that some sequences were excluded from the original possible clusters (1, 3, 6, 9, 10, 12, and 14). The possible clusters 1 and 6 give rise to two new transmission clusters (1a,b and 6a,b). The original possible clusters (Figure 2A,B) and the confirmed transmission clusters (Figure 3, 4) were summarized in Table 3. Since the clusters BD (22) and BF1 (23 and 24) were unique recombinant forms, we did not perform an additional ML phylogenetic tree.
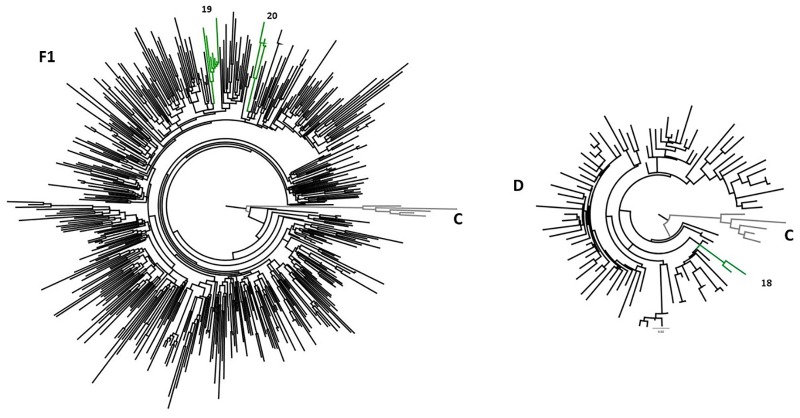
FIGURE 4.
ML phylogenetic tree showing the transmission clusters among HIV-1 subtypes of D and F1 sequences. The confirmed HIV-1 transmission clusters were highlighted in green and numbered according to the previous grouping from Figure 1. All clusters present aLRT ≥ 0.90 and low mean pairwise genetic distances (≤4.5). The sub-subtype F1 analysis involved 493 PR/RT sequences (15 sequences from the present study, 467 Brazilian reference sequences, 6 non-Brazilian reference sequences, and 5 HIV-1 Subtype C sequences as outgroup). From subtype D analysis, 94 sequences were used as follows: 2 detected in the present study, 17 HIV-1 Brazilian reference sequences, 72 non-Brazilian reference sequences and HIV-1 Subtype C sequences as outgroup).
Table 3.
Cluster confirmation of cART-naïve HIV-1 sequences according to aLRT and genetic distance.
| Figure 2 | Figure 3 | Transmission cluster confirmation | |||||
|---|---|---|---|---|---|---|---|
| Sequence | Cluster number | aLRT | Sequence | Cluster number | aLRT | Genetic distance | |
| Subtype HIV-1 B | |||||||
| HSH645 | 1 | 0.98 | HSH645 | 1a | 0.99 | 1.8 | Confirmed |
| HSH684 | HSH684 | ||||||
| HSH876 | BRMS57 | ||||||
| HSH878 | HSH878 | 1b | 0.93 | Confirmed | |||
| HSH543 | HSH876 | 3.5 | |||||
| HET220 | BRMS171 | ||||||
| HET673 | HSH543 | 1c | 0.9 | 6.0 | Not confirmed | ||
| HET075 | HET220 | ||||||
| HET673 | not confirmed | ||||||
| HET075 | not confirmed | ||||||
| HET141 | 2 | 0.95 | HET141 | 2 | 0.94 | Confirmed | |
| HET642 | HET642 | 4.2 | |||||
| HET737 | HET737 | ||||||
| HET362 | 3 | 0.91 | HET362 | 3 | 0.93 | 5.8 | Not confirmed |
| HET622 | HET622 | ||||||
| HET219 | HET219 | not confirmed | |||||
| HSH187 | 4 | 0.93 | HSH187 | 4 | 0.95 | 3.4 | Confirmed |
| HSH595 | HSH595 | ||||||
| HET607 | BRMS58 | ||||||
| BRMS14_10 | |||||||
| HET607 | |||||||
| HSH152 | 5 | 1 | HSH152 | 5 | 1 | Confirmed | |
| HSH430 | HSH430 | 3.9 | |||||
| BRMS40 | |||||||
| HSH408 | 6 | 0.92 | HSH408 | 6a | 1 | 1.3 | Confirmed |
| HSH862 | HSH862 | ||||||
| HSH871 | HSH871 | 6b | 0.99 | 1.4 | |||
| HET850 | BRMS97 | Confirmed | |||||
| BRMS99 | |||||||
| HET850 | not confirmed | ||||||
| HET671 | 7 | 1 | HET671 | 7 | 1 | 2.6 | Confirmed |
| HET438 | HET438 | ||||||
| HET517 | 8 | 1 | HET517 | 8 | 1 | 3.9 | |
| HET797 | HET797 | Confirmed | |||||
| HSH703 | HSH703 | ||||||
| HET562 | 9 | 0.93 | HET562 | 9 | 0.96 | 3.6 | |
| HET188 | BRMS55 | Confirmed | |||||
| BRMS05 | |||||||
| HET188 | not confirmed | ||||||
| HSH143 | 10 | 0.93 | HSH143 | not confirmed | Not confirmed | ||
| HET437 | HET437 | not confirmed | |||||
| HSH239 | 11 | 0.92 | HSH239 | 11 | 0.95 | Confirmed | |
| HSH066 | HSH066 | 4.4 | |||||
| HET694 | HET694 | ||||||
| HSH881 | 12 | 0.96 | HSH881 | 12 | 0.93 | 3.3 | Confirmed |
| HET249 | HET249 | ||||||
| HET814 | HET814 | not confirmed | |||||
| HSH874 | HSH874 | not confirmed | |||||
| HET367 | HET367 | not confirmed | |||||
| HET757 | 13 | 1 | HET757 | 13 | 1 | 1.3 | |
| HSH867 | HSH867 | ||||||
| HSH875 | HSH875 | Confirmed | |||||
| HSH716 | HSH716 | ||||||
| HSH717 | HSH717 | ||||||
| HSH53 | 14 | 0.98 | HSH53 | 14 | 0.99 | 1.9 | Confirmed |
| HSH94 | HSH94 | ||||||
| HSH240 | HSH240 | not confirmed | |||||
| HET111 | 15 | 0.98 | HET111 | 15 | 0.96 | 3.6 | |
| HSH109 | HSH109 | Confirmed | |||||
| MS34 | |||||||
| HET38 | 16 | 0.98 | HET38 | 16 | 0.94 | 3.7 | Confirmed |
| HET510 | HET510 | ||||||
| HET462 | 17 | 1 | HET462 | 17 | 0.98 | 3.4 | Confirmed |
| HET631 | HET631 | ||||||
| HET801 | HET801 | ||||||
| MS02 | |||||||
| MS46 | |||||||
| Figure 2 | Figure 4 | Transmission cluster confirmation | |||||
| Sequence | Cluster number | aLRT | Sequence | Cluster number | aLRT | Genetic distance | |
| Subtype D | |||||||
| HET463 | 18 | 1 | HET463 | 18 | 1 | 3.4 | Confirmed |
| HSH761 | HSH761 | ||||||
| Sub-subtype F1 | |||||||
| HSH006 | 19 | 1 | HSH006 | 19 | 0.97 | 4.0 | Confirmed |
| HSH264 | HSH264 | ||||||
| HSH265 | HSH265 | ||||||
| HSH387 | HSH387 | ||||||
| HSH534 | HSH534 | ||||||
| BR07SP153 | |||||||
| HET256 | 20 | 0.99 | HET256 | 20 | 1 | 3.6 | Confirmed |
| HET318 | HET318 | ||||||
| HET361 | HET361 | ||||||
| BRMS38 | |||||||
| HET140 | 21 | 0.92 | HET140 | 21 | 1 | 4.8 | Not confirmed |
| HSH739 | HSH739 | ||||||
| HET796 | HET796 | ||||||
| BRGO4074 | |||||||
| BRGO6051 | |||||||
| Recombinant BD | |||||||
| HET45 | 22 | 1 | |||||
| HET46 | |||||||
| Recombinant BF | |||||||
| HET643 | 23 | 1 | |||||
| HET122 | |||||||
| HET483 | |||||||
| HET793 | 24 | 0.99 | |||||
| HET353 | |||||||
HET: sample obtained from heterosexual individual; HSH: sample obtained from men who have sex with men.
All subtype B sequences were classified as pandemic B. Among subtype B confirmed clusters, twelve (12/17; 70.6%) had more than two sequences, and five (5/17; 29.4%) were composed of two sequences. Five clusters comprised MSM samples of this study with or without other Brazilian sequences (clusters 1a, 1b, 5, 6a, and 14), four with HET samples (clusters 2, 7, 16, and 17), six, mixed HET, and MSM sequences (clusters 4, 8, 11, 12, 13, and 15). Two clusters (6b and 9) were formed by one sequence from our study and two other Brazilian sequences from MS state, retrieved from Genbank (Table 3).
Individuals from ten clusters of subtype B were positive for lifetime syphilis and/or Hepatitis B and C infections. Four (4/17; 23.5%) contained sequences with TDRM, and two of them (clusters 1b and 4) were composed by MSM sharing the same TDRM. Cluster 1b included two MSM who had a history of Treponema pallidum infection and K103N mutation, and one of them reported being a sex worker and bisexual. Cluster 4 grouped two sequences from MSM (HSH187 and HSH595), one from a male HET, and sequences BRMS58 and BRMS14_10, both from males (da Silveira et al., 2012), all of them had the V75M substitution, associated with resistance to NRTI inhibitors.
Samples belonging to non-B subtypes were grouped into three clusters (Figure 4). Two of them (19 and 20), belonging to F1 subtype, contained more than two samples. The cluster 19 contained five sequences from MSM, three of which reported the use of illicit drugs and two were positive for syphilis (anti-T. pallidum). Additionally, cluster 19 also grouped a sequence from São Paulo (Brígido et al., 2011). The two samples characterized as subtype D clustered together (cluster 18).
Discussion
This phylogenetic study combined detailed clinical and epidemiological data, providing valuable data for surveillance, which allowed the monitoring of HIV-1 variants, TDRM, and associations between sociodemographic characteristics and behavioral sexual groups. It is noteworthy that the study subjects were antiretroviral naïve, and therefore, they were not in virologic suppression at the time of sample collection. This fact, associated with unprotected sexual practices, a multiplicity of sexual partners and a history of sexually transmitted infections (STIs), may be crucial for the maintenance of high HIV transmission rates.
In this study, HIV-1 B subtype was identified in 67.3% of the isolates, followed by recombinant forms, subtypes F1, C, and D. This distribution reflects that found in most Brazilian regions (da Silveira et al., 2012; de Moraes Soares et al., 2014). The frequency of 13.3% (95% CI: 7.9 to 18.8%) of recombinant forms found in this study was similar to that found in previous studies conducted in Central Brazil (16.3% and 14.5%) (Stefani et al., 2007; da Silveira et al., 2012). The absence of the Caribbean non-pandemic subtype B (BCAR) differs from the previous study by Divino et al. (2016), where a frequency of 5.5% from BCAR were detected in Mato Grosso do Sul. Previous studies conducted in a southern region of Brazil identified differences in the distribution of subtypes according to sex and exposure category (De Sa Filho et al., 2005; Dias et al., 2009). The present study is the first conducted in MS addressing this issue, and the lack of association herein can be justified by the high frequency of bisexual behavior (33.9%) reported by homosexual individuals from our cohort, suggesting that the differential transmission of subtypes according to the exposure category is not restricted to the MSM.
In the present study, an intermediate prevalence (12.0%) of TDRM was found, according to the WHO classification (Bennett et al., 2009), which is higher than that found in Northern Brazil (1.0%) (dos Anjos Silva et al., 2016) and is consistent with those found in previous Brazilian studies using similar sequencing technologies (6.8% to 17.2%) (Brindeiro et al., 2003; De Sa Filho et al., 2005; Cardoso et al., 2009; Sprinz et al., 2009; Alencar et al., 2013; Pessôa et al., 2015; Arruda et al., 2018). Recently, among crack cocaine users in Central Brazil, a high prevalence of TDRM was found (58.3%). It is worth noting that only 12 HIV-positive individuals were investigated (Da Silva França et al., 2018).
Recently, one study using massive parallel sequences of Brazilian blood donors found an overall prevalence of TDRM in PR and RT regions of the HIV-1 pol gene of 44.5% (Pessôa and Sanabani, 2017). Insufficient data to evaluate the time of HIV-1 infection and conventional sequencing usage may have caused an underestimation of TDRM prevalence (Palmer et al., 2005; Jain et al., 2011; Mohamed et al., 2014). Besides, it has been reported that significant inequalities in access to treatment persists in Brazil, resulting in different impacts on mortality in some groups, such as non-white individuals, or those with poor formal education (Lima et al., 2018).
It is remarkable that 4.0% of virus isolates obtained in this study had multiple mutations that may further influence the response to treatment. K103N, the most frequent resistance mutation observed, is commonly related to decreased susceptibility to efavirenz and nevirapine and the V75M mutation was associated with lamivudine and/or stavudine use (NNRTI). Some studies point out that genotyping tests before initiation of cART for all patients could be cost-effective in Brazil (Sanabani et al., 2011; Luz et al., 2015). However, these tests are still available only to specific populations, such as serodiscordant partners and HIV infected pregnant women.
Although HIV prevalence among MSM increased beyond expectations in Brazil, no difference between TDRM prevalence in homosexuals and heterosexuals was observed in this study. This result may reflect trends of feminization and the increase in heterosexual transmissions observed in Brazil (Brasil, 2017). In contrast, (Bermúdez-Aza et al., 2011) found higher TDRM prevalence in MSM (21.4%) recruited in Brazil by respondent-driven sampling, a particular sampling technique for hard-to-reach populations. As a result, transmission networks of resistance variants may have been selected among these MSM, thus reflecting this prevalence. Due to the higher prevalence of HIV infection in MSM (Kerr et al., 2018) and transgender women in Brazil (Grinsztejn et al., 2017), pre-exposure prophylaxis is recommended by the Brazilian Ministry of Health, who have made efforts to implement it and suggest it may be cost-effective (Luz et al., 2018).
Transmission clusters are frequently defined by low genetic distance (1.0%-4.5%) within cluster sequences and high support phylogenetic clusters (Lewis et al., 2008; Bezemer et al., 2010), herein employing both resources we were able to determine nineteen transmission clusters. However, more recently, transmission network approaches have also been used to this purpose, such as HIV clustering (Wertheim et al., 2014), Cluster picker and Cluster Matcher (Ragonnet-Cronin et al., 2013).
Seventeen transmission clusters were confirmed among subtype B isolates, some of them grouped patients with co-infections. Further evidence suggests that unprotected sexual intercourse and the presence of STIs that cause ulcerative lesions such as syphilis play important roles as cofactors in HIV transmission (Lynn and Lightman, 2004; Karp et al., 2009). This emphasizes the importance of prevention and treatment interventions.
Preventive actions regarding HIV-1 transmission are needed to disrupt the network and to reduce the spread of TDRM, since 29.4% of the clusters (5/17) contained samples with TDRM. Two of these groups were sharing the same substitution, showing the possibility of transmission of resistance between these individuals. Therefore, since 2013 the Brazilian Health Ministry recommendation, following the WHO recommendation, established that all HIV infected individuals should start the treatment to accomplish viral suppression, this being an effective way to reduce the HIV transmission (Brasil, 2013).
Clusters containing sequences from individuals with different sexual behaviors, including homosexual and bisexual contacts, were found in HIV-1 B (clusters 4, 8,11,12, 13, and 15) and D subtypes (cluster 18). Thus, factors such as being a sex worker, having multiple sexual partners, inconsistent condom use, and bisexual behavior may increase exposure to resistant HIV-1 isolates, both in heterosexual and homosexual networks.
The detection of clusters containing Brazilian samples from previous studies in Central-Western and Southeastern Brazil (Brígido et al., 2011; Cardoso et al., 2011; da Silveira et al., 2012) can be explained by the high mobility of the population, reinforcing the possibility of the spreading of infection despite great geographic distances, thus influencing local dynamics of diseases. Therefore, transmission networks and potential links with the different exposure categories should be further investigated in Brazil.
The study has some limitations. We interviewed all individuals face-to-face; consequently, risk behaviors may have been under-reported, leading to potential underestimation of associations with these variables and TDRM prevalence. Moreover, due to the study design, sample composition may not be representative of Campo Grande-MS epidemic and the absence of time of HIV-1 infection or diagnosis can portray an older epidemic. Even using a very limited number (1.4% from the total number of AIDS cases in Mato Grosso do Sul) of HIV-1 sequences from Mato Grosso do Sul, we were able to detect transmission clusters. However, we could not obtain detailed epidemiological information about the sequences from other Brazilian studies that were in some clusters. On the other hand, these findings enhance the understanding of the HIV-1 genetic characteristics, transmitted drug resistance, and transmission networks, as the research comprises not only individuals with epidemiological features in common but also the spread of strains between homosexuals and heterosexuals.
We highlight the urgent need for increased transmission monitoring of antiretroviral-resistant isolates, aiming for the selection of more effective therapeutic regimens, viral suppression, and hence the interruption of HIV-1 transmission networks. Improved understandings of risks, including potential linkages between sexual exposures among MSM, may contribute to designing preventive interventions and for improving HIV surveillance regarding TDRM in the largest country in Latin America.
Author Contributions
MLG and AM-C conceived the presented idea. TT, TFL, MLG, and AM-C discussed the results and wrote the manuscript. TT, SF, GC, and GR collected blood samples and also performed DNA extraction. AL provided medical support. TT, TFL, and MLG performed the experiments. TT, TFL, MLG, and AM-C analyzed the data. All the authors contributed to the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Priscila Brunini Zanini, GR and SF who started studying this population in Campo Grande, MS, and recruited a large number of subjects for this research. TT and AM-C also thank MLG for having welcomed us in the Fiocruz laboratory.
Funding. The authors acknowledge CNPq, Fundect-MS 0020/10 (Number 23/200.283/2009), and IOC for providing some financial support. MLG and AM-C are recipient of a CNPq fellowship. TT and TFL are funded by a CAPES Ph.D. fellowship.
References
- Alencar C. S., Sabino E. C., Carvalho S. M. F., Leao S. C., Carneiro-Proietti A. B., Capuani L., et al. (2013). HIV genotypes and primary drug resistance among HIV-seropositive blood donors in Brazil: role of infected blood donors as sentinel populations for molecular surveillance of HIV. J. Acquir. Immune Defic. Syndr. 63 387–392. 10.1097/QAI.0b013e31828ff979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55 539–552. 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- Arruda M. B., Boullosa L. T., Cardoso C. C., da Costa C. M., Alves C. R., de Lima S. T., et al. (2018). Brazilian network for HIV Drug Resistance Surveillance (HIV-BresNet): a survey of treatment-naive individuals. J. Int. AIDS Soc. 21:e25032. 10.1002/jia2.25032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D. E., Camacho R. J., Otelea D., Kuritzkes D. R., Fleury H., Kiuchi M., et al. (2009). Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4:e4724. 10.1371/journal.pone.0004724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-Aza E. H., Kerr L. R. F. S., Kendall C., Pinho A. A., de Mello M. B., Mota R. S., et al. (2011). Antiretroviral drug resistance in a respondent-driven sample of HIV-infected men who have sex with men in Brazil. J. Acquir. Immune Defic. Syndr. 57(Suppl. 3), S186–S192. 10.1097/QAI.0b013e31821e9c36 [DOI] [PubMed] [Google Scholar]
- Bezemer D., Van Sighem A., Lukashov V. V., Van Der Hoek L., Back N., Schuurman R., et al. (2010). Transmission networks of hiv-1 among men having sex with men in the Netherlands. AIDS 24 271–282. 10.1097/QAD.0b013e328333ddee [DOI] [PubMed] [Google Scholar]
- Booth C. L., Geretti A. M. (2007). Prevalence and determinants of transmitted antiretroviral drug resistance in HIV-1 infection. J. Antimicrob. Chemother. 59 1047–1056. 10.1093/jac/dkm082 [DOI] [PubMed] [Google Scholar]
- Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde. Departamento de DST, Aids e Hepatites Virais (2013). Protocolo clínico e Diretrizes Terapêuticas para Manejo da Infecção Pelo HIV em Adultos. [Google Scholar]
- Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde. Departamento de DST, Aids e Hepatites Virais (2017). Boletim Epidemiologico HIV/Aids. [Google Scholar]
- Brígido L. F. M., Ferreira J. L. P., Almeida V. C., Rocha S. Q., Ragazzo T. G., Estevam D. L., et al. (2011). Southern Brazil HIV type 1 C expansion into the state of São Paulo, Brazil. AIDS Res. Hum. Retrovir. 27 339–344. 10.1089/aid.2010.0157 [DOI] [PubMed] [Google Scholar]
- Brindeiro R. M., Diaz R. S., Sabino E. C., Morgado M. G., Pires I. L., Brigido L., et al. (2003). Brazilian network for HIV drug resistance surveillance (HIV-BResNet): a survey of chronically infected individuals. AIDS 17 1063–1069. 10.1097/01.aids.0000060345.12269.d7 [DOI] [PubMed] [Google Scholar]
- Cardoso L. P., Queiroz B. B., Stefani M. M. (2011). Molecular characteristics of HIV type 1 infection among prisoners from central western Brazil. AIDS Res. Hum. Retrovir. 27 1349–1353. 10.1089/aid.2011.0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso L. P. V., Queiroz B. B., Stefani M. M. (2009). HIV-1 pol phylogenetic diversity and antiretroviral resistance mutations in treatment naïve patients from Central West Brazil. J. Clin. Virol. 46 134–139. 10.1016/j.jcv.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Da Silva França D. D., Del-Rios N. H. A., Dos Santos Carneiro M. A., Guimarães R. A., Caetano K. A. A., Da Guarda Reis M. N., et al. (2018). HIV-1 infection among crack cocaine users in a region far from the epicenter of the HIV epidemic in Brazil: prevalence and molecular characteristics. PLoS One 13:e0199606. 10.1371/journal.pone.0199606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira A. A., Cardoso L. P. V., Francisco R. B. L., de Araújo Stefani M. M. (2012). HIV type 1 molecular epidemiology in pol and gp41 genes among naive patients from Mato Grosso do Sul State, Central Western Brazil. AIDS Res. Hum. Retrovir. 28 304–307. 10.1089/aid.2011.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Medeiros R. M., Junqueira D. M., Matte M. C. C., Barcellos N. T., Chies J. A. B., Almeida S. E. D. M. (2011). Co-circulation HIV-1 subtypes B, C and CRF31_BC in a drug-naïve population from southernmost Brazil: analysis of primary resistence mutations. J. Med. Virol. 83 1682–1688. 10.1002/jmv.22188 [DOI] [PubMed] [Google Scholar]
- de Moraes Soares C. M. P., Vergara T. R. C., Brites C., Brito J. D. U., Grinberg G., Caseiro M. M., et al. (2014). Prevalence of transmitted HIV-1 antiretroviral resistance among patients initiating antiretroviral therapy in Brazil: a surveillance study using dried blood spots. J. Int. AIDS Soc. 17:19042. 10.7448/IAS.17.1.19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sa Filho D. J., Sanabani S., Diaz R. S., Munerato P., Brunstein A., Fusuma E., et al. (2005). Analysis of full-length human immunodeficiency virus type 1 genome reveals a variable spectrum of subtypes B and F recombinants in São Paulo, Brazil. AIDS Res. Hum. Retrovir. 21 145–151. 10.1089/aid.2005.21.145 [DOI] [PubMed] [Google Scholar]
- Delatorre E., Silva-de-Jesus C., Couto-Fernandez J. C., Pilotto J. H., Morgado M. G. (2017). High HIV-1 diversity and prevalence of transmitted drug resistance among antiretroviral-naive HIV-infected pregnant women from Rio de Janeiro, Brazil. AIDS Res. Hum. Retrovir. 33 68–73. 10.1089/aid.2016.0159 [DOI] [PubMed] [Google Scholar]
- Dias C. F., Nunes C. C., Freitas I. O., Lamego I. S., De Oliveira I. M. R., Gilli S., et al. (2009). High prevalence and association of HIV-1 non-B subtype with specific sexual transmission risk among antiretroviral naïve patients in Porto Alegre, RS, Brazil. Rev. Inst. Med. Trop. Sao Paulo 51 191–196. 10.1590/S0036-46652009000400003 [DOI] [PubMed] [Google Scholar]
- Divino F., De Corado A. L. G., Naveca F. G., Stefani M. M. A., Bello G. (2016). High prevalence and onward transmission of non-pandemic HIV-1 subtype B clades in northern and northeastern Brazilian regions. PLoS One 11:e0162112. 10.1371/journal.pone.0162112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Anjos Silva L., Divino F., da Silva Rêgo M. O., Lima Lopes I. G., Nóbrega Costa C. M., da Silva Pereira F. C., et al. (2016). HIV-1 genetic diversity and transmitted drug resistance in antiretroviral treatment-naive individuals from Amapá State, Northern Brazil. AIDS Res. Hum. Retrovir. 32 373–376. 10.1089/aid.2015.0280 [DOI] [PubMed] [Google Scholar]
- Fernandes F. R. P., Zanini P. B., Rezende G. R., Castro L. S., Bandeira L. M., Puga M. A., et al. (2015). Syphilis infection, sexual practices and bisexual behaviour among men who have sex with men and transgender women: a cross-sectional study. Sex. Transm. Infect. 91 142–149. 10.1136/sextrans-2014-051589 [DOI] [PubMed] [Google Scholar]
- Freitas S. Z., Soares C. C., Tanaka T. S. O., Lindenberg A. S. C., Teles S. A., Torres M. S., et al. (2014). Prevalence, risk factors and genotypes of hepatitis B infection among HIV-infected patients in the State of MS, Central Brazil. Braz. J. Infect. Dis. 18 473–480. 10.1016/j.bjid.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R. J., Liu T. F., Rhee S. Y., Kiuchi M., Hue S., Pillay D., et al. (2009). The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 25 1197–1198. 10.1093/bioinformatics/btp134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräf T., Passaes C. P. B., Ferreira L. G. E., Grisard E. C., Morgado M. G., Bello G., et al. (2011). HIV-1 genetic diversity and drug resistance among treatment naïve patients from Southern Brazil: an association of HIV-1 subtypes with exposure categories. J. Clin. Virol. 51 186–191. 10.1016/j.jcv.2011.04.011 [DOI] [PubMed] [Google Scholar]
- Gräf T., Pinto A. R. (2013). The increasing prevalence of HIV-1 subtype C in Southern Brazil and its dispersion through the continent. Virology 435 170–178. 10.1016/j.virol.2012.08.048 [DOI] [PubMed] [Google Scholar]
- Grinsztejn B., Jalil E. M., Monteiro L., Velasque L., Moreira R. I., Garcia A. C. F., et al. (2017). Unveiling of HIV dynamics among transgender women: a respondent-driven sampling study in Rio de Janeiro, Brazil. Lancet HIV 4 e169–e176. 10.1016/S2352-3018(17)30015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães M. L., Marques B. C. L., Bertoni N., Teixeira S. L. M., Morgado M. G., Bastos F. I., et al. (2015). Assessing the HIV-1 epidemic in Brazilian drug users: a molecular epidemiology approach. PLoS One 10:e0141372. 10.1371/journal.pone.0141372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and Methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 2.0. Syst. Biol. 59 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Jain V., Sucupira M. C., Bacchetti P., Hartogensis W., Diaz R. S., Kallas E. G., et al. (2011). Differential persistence of transmitted HIV-1 drug resistance mutation classes. J. Infect. Dis. 203 1174–1181. 10.1093/infdis/jiq167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp G., Schlaeffer F., Jotkowitz A., Riesenberg K. (2009). Syphilis and HIV co-infection. Eur. J. Intern. Med. 20 9–13. 10.1016/j.ejim.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Kerr L., Kendall C., Guimarães M. D. C., Mota R. S., Veras M. A., Dourado I., et al. (2018). HIV prevalence among men who have sex with men in Brazil: results of the 2nd national survey using respondent-driven sampling. Medicine 97(1S Suppl. 1), S9–S15. 10.1097/MD.0000000000010573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V., Longueville J. E., Gascuel O. (2017). SMS: smart model selection in PhyML. Mol. Biol. Evol. 34 2422–2424. 10.1093/molbev/msx149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis F., Hughes G. J., Rambaut A., Pozniak A., Leigh Brown A. J. (2008). Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 5:e50. 10.1371/journal.pmed.0050050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima T. A., Beyrer C., Golub J. E., Mota J. C. D., Malta M. S., Silva C. M. F. P. D., et al. (2018). Inequalities in HAART uptake and differential survival according to exposure category in Rio de Janeiro, Brazil. Cad. Saude Publica. 34:e00009617. 10.1590/0102-311x00009617 [DOI] [PubMed] [Google Scholar]
- Lole K. S., Bollinger R. C., Paranjape R. S., Gadkari D., Kulkarni S. S., Novak N. G., et al. (1999). Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz P. M., Morris B. L., Grinsztejn B., Freedberg K. A., Veloso V. G., Walensky R. P., et al. (2015). Cost-effectiveness of genotype testing for primary resistance in Brazil. J. Acquir. Immune Defic. Syndr. 68 152–161. 10.1097/QAI.0000000000000426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz P. M., Osher B., Grinsztejn B., Maclean R. L., Losina E., Stern M. E., et al. (2018). The cost-effectiveness of HIV pre-exposure prophylaxis in men who have sex with men and transgender women at high risk of HIV infection in Brazil. J. Int. AIDS Soc. 21:e25096. 10.1002/jia2.25096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn W. A., Lightman S. (2004). Syphilis and HIV: a dangerous combination. Lancet Infect. Dis. 4 456–466. 10.1016/S1473-3099(04)01061-8 [DOI] [PubMed] [Google Scholar]
- Machado L. F. A., Ishak M. O. G., Vallinoto A. C. R., Lemos J. A. R., Azevedo V. N., Moreira M. R. C., et al. (2009). Molecular epidemiology of HIV type 1 in Northern Brazil: identification of subtypes C and D and the introduction of CRF02_AG in the Amazon region of Brazil. AIDS Res. Hum. Retrovir. 25 961–966. 10.1089/aid.2009.0027 [DOI] [PubMed] [Google Scholar]
- Mohamed S., Penaranda G., Gonzalez D., Camus C., Khiri H., Boulmé R., et al. (2014). Comparison of ultra-deep versus Sanger sequencing detection of minority mutations on the HIV-1 drug resistance interpretations after virological failure. AIDS 28 1315–1324. 10.1097/QAD.0000000000000267 [DOI] [PubMed] [Google Scholar]
- Palmer S., Kearney M., Maldarelli F., Halvas E. K., Bixby C. J., Bazmi H., et al. (2005). Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43 406–413. 10.1128/JCM.43.1.406-413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso C., Queiroz A. T. L., Alcĝntara L. C., Drexler J. F., Diaz R. S., Weyll N., et al. (2007). High prevalence of primary antiretroviral resistance among HIV-1-infected adults and children in Bahia, a northeast state of Brazil [3]. J. Acquir. Immune Defic. Syndr. 45 251–253. 10.1097/QAI.0b013e318050d8b0 [DOI] [PubMed] [Google Scholar]
- Pessôa R., Sanabani S. S. (2017). High prevalence of HIV-1 transmitted drug-resistance mutations from proviral DNA massively parallel sequencing data of therapy-naïve chronically infected Brazilian blood donors. PLoS One 12:e0185559. 10.1371/journal.pone.0185559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessôa R., Watanabe J. T., Calabria P., Alencar C. S., Loureiro P., Lopes M. E., et al. (2015). Enhanced detection of viral diversity using partial and near full-length genomes of human immunodeficiency virus Type 1 provirus deep sequencing data from recently infected donors at four blood centers in Brazil. Transfusion 55 980–990. 10.1111/trf.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragonnet-Cronin M., Hodcroft E., Hué S., Fearnhill E., Delpech V., Brown A. J. L., et al. (2013). Automated analysis of phylogenetic clusters. BMC Bioinformatics 14:317. 10.1186/1471-2105-14-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabani S. S., de Pastena É. R. S., da Costa A. C., Martinez V. P., Kleine-Neto W., de Oliveira A. C. S., et al. (2011). Characterization of partial and near full-length genomes of HIV-1 strains sampled from recently infected individuals in São Paulo, Brazil. PLoS One 6:e25869. 10.1371/journal.pone.0025869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. M. G., Telles F. Q., da Cunha C. A., Rhame F. S. (2010). HIV subtype, epidemiological and mutational correlations in patients from Paraná, Brazil. Braz. J. Infect. Dis. 14 495–501. 10.1590/S1413-86702010000500012 [DOI] [PubMed] [Google Scholar]
- Souza Junior P. R. B., Borges De Souza Junior P. R., Szwarcwald C. L., Ayres De Castilho E. (2011). Self-rated health by HIV-infected individuals undergoing antiretroviral therapy in Brazil. Cad. Saude Publica 2011(27 Suppl. 1), S56–S66. [DOI] [PubMed] [Google Scholar]
- Sprinz E., Netto E. M., Patelli M., Lima J. S., Furtado J. J. D., da Eira M., et al. (2009). Primary antiretroviral drug resistance among HIV type 1-infected individuals in Brazil. AIDS Res. Hum. Retrovir. 25 861–867. 10.1089/aid.2009.0012 [DOI] [PubMed] [Google Scholar]
- Stefani M. M. A., Pereira G. A. S., Lins J. A. B., Alcantara K. C., Silveira A. A., Viegas A. A., et al. (2007). Molecular screening shows extensive HIV-1 genetic diversity in Central West Brazil. J. Clin. Virol. 39 205–209. 10.1016/j.jcv.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS (2018). AIDS info [Internet]. The Joint United Nations Programme on HIV/AIDS. Available at: http://aidsinfo.unaids.org/# [Google Scholar]
- Wertheim J. O., Leigh Brown A. J., Hepler N. L., Mehta S. R., Richman D. D., Smith D. M., et al. (2014). The global transmission network of HIV-1. J. Infect. Dis. 209 304–313. 10.1093/infdis/jit524 [DOI] [PMC free article] [PubMed] [Google Scholar]