Abstract
The poly(A) RNA binding Zn finger ribonucleoprotein Nab2 functions to control the length of 3′ poly(A) tails in Saccharomyces cerevisiae as well as contributing to the integration of the nuclear export of mature mRNA with preceding steps in the nuclear phase of the gene expression pathway. Nab2 is constructed from an N‐terminal PWI‐fold domain, followed by QQQP and RGG motifs and then seven CCCH Zn fingers. The nuclear pore‐associated proteins Gfd1 and Mlp1 bind to opposite sides of the Nab2 N‐terminal domain and function in the nuclear export of mRNA, whereas the Zn fingers, especially fingers 5–7, bind to A‐rich regions of mature transcripts and function to regulate poly(A) tail length as well as mRNA compaction prior to nuclear export. Nab2 Zn fingers 5–7 have a defined spatial arrangement, with fingers 5 and 7 arranged on one side of the cluster and finger 6 on the other side. This spatial arrangement facilitates the dimerization of Nab2 when bound to adenine‐rich RNAs and regulates both the termination of 3′ polyadenylation and transcript compaction. Nab2 also functions to coordinate steps in the nuclear phase of the gene expression pathway, such as splicing and polyadenylation, with the generation of mature mRNA and its nuclear export. Nab2 orthologues in higher Eukaryotes have similar domain structures and play roles associated with the regulation of splicing and polyadenylation. Importantly, mutations in the gene encoding the human Nab2 orthologue ZC3H14 and cause intellectual disability.
Keywords: RNA nuclear export, Nab2, polyadenylation, mRNA nuclear processing, poly(A) tail
Introduction
In eukaryotes, the separation by the nuclear envelope of transcription from translation enables mRNAs to be modified by capping, splicing, and polyadenylation. These processing steps are mediated by a large number of different proteins that interact with transcripts as they pass through the nuclear phase of the gene expression pathway before they are finally exported to the cytoplasm through nuclear pores.1, 2, 3 In budding yeast, Nab2, which is the founding member of an evolutionarily conserved family of polyadenosine RNA binding Zn finger proteins,4 functions in regulating the length of poly(A) tails, compacting mature transcripts, and coordinating key nuclear RNA processing steps with RNA export (reviewed in Reference 5). Saccharomyces cerevisiae Nab2 (ScNab2) accompanies mature transcripts as they move through nuclear pores to the cytoplasm, after which ScNab2 is thought to be removed from the mRNA by the DEAD‐box RNA helicase, Dbp56 that is located at the cytoplasmic face of the pore. ScNab2 is then recycled back to the nucleus through nuclear pores using the transport factor, karyopherin β2 (also termed Kap104 or transportin).7, 8 In addition, ScNab2 shows genetic interactions with the splicing machinery9 and also appears to function in mRNA quality control10 and Pol‐III transcription.11
In higher eukaryotes, the human Nab2 orthologue, ZC3H14, and Drosophila orthologue, DmNab2 are also required for proper poly(A) tail length control,12, 13 in addition to the well‐characterized poly(A) tail length regulator and nuclear poly(A) binding protein, PABPN1.14, 15 Importantly, mutations in the human ZC3H14 gene have been linked to a nonsyndromic form of autosomal recessive intellectual disability,12, 16 linking Nab2/ZC3H14 to proper neuronal function. In strong support of a role for Nab2 in the brain, DmNab2 mutant flies exhibit impaired short‐term memory and defects in neuronal patterning in the learning and memory center (mushroom body) of the fly brain.12, 17 Notably, neuronal expression of human ZC3H14 in DmNab2 mutant flies rescues function, indicating that ZC3H14 is a functional orthologue of DmNab2.13
Molecular Architecture of Nab2 Family Members
Members of the Nab2 protein family share a common architecture (Fig. 1) exemplified by the founding member ScNab2 in S. cerevisiae. ScNab2 contains an N‐terminal domain that has a Proline‐Tryptophan‐Isoleucine (PWI)‐like fold,18 followed by a Glutamine (Q)‐rich region of variable length, an Arginine‐Glycine (RGG) domain that functions as a nuclear targeting sequence in budding yeast, and finally a C‐terminal domain that contains seven tandem Cysteine‐Cysteine‐Cysteine‐Histidine (CCCH) Zn fingers that mediates high‐affinity binding to polyadenosine RNA.4, 19 These ScNab2 Zn fingers (ZnFs) are arranged into three groups: ZnF12; ZnF34; and ZnF567. The Nab2 orthologues in other species—S. pombe Nab2 (SpNab2); C. thermophilum Nab2 (CtNab2); D. melanogaster Nab2 (DmNab2); C. elegans SUT‐2 (CeSUT‐2); H. sapiens ZC3H14 (HsZC3H14)—have a similar overall domain architecture (Fig. 1). Although the N‐terminal domain (PWI fold) and Zn finger domain are highly conserved in all Nab2 orthologues, the number of Zn fingers varies in Nab2 orthologues with SpNab2 having only three and human ZC3H14 and most other Nab2 orthologues having five (Fig. 1).
Figure 1.
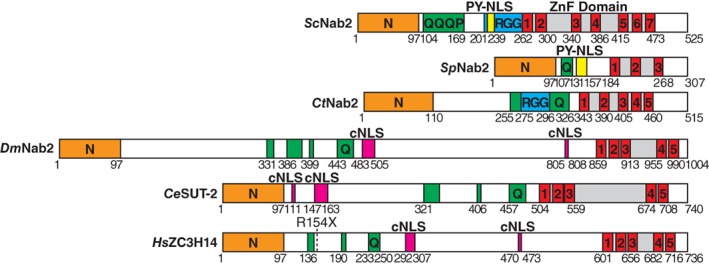
Members of the Nab2/ZC3H14 zinc finger polyadenosine RNA binding protein family have a similar domain architecture. Nab2 orthologues – S. cerevisiae Nab2 (ScNab2; Uniprot ID: P32505), S. pombe Nab2 (SpNab2; Uniprot ID: O13713), C. thermotolerans Nab2 (CtNab2; Uniprot ID: G0SCL7), Drosophila melanogaster Nab2 (DmNab2; Uniprot ID: Q9V3H9), Caenorhabditis elegans SUT‐2 (CeSUT‐2; Uniprot ID: Q95XU6), and Homo sapiens (HsZC3H14; Uniprot ID: Q6PJT7) share a common domain structure based on a N‐terminal PWI‐fold domain (orange), which serves as a protein–protein interaction domain in ScNab2, followed by a Q‐rich region (green), and a C‐terminal Zn finger (ZnF) domain (gray) containing a series of Zn fingers (red), which bind to polyadenosine RNA. ScNab2 and CtNab2 contain an RGG domain (blue), which functions in karyopherin‐based nuclear import in ScNab2. In addition, ScNab2 contains a Pro‐Tyr nuclear localization signal (PY‐NLS) (yellow) and SpNab2 contains a predicted PY‐NLS that functions in nuclear import in ScNab2. Nab2 orthologues from higher Eukaryotes contain two predicted classical nuclear localization signals (cNLS) (magenta) that function in karyopherin‐based nuclear import. The nonsense mutation R154X identified in ZC3H14 in individuals with autosomal recessive intellectual disability is highlighted.
Although the steady‐state localization for the Nab2 family members that have been studied is nuclear,4, 12, 19, 20, 21, 22, 23 there is evidence that ScNab2 can shuttle into and out of the nucleus24, 25 and in higher eukaryotes Nab2/ZC3H14 can be detected in the cytoplasm of neuronal cells.26 As shown in Figure 1, the nuclear localization signals (NLSs) that target Nab2 family members to the nucleus vary between species. The RGG nuclear targeting signal within ScNab2 has been defined experimentally,7 as has a PY‐NLS motif in ScNab2.27 In higher eukaryotes, the RGG nuclear targeting domain is replaced by two predicted classical lysine‐rich nuclear localization signals (cNLSs).
Binding to polyadenosine RNA, a primary function of the Nab2 protein family, is mediated by the Zn finger motifs.4, 19, 22 Functional studies of the Zn fingers have been performed most extensively in ScNab2 and have been complemented with structural studies that provide insight into recognition of poly(A) RNA using CtNab2 and ScNab2.28, 29, 30 The grouping of the Zn fingers varies somewhat among species (Fig. 1). ScNab2 Zn fingers 5, 6, and 7 (ZnF567) are critical for function, and studies demonstrate that a nab2 mutant lacking ZnF567 is not functional in budding yeast.19 Structure–function studies28, 30 have also defined key conserved residues in ZnF567 that are important for the proper function of ScNab2.
Structure and Function of the Nab2 N‐Terminal Domain
The crystal and solution structures of the ScNab2 N‐terminal domain (Nab2‐N)31, 32 showed that Nab2‐N has a PWI fold that is based on five α‐helices [Fig. 2(a)]. A budding yeast nab2 mutant in which the N‐terminal domain has been deleted (nab2‐ΔN) exhibits severely impaired growth and nuclear accumulation of poly(A) RNA.19, 33 Moreover, bulk poly(A) tails are substantially longer in nab2‐ΔN cells, indicating that the N‐terminal domain might contribute to the control of poly(A) tail length.34 The ScNab2 N‐terminal domain interacts physically with both Mlp1, a component of the nuclear basket that is located on the nuclear face of nuclear pores,31, 35, 36 and Gfd1, which is thought to reinforce the function of the RNA export factor, Gle1.31, 35, 36 Mutagenesis studies have indicated that ScNab2 Phe73, which is located on a hydrophobic surface patch on the Nab2 N‐terminal domain [Fig. 2(b)], is important for the interaction with Mlp1, since ScNab2 F73A and F73D variants show impaired binding to Mlp1 in yeast lysates and in vitro.31, 37 The Mlp1‐Nab2 interaction could function to concentrate mature polyadenylated transcripts at the nuclear face of nuclear pores to facilitate their export to the cytoplasm37, 38 and also plays an important role in mRNP quality control.39
Figure 2.
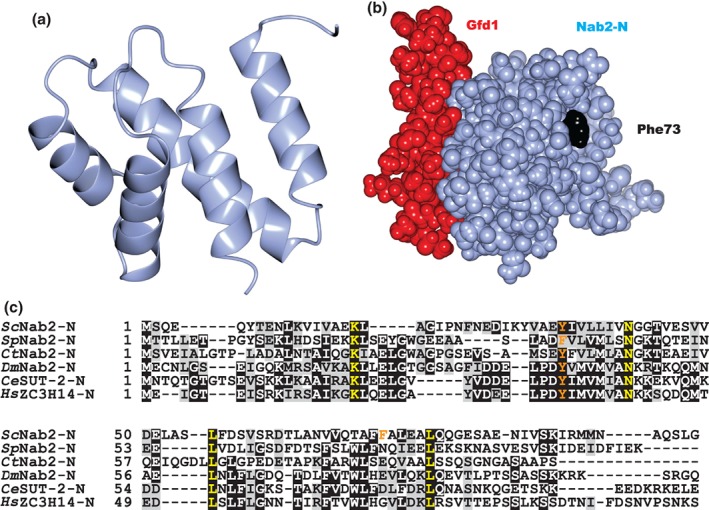
Structure of the S. cerevisiae Nab2 N‐terminal domain. (a) The Nab2 N‐terminal domain (Nab2‐N) has a PWI fold based on five α‐helices. (b) Interaction interfaces between Nab2‐N and the nuclear pore‐associated proteins Mlp1 (centred on Phe73 – Black) and Gfd1 (red) are on opposite sides of the Nab2‐N domain (based on PDB 3LCN). (c) The Nab2 N‐terminal domain is highly conserved between different Nab2 orthologues. Identical residues in all Nab2 orthologues are highlighted in yellow. ScNab2 residue Phe73 that is important for interaction with Mlp1, and ScNab2 residue Tyr34 that is important for interaction with Gfd1 are highlighted in orange. Identical residues (black) and similar residues (gray) are highlighted.
The ScNab2 N‐terminal domain also interacts with Gfd1,32 which is a multicopy suppressor of the dbp5(rat8‐2) RNA helicase mutant40 and the gle1‐8 RNA export factor mutant.41 Crystallography, supported by solution NMR studies, showed that Gfd1 residues 126–150 form a helix when bound to the ScNab2 N‐terminal domain and identified a key contribution made by ScNab2 Tyr34, which is located on the opposite side of the N‐terminal domain to ScNab2 Phe73, which is important for Mlp1 binding32 [Fig. 2(b,c)]. Critically, a nab2‐Y34A dbp5(rat8‐2) double mutant shows a synthetic slow growth phenotype.32 Together, these results support the importance of the Nab2‐Gfd1 interaction for Dbp5 function, which is crucial for remodeling mRNPs following nuclear export in vivo.
Although the N‐terminal domain is conserved in Nab2 orthologues [Fig. 2(c)], there is currently little information regarding the function of this domain in higher eukaryotes. However, deletion of the N‐terminal domain of human ZC3H14 (ZC3H14‐N) does not impair its nuclear localization,42 consistent with the observation that the predicted cNLSs of ZC3H14 are C‐terminal of ZC3H14‐N (Fig. 1). The observation that ScNab2‐N interacts with the nuclear pore associated proteins, Mlp1 and Gfd1, suggests that Nab2‐N PWI domain could also function as a protein–protein interaction module in higher eukaryotes, although no partners have currently been identified.
Structure of Nab2 Zn Fingers
The structures of several Nab2 Zn finger clusters have been established using both X‐ray crystallography and NMR.28, 29, 30, 43 In contrast to many other Zn finger proteins, all the Nab2 structures show that the Zn fingers interact with one another to varying extents and so have defined orientations to one another. ScNab2 ZnF56728, 30 and the corresponding ZnF345 from CtNab229 form single structural units, in which the Zn fingers are arranged so that the first and third Zn finger are arranged on one side of the unit, with the middle Zn finger directed toward the opposite side. This 3‐dimensional spatial relationship between the Zn fingers precludes a single poly(A) RNA chain binding to all of them simultaneously. Similarly, NMR has demonstrated that ScNab2 ZnF1 and ZnF2 interact with one another as do ScNab2 ZnF3 and ZnF4 to form defined structural units.43
In ScNab2 ZnF567, NMR chemical shift perturbations associated with binding either AMP or A3 identified a series of basic and aromatic residues associated with RNA binding28 [Fig. 3(b)]. These ScNab2 ZnF567 residues are strongly conserved between Nab2 orthologues [Fig. 4(d)] and ScNab2 variants in which these ZnF residues were substituted showed reduced affinity for A9 RNA.28 Although nab2 ZnF567 mutants containing substitutions of these basic and aromatic residues had growth rates similar to wild‐type cells, the nab2 ZnF567 mutants generated longer poly(A) tails in vivo and also showed genetic interactions with both Dbp5 and Yra1, consistent with their also influencing the generation of mature mRNPs.28 Moreover, in ScNab2, structural coherence between ZnF567 was lost in the ScNab2 RNA‐binding mutant, nab2‐C437S, in which a Ser was substituted for the first Zn‐coordinated Cys in ZnF6 [Fig. 3].28 Importantly, the nab2‐C437S yeast mutant exhibited cold‐sensitive growth and hyperadenylation of bulk poly(A) tails.28 Furthermore, combining the nab2‐C437S mutant with the dbp5(rat8‐2) RNA helicase mutant suppressed the growth defect of the dbp5(rat8‐2) mutant.28 Analysis of additional structure‐guided nab2 ZnF mutants in combination with the dbp5(rat8‐2) mutant indicated that dbp5(rat8‐2) suppression by nab2 ZnF mutants was more closely linked to hyperadenylation and suppression of mutant alleles of the nuclear RNA export adaptor, Yra1, than to the affinity of the mutant Nab2 for poly(A) RNA.28 Overall, these results indicate that, in addition to modulating poly(A) tail length, ScNab2 has an unanticipated function associated with generating export‐competent mRNPs, and that changes within ZnF567 lead to suboptimal assembly of mRNP export complexes that are more easily disassembled by Dbp5 upon reaching the cytoplasm.
Figure 3.
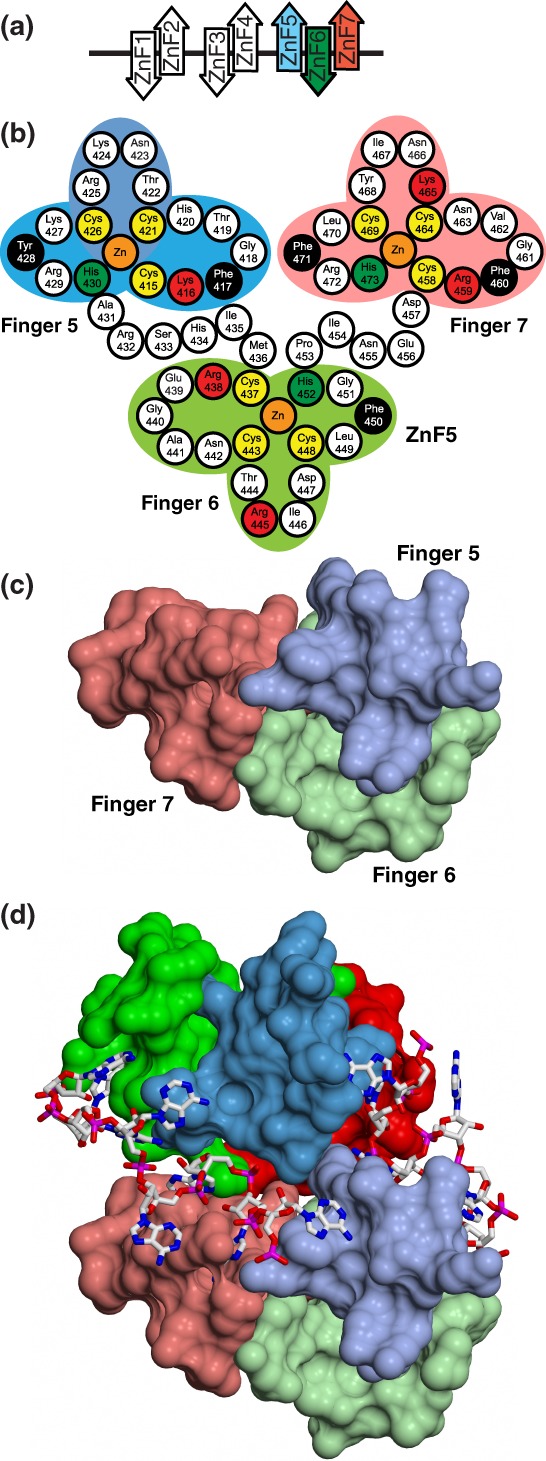
Nab2 Zn fingers. (a) Schematic illustration of the structures of the S. cerevisiae Nab2 Zn fingers determined by NMR (28, 43) and X‐ray crystallography.29, 30 The Zn fingers are clustered into three groups, and the fingers within each group have a defined orientation that influences their interaction with adenine‐rich RNA chains. (b) Arrangement of residues in ScNab2 Zn fingers 5–7. Key hydrophobic and basic residues that are important for binding to adenines (see also Fig. 4) are black and red, respectively. (c) Structure of ScNab2 Zn fingers 567. Zn finger 6 (green) projects on the opposite of the module to Zn fingers 5 (blue) and 7 (red). (d) Heterotetramer produced by A11G binding to ScNab2 Zn fingers 567 in which each RNA chain binds to both protein chains. Based on PDB 5L2L Panel (B) is derived from C. Brockmann et al. (2012) “Structural basis for polyadenosine‐RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export.” Structure, 20:1007–1018.
Figure 4.
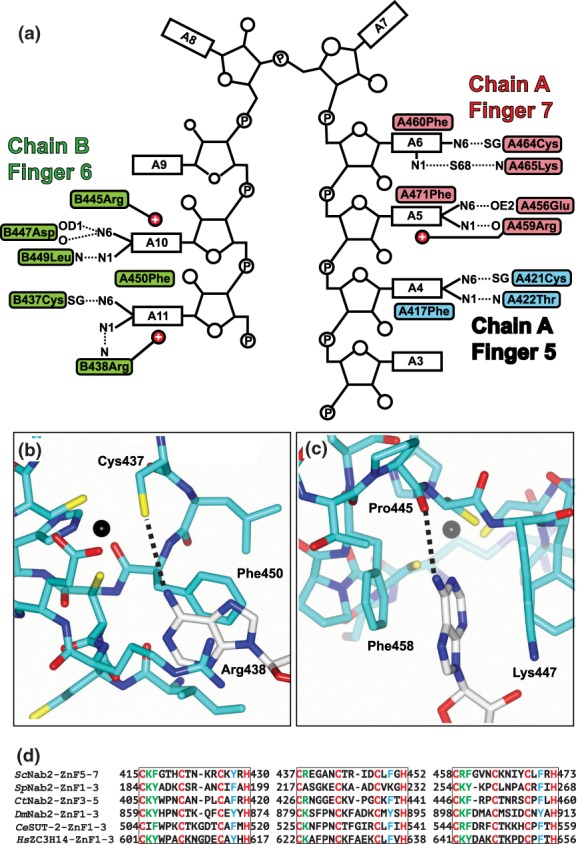
Interaction of ScNab2 Zn fingers 567 with adenine‐rich RNA. (a) Interactions between a single A11G RNA chain and the two Nab2 chains within a heterotetramer. Adenine rings are bound into surface clefts and are sandwiched between an aromatic residue on one side and a basic residue on the other that participates in a cation‐π interaction. The specificity of the interaction derives from H‐bonds formed by the adenine N6 and either a cysteine SG bound to Zn (b) or a main chain carbonyl (c). In each case, guanine cannot interact in this way because its O6 lacks hydrogen. Based on PDB 5L2L. (d) ScNab2 Zn fingers 567 are evolutionarily highly conserved between different Nab2 orthologues. ScNab2 ZnF567 are most similar to ZnF123 in most Nab2 orthologues and ZnF345 in CtNab2. Key conserved basic and aromatic residues (green) follow the first cysteine in the Zn fingers and conserved aromatic residues (blue) follow the third cysteine in Zn fingers. These conserved basic and aromatic residues interact specifically with polyadenosine RNA in S. cerevisiae and C. thermophilum Nab2. Panels (a), (b), and (c) reproduced from S. Aibara et al. “Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae.” Nucleic Acids Res. 45:1529–1538 (2017) by permission of Oxford University Press. Available at DOI: 10.1093/nar/gkw1224.
Structural Basis for the Interaction of the Zn Fingers with Poly(A) RNA
The crystal structures of ScNab2 ZnF56730 and CtNab2 ZnF34529 bound to A11G and A8 RNA, respectively, indicated the basis for selective binding of Nab2 Zn fingers to adenine and identified the importance of H‐bonds formed by adenine N6 (Figs. 3 and 4). In both structures of Nab2 Zn fingers bound to poly(A) RNA, the purine ring binds in a surface groove, where it stacks against an aromatic side chain on one side with a basic residue forming a cation‐π interaction on the other. These ScNab2 aromatic and basic Zn finger residues are strongly conserved in other Nab2 orthologues [Fig. 4(d)], and interactions between these ScNab2 residues and adenines were also seen in NMR studies.28 In addition, ScNab2 Zn finger variants in which these aromatic and basic residues were substituted showed reduced affinity for poly(A) RNA in vitro and generated longer bulk poly(A) tails in vivo.28 Specificity for the interaction is provided by a novel pattern of H‐bonds, most commonly between purine N6 and a Zn‐coordinated Cys residue or a main‐chain carbonyl, supplemented by H‐bonds between purine N7 and backbone amides. In both interactions involving adenine N6, the H‐bond formed either to a Cys SG thiol or a main‐chain carbonyl cannot be formed with guanine because its O6 does not have a donor hydrogen [Fig. 4(b,c)].
In the structures of both ScNab2 Zn fingers 567 and CtNab2 Zn fingers 345 complexed with poly(A) RNA, the spatial arrangement of the fingers precludes them from all binding to the same RNA chain, so that the first and third Zn fingers of the module were bound to one RNA chain, whereas the middle Zn finger was bound to a second RNA chain. Moreover, the crystal structure of ScNab2 ZnF567 complexed with A11G30 showed that binding RNA generated a distinctive heterotetramer that contained two protein chains and two RNA chains (Figs. 3(d) and 4(a)). In vitro binding studies30 demonstrated that this heterotetramer was also formed in solution between ScNab2 ZnF567 and either A12 or A11G, indicating that it was not a crystallization artifact and also that the 3′ terminal G was not necessary for its formation. Unusually, the dimerization of the ScNab2 protein chains was mediated almost entirely by each RNA chain binding to both protein chains and not by specific interactions between residues on the protein chains themselves, precluding the engineering of ScNab2 variants in which the Nab2‐Nab2 interaction was impaired. However, ScNab2 dimerization was impaired in Nab2 variants in which RNA binding was disturbed, with substitution of Phe450 in ZnF6, that is important for the interaction with the two adenines that bind ZnF6 (Fig. 4), showing the greatest decrease.30 Compared with other ScNab2 ZnF variants, ScNab2 F450A also showed one of the largest decreases in affinity for A8 RNA in vitro coupled with one of the largest increases in poly(A) tail length in vivo.28 Furthermore, the ScNab2 F450A variant exhibited some of the strongest genetic interactions with Yra1 and Dbp5.28 Combined, these data underscore the importance for Nab2 function of the dimerization induced by its binding to adenine‐rich RNA.
In the ScNab2 ZnF567‐A11G heterotetramer, not all the adenines interact directly with the Nab2 protein chains, suggesting that Nab2 could also form analogous dimers with RNA sequences that only contain adenines in key positions [Fig. 5(a)]. Binding to such A‐rich RNA sequences would be consistent with the observation that ScNab2 binds along the coding region of transcripts as well as to the poly(A) tail,44, 45 lending weight to the proposal46 that Nab2 could also have a function in mRNP compaction. Support for this hypothesis was provided by assessing the function of ScNab2 ZnF567 in mediating the compaction of GAL1 transcripts in vitro [Fig. 5(b,c)]. Negatively stained electron micrographs indicated that wild‐type ScNab2 ZnF567, which were able to form dimers, resulted in much more compact complexes than those formed by the ScNab2 F450A variant, in which dimerization was impaired.30
Figure 5.
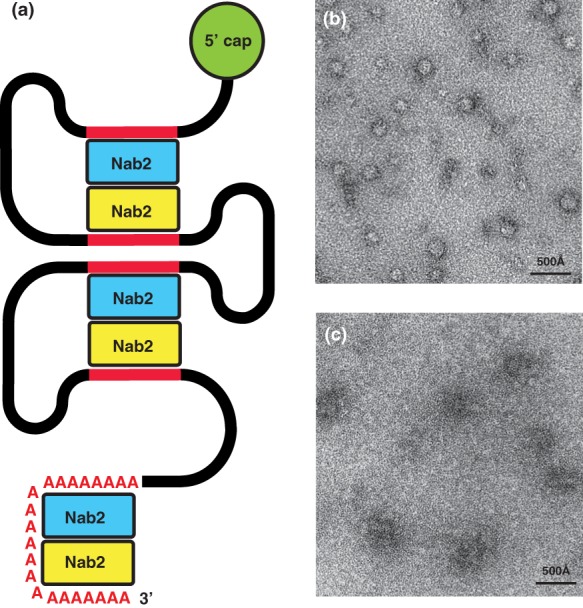
(a) The ScNab2 dimerization generated by its binding to two adenine‐rich regions of a transcript has the potential to both terminate 3′ polyadenylation and to facilitate compaction of the mRNP. (b) Electron micrograph of negatively stained GAL1 RNA complexed with ScNab2 Zn fingers 567 shows that compact spherical particles were generated, whereas a more open, less condensed complex was obtained with the F460A variant (c) in which Nab2 dimerization is impaired. Panels (a), (b), and (c) are reproduced from S. Aibara et al. “Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae.” Nucleic Acids Res. 45:1529–1538 (2017) by permission of Oxford University Press. Available at DOI: 10.1093/nar/gkw1224.
ScNab2 Regulation of Poly(A) Tail Length
Polyadenylation is the final processing step in the nuclear phase of the gene expression pathway. In S. cerevisiae, the 3′‐end processing machinery comprises cleavage factors IA and 1B and the cleavage and polyadenylation factor (CPF), a complex including the riboendonuclease, Ysh1/Brr5, the poly(A) polymerase, Pap1, and the Pap1 regulation factor, Fip1.47 Rna15, associated with the scaffold protein, Rna14, and other CFIA subunits, Pcf11 and Clp1, positions the CPF to cleave the poly(A) site. After the cleavage reaction, poly(A) polymerase, Pap1, synthesizes a poly(A) tail of ~60–80 adenosines in a template‐independent manner. Because isolated Pap1 is an inefficient distributive enzyme, Pap1 processivity requires stimulation/regulation by factors that bind Pap1 and tether it to the RNA.14, 47, 48, 49 In mammals, the RRM‐containing nuclear poly(A) RNA binding protein, PABPN1, enhances the processivity of PAP and, in S. cerevisiae, the CPF component, Fip1, binds Pap1 directly and tethers it to CPF to stimulate/regulate Pap1 activity.48, 49 Figure 6 illustrates a possible mechanism by which Nab2 could control poly(A) tail length in which Nab2 binding to the growing poly(A) chain results in the dissociation of poly(A) polymerase Pap1 from the CPF, analogous to that generated by PABPN1 in higher eukaryotes.5, 14, 50, 51 As polyadenylation proceeds, a growing loop of poly(A) RNA is generated because the poly(A) tail is held both by the CPF and Pap1. This loop can be accommodated both by the inherent flexibility of the poly(A) RNA and by the flexibility of Fip149 to which Pap1 is attached (Fig. 6). However, when the poly(A) tail becomes sufficiently long, a complex could be formed in which Nab2 could bind and generate a dimer by the RNA wrapping around two protein chains, analogous to the heterotetramer seen with ZnF567.30 Formation of this Nab2‐RNA complex could generate sufficient stiffening of the poly(A) RNA chain to result in the dissociation of Pap1 from the CPF and so terminate polyadenylation in a manner analogous to that proposed for PABPN1 in Metazoans.14, 50, 51 S. cerevisiae poly(A) tails have a length of ~60–80 nucleotides (reviewed by Reference 14), but it is not clear how many adenines are bound to a Nab2 dimer. A nuclease digestion study52 has indicated that Nab2 may bind ~25–30 nucleotides in vitro, but it is not clear whether this result reflects binding to a dimer or whether it might reflect digestion of the poly(A) RNA as it loops between Nab2 chains. Consequently, it is not clear whether generating a single Nab2 dimer is sufficient to terminate polyadenylation or whether instead it is necessary to form two dimers. Further work will be required to address this question.
Figure 6.
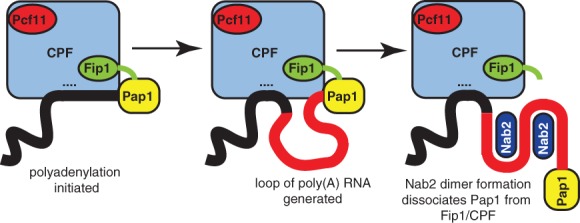
Function of ScNab2 in polyadenylation termination. Illustration of how the formation of an ScNab2 dimer could terminate polyadenylation by decreasing the flexibility of the poly(A) chain (red) so that poly(A) polymerase (Pap1—yellow) is dissociated from the cleavage and polyadenylation factor (CPF) so that it ceases to function processively and dissociates. The growing poly(A) chain (red) is held at its 5′ end by the transcript that is bound to the CPF (….) and at its 3′ end by Pap1 so that a flexible loop of poly(A) RNA is formed as polyadenylation progresses. When the poly(A) tail is sufficiently long, Nab2 binds and the resulting dimerization reduces the flexibility of the RNA so that Pap1 is forced to dissociate from the CPF and Fip1, after which it ceases to be processive and so dissociates from the poly(A) tail, terminating polyadenylation.
In addition to terminating polyadenylation by dissociating Pap1 from the CPF, Nab2 may also contribute to regulating poly(A) tail length through interactions with the 3′‐5′ riboexonuclease exosome complex,5 which is known to trim back poly(A) tails. Binding of Nab2 to the poly(A) tail may protect a certain length of poly(A) RNA from digestion by the RNA exosome. This hypothesis is supported by the physical and genetic interactions seen between Nab2 and the nuclear exosome catalytic subunit, Rrp6, which has been shown to restrict the length of poly(A) tails.44, 53, 54 Furthermore, both recombinant ScNab2 and SpNab2 can protect poly(A) RNA from degradation by the RNA exosome in vitro.20, 55 Critically, in the absence of nuclear Nab2, nuclear mRNA is rapidly degraded by the RNA exosome.56 Moreover, if nuclear export of mature mRNPs is blocked, newly synthesized mRNA transcripts are quickly degraded and this has been proposed to be because of the reduced availability of Nab2, which is unavailable because it is bound to the older previously generated transcripts.57
Overall, extensive analyses of ScNab2 Zn finger variants has indicated that the ScNab2 ZnF567 interaction with polyadenosine RNA plays a central role in the regulation of poly(A) tail length and mRNA compaction in S. cerevisiae. In this context, the observation that the loss of DmNab2 in Drosophila and ZC3H14 in mice and humans results in longer poly(A) tails12, 13, 55 suggests that DmNab2 ZnF123 and ZC3H14 ZnF123, which are most similar to ScNab2 ZnF567 [Fig. 4(a)], could interact with polyadenosine RNA in a similar manner to ScNab2 to contribute to the control of poly(A) tail length. In the future, it will be informative to assess the functional consequences of specific DmNab2/ZC3H14 Zn finger variants, such as DmNab2 C879S and HsZC3H14 C622S in ZnF2 that are equivalent to ScNab2 C437S in ZnF6 [Fig. 3(b)].
Interactions between Nab2 and the mRNA Nuclear Export Machinery
In addition to its function in controlling poly(A) tail length in S. cerevisiae, Nab2 also shows genetic interactions with components of the mRNA nuclear export machinery, such as Yra1, Sub2, and Mex67.28, 37, 58, 59 These data would be consistent with Nab2 having a role in signaling that polyadenylation had been completed and that the resultant mRNP was now suitable for export to the cytoplasm. Although ScNab2 may also interact directly with Mex67, one possible mechanism for signaling the completion of polyadenylation could be mediated through Pcf11 and the THO complex as a result of the dissociation of poly(A) polymerase Pap1 from the CPF complex.58, 60 In such a mechanism, Pcf11 could initiate the Sub2‐mediated mRNP remodeling that results in the dissociation of Yra1 and the generation of a Mex67:Mtr2‐bound export‐competent mature mRNP.
Questions Outstanding
Although Nab2 dimerization following binding to the growing poly(A) tail could provide a mechanism by which Nab2 terminates polyadenylation in S. cerevisiae, direct evidence for such a dimer containing full‐length Nab2 or the way in which the protein chains are arranged in the Nab2 dimer has not yet been obtained either in vitro or in vivo. It is also unclear whether one or two Nab2 dimers are needed to terminate polyadenylation. Although CPF components, such as Pcf11, appear to participate in signaling the termination of polyadenylation to the mRNA export machinery (TREX/Yra1/Sub2/Mex67:Mtr2) that generates an export‐competent mRNP, precise details of the signaling pathway and how polyadenylation termination is transmitted to Pcf11 remain to be established. Similarly, although there is clearly crosstalk between Nab2/ZC3H14 and the splicing machinery, most notably involving genetic interactions between ScNab2 and the splicosome component, Mud2,5, 9 again the precise mechanism by which this information is transferred is unclear. In addition, although binding of Nab2 to nuclear basket component, Mlp1, appears to be important to localizing Nab2‐bound transcripts to nuclear pores to facilitate both export and processing, it is not clear how the Nab2‐containing mRNPs are then released from Mlp1 to allow mature mRNPs to be exported.
In summary, a combination of functional and structural studies has provided a wealth of insight into the way in which Nab2 regulates mRNA poly(A) tail length, contributes to mRNA compaction and the integration of nuclear steps in the gene expression pathway with nuclear export in S. cerevisiae. Nab2 orthologues also contribute to these functions in higher Eukaryotes, however, the greater complexity of these systems has made establishing precise molecular mechanisms and signaling pathways more difficult. The spatial arrangement of Nab2 Zn fingers facilitates dimerization when bound to adenine‐rich RNAs that is important for the termination of 3′ polyadenylation and transcript compaction, albeit the precise structure of the Nab2 dimers generated in vivo remains to be established. Overall, the wealth of information that has been generated about the structure of Nab2 and its interactions with other components of the nuclear gene expression machinery has laid the foundation for beginning to define the precise ways in which these pathways are coordinated and also provides insight into the contribution made by Nab2 orthologues to these processes in higher Eukaryotes.
Acknowledgments
Supported in part by MRC grants MC_U105178939, MC‐A025‐5PL41 and MC_UP_1201/6 and a Leverhulme Emeritus Fellowship (EM‐2016‐062) to MS and National Institutes of Health grants 5R01GM058728 and 1R21AG054206 to AHC.
References
- 1. Kelly SM, Corbett AH (2009) Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic 10:1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nino CA, Herissant L, Babour A, Dargemont C (2013) mRNA nuclear export in yeast. Chem Rev 113:8523–8545. [DOI] [PubMed] [Google Scholar]
- 3. Katahira J (2015) Nuclear export of messenger RNA. Genes 6:163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson JT, Wilson SM, Datar KV, Swanson MS (1993) NAB2: a yeast nuclear polyadenylated RNA‐binding protein essential for cell viability. Mol Cell Biol 13:2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Soucek S, Corbett AH, Fasken MB (2012) The long and the short of it: the role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim Biophys Acta 1819:546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tran EJ, Zhou Y, Corbett AH, Wente SR (2007) The DEAD‐box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell 28:850–859. [DOI] [PubMed] [Google Scholar]
- 7. Aitchison JD, Blobel G, Rout MP (1996) Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624–627. [DOI] [PubMed] [Google Scholar]
- 8. Truant R, Fridell RA, Benson ER, Herold A, Cullen BR (1998) Nucleocytoplasmic shuttling by protein nuclear import factors. Eur J Cell Biol 77:269–275. [DOI] [PubMed] [Google Scholar]
- 9. Soucek S, Zeng Y, Bellur DL, Bergkessel M, Morris KJ, Deng Q, Duong D, Seyfried NT, Guthrie C, Staley JP, Fasken MB, Corbett AH (2016) The evolutionarily‐conserved polyadenosine RNA binding protein, Nab2, cooperates with splicing machinery to regulate the fate of pre‐mRNA. Mol Cell Biol 36:L2697–L2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fasken MB, Corbett AH (2005) Process or perish: quality control in mRNA biogenesis. Nat Struct Mol Biol 12:482–488. [DOI] [PubMed] [Google Scholar]
- 11. Reuter LM, Meinel DM, Strasser K (2015) The poly(A)‐binding protein Nab2 functions in RNA polymerase III transcription. Genes Develop 29:1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pak C, Garshasbi M, Kahrizi K, Gross C, Apponi LH, Noto JJ, Kelly SM, Leung SW, Tzschach A, Behjati F, Abedini SS, Mohseni M, Jensen LR, Hu H, Huang B, Stahley SN, Liu G, Williams KR, Burdick S, Feng Y, Sanyal S, Bassell GJ, Ropers HH, Najmabadi H, Corbett AH, Moberg KH, Kuss AW (2011) Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in drosophila and humans. Proc Natl Acad Sci U S A 108:12390–12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelly SM, Leung SW, Pak C, Banerjee A, Moberg KH, Corbett AH (2014) A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA 20:681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eckmann CR, Rammelt C, Wahle E (2011) Control of poly(A) tail length. Wiley Interdiscip Rev RNA 2:348–361. [DOI] [PubMed] [Google Scholar]
- 15. Kuhn U, Buschmann J, Wahle E (2017) The nuclear poly(A) binding protein of mammals, but not of fission yeast, participates in mRNA polyadenylation. RNA 23:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kelly S, Pak C, Garshasbi M, Kuss A, Corbett AH, Moberg K (2012) New kid on the ID block: neural functions of the Nab2/ZC3H14 class of Cys(3)his tandem zinc‐finger polyadenosine RNA binding proteins. RNA Biol 9:555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelly SM, Bienkowski R, Banerjee A, Melicharek DJ, Brewer ZA, Marenda DR, Corbett AH, Moberg KH (2016) The drosophila ortholog of the Zc3h14 RNA binding protein acts within neurons to pattern axon projection in the developing brain. Dev Neurobiol 76:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szymczyna BR, Bowman J, McCracken S, Pineda‐Lucena A, Lu Y, Cox B, Lambermon M, Graveley BR, Arrowsmith CH, Blencowe BJ (2003) Structure and function of the PWI motif: a novel nucleic acid‐binding domain that facilitates pre‐mRNA processing. Genes Develop 17:461–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marfatia KA, Crafton EB, Green DM, Corbett AH (2003) Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p‐facilitated poly(A) RNA export. J Biol Chem 278:6731–6740. [DOI] [PubMed] [Google Scholar]
- 20. Grenier St‐Sauveur V, Soucek S, Corbett AH, Bachand F (2013) Poly(A) tail‐mediated gene regulation by opposing roles of Nab2 and Pab2 nuclear poly(A)‐binding proteins in pre‐mRNA decay. Mol Cell Biol 33:4718–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guthrie CR, Schellenberg GD, Kraemer BC (2009) SUT‐2 potentiates tau‐induced neurotoxicity in Caenorhabditis elegans . Hum Mol Genet 18:1825–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelly SM, Pabit SA, Kitchen CM, Guo P, Marfatia KA, Murphy TJ, Corbett AH, Berland KM (2007) Recognition of polyadenosine RNA by zinc finger proteins. Proc Natl Acad Sci U S A 104:12306–12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leung SW, Apponi LH, Cornejo OE, Kitchen CM, Valentini SR, Pavlath GK, Dunham CM, Corbett AH (2009) Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene 439:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duncan K, Umen JG, Guthrie C (2000) A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr Biol 10:687–696. [DOI] [PubMed] [Google Scholar]
- 25. Lee DC, Aitchison JD (1999) Kap104p‐mediated nuclear import. Nuclear localization signals in mRNA‐binding proteins and the role of ran and Rna. J Biol Chem 274:29031–29037. [DOI] [PubMed] [Google Scholar]
- 26. Bienkowski RS, Banerjee A, Rounds JC, Rha J, Omotade OF, Gross C, Morris KJ, Leung SW, Pak C, Jones SK, Santoro MR, Warren ST, Zheng JQ, Bassell GJ, Corbett AH, Moberg KH (2017) The conserved, disease‐associated RNA binding protein dNab2 interacts with the fragile X protein ortholog in drosophila neurons. Cell Rep 20:1372–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soniat M, Sampathkumar P, Collett G, Gizzi AS, Banu RN, Bhosle RC, Chamala S, Chowdhury S, Fiser A, Glenn AS, Hammonds J, Hillerich B, Khafizov K, Love JD, Matikainen B, Seidel RD, Toro R, Rajesh Kumar P, Bonanno JB, Chook YM, Almo SC (2013) Crystal structure of human Karyopherin beta2 bound to the PY‐NLS of Saccharomyces cerevisiae Nab2. J Struct Funct Genomics 14:31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brockmann C, Soucek S, Kuhlmann SI, Mills‐Lujan K, Kelly SM, Yang JC, Iglesias N, Stutz F, Corbett AH, Neuhaus D, Stewart M (2012) Structural basis for polyadenosine‐RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export. Structure 20:1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhlmann SI, Valkov E, Stewart M (2014) Structural basis for the molecular recognition of polyadenosine RNA by Nab2 Zn fingers. Nucleic Acids Res 42:672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aibara S, Gordon JM, Riesterer AS, McLaughlin SH, Stewart M (2017) Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(a) tail length determination and transcript compaction in Saccharomyces cerevisiae . Nucleic Acids Res 45:1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grant RP, Marshall NJ, Yang JC, Fasken MB, Kelly SM, Harreman MT, Neuhaus D, Corbett AH, Stewart M (2008) Structure of the N‐terminal Mlp1‐binding domain of the Saccharomyces cerevisiae mRNA‐binding protein, Nab2. J Mol Biol 376:1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng C, Fasken MB, Marshall NJ, Brockmann C, Rubinson ME, Wente SR, Corbett AH, Stewart M (2010) Structural basis for the function of the Saccharomyces cerevisiae Gfd1 protein in mRNA nuclear export. J Biol Chem 285:20704–20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH (2002) Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem 277:7752–7760. [DOI] [PubMed] [Google Scholar]
- 34. Kelly SM, Leung SW, Apponi LH, Bramley AM, Tran EJ, Chekanova JA, Wente SR, Corbett AH (2010) Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA‐binding protein 2 (Nab2) is required for correct mRNA 3′‐end formation. J Biol Chem 285:26022–26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kolling R, Nguyen T, Chen EY, Botstein D (1993) A new yeast gene with a myosin‐like heptad repeat structure. Mol Gen Genet 237:359–369. [DOI] [PubMed] [Google Scholar]
- 36. Strambio‐de‐Castillia C, Blobel G, Rout MP (1999) Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol 144:839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fasken MB, Stewart M, Corbett AH (2008) Functional significance of the interaction between the mRNA‐binding protein, Nab2, and the nuclear pore‐associated protein, Mlp1, in mRNA export. J Biol Chem 283:27130–27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Green DM, Johnson CP, Hagan H, Corbett AH (2003) The C‐terminal domain of myosin‐like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci U S A 100:1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonnet A, Bretes H, Palancade B (2015) Nuclear pore components affect distinct stages of intron‐containing gene expression. Nucleic Acids Res 43:4249–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hodge CA, Colot HV, Stafford P, Cole CN (1999) Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1‐1 cells. EMBO J 18:5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, Stutz F (1999) The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG‐nucleoporin Rip1p, the DEAD‐box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J 18:5761–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miyakura S, Hara M (2015) Molecular characterization of UKp83/68, a widespread nuclear proteins that bind poly(A) and colocalize with a nuclear speckle's component. J Med Dent Sci 62:43–56. [DOI] [PubMed] [Google Scholar]
- 43. Martinez‐Lumbreras S, Santiveri CM, Mirassou Y, Zorrilla S, Perez‐Canadillas JM (2013) Two singular types of CCCH tandem zinc finger in Nab2p contribute to polyadenosine RNA recognition. Structure 21:1800–1811. [DOI] [PubMed] [Google Scholar]
- 44. Gonzalez‐Aguilera C, Tous C, Babiano R, de la Cruz J, Luna R, Aguilera A (2011) Nab2 functions in the metabolism of RNA driven by polymerases II and III. Mol Biol Cell 22:2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tuck AC, Tollervey D (2013) A transcriptome‐wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154:996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Batisse J, Batisse C, Budd A, Bottcher B, Hurt E (2009) Purification of nuclear poly(A)‐binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem 284:34911–34917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Casanal A, Kumar A, Hill CH, Easter AD, Emsley P, Degliesposti G, Gordiyenko Y, Santhanam B, Wolf J, Wiederhold K, Dornan GL, Skehel M, Robinson CV, Passmore LA (2017) Architecture of eukaryotic mRNA 3′‐end processing machinery. Science 358:1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ezeokonkwo C, Zhelkovsky A, Lee R, Bohm A, Moore CL (2011) A flexible linker region in Fip1 is needed for efficient mRNA polyadenylation. RNA 17:652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaufmann I, Martin G, Friedlein A, Langen H, Keller W (2004) Human Fip1 is a subunit of CPSF that binds to U‐rich RNA elements and stimulates poly(A) polymerase. EMBO J 23:616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wahle E (1995) Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem 270:2800–2808. [DOI] [PubMed] [Google Scholar]
- 51. Wahle E, Ruegsegger U (1999) 3′‐end processing of pre‐mRNA in eukaryotes. FEMS Microbiol Rev 23:277–295. [DOI] [PubMed] [Google Scholar]
- 52. Viphakone N, Voisinet‐Hakil F, Minvielle‐Sebastia L (2008) Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res 36:2418–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Klass DM, Scheibe M, Butter F, Hogan GJ, Mann M, Brown PO (2013) Quantitative proteomic analysis reveals concurrent RNA‐protein interactions and identifies new RNA‐binding proteins in Saccharomyces cerevisiae . Genome Res 23:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH (2012) Rrp6p controls mRNA poly(A) tail length and its decoration with poly(a) binding proteins. Mol Cell 47:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rha J, Jones SK, Fidler J, Banerjee A, Leung SW, Morris KJ, Wong JC, Inglis GAS, Shapiro L, Deng Q, Cutler AA, Hanif AM, Pardue MT, Schaffer A, Seyfried NT, Moberg KH, Bassell GJ, Escayg A, Garcia PS, Corbett AH (2017) The RNA‐binding protein, ZC3H14, is required for proper poly(A) tail length control, expression of synaptic proteins, and brain function in mice. Hum Mol Genet 26:3663–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmid M, Olszewski P, Pelechano V, Gupta I, Steinmetz LM, Jensen TH (2015) The nuclear polyA‐binding protein Nab2p is essential for mRNA production. Cell Rep 12:128–139. [DOI] [PubMed] [Google Scholar]
- 57. Tudek A, Schmid M, Makaras M, Barrass JD, Beggs JD, Jensen TH (2018) A nuclear export block triggers the decay of newly synthesized polyadenylated RNA. Cell Rep 24:2457–2467 e2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F (2010) Ubiquitin‐mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 24:1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jimeno S, Luna R, Garcia‐Rubio M, Aguilera A (2006) Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants. Mol Cell Biol 26:4387–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnson SA, Cubberley G, Bentley DL (2009) Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]


