Abstract
Poor electronic medical record (EMR) usability is detrimental to both clinicians and patients. A better EMR would provide concise, context sensitive patient data, but doing so entails the difficult task of knowing which data are relevant. To determine the relevance of patient data in different contexts, we collect and model the information seeking behavior of clinicians using a learning EMR (LEMR) system. Sufficient data were collected to train predictive models for 80 different targets (e.g., glucose level, heparin administration) and 27 of them had AUROC values of greater than 0.7. These results are encouraging considering the high variation in information seeking behavior (intraclass correlation 0.40). We plan to apply these models to a new set of patient cases and adapt the LEMR interface to highlight relevant patient data, and thus provide concise, context sensitive data.
Introduction
Clinicians and other health care providers remain dissatisfied with electronic medical records (EMRs).1 Poor EMR usability results in substantial amounts of time spent using them (up to two-thirds of clinician time2), takes attention away from patients,3 and impacts patient satisfaction.4 EMR usability challenges are highlighted by the American Medical Association in a published list of priorities for improving EMR usability.5 A top priority is to reduce the cognitive workload of EMRs by providing “concise, context sensitive” data.
Clinicians typically navigate the EMR laboriously to seek data they need to evaluate and make decisions for their patients. While navigating the EMR, they are burdened with viewing and filtering out extraneous data. A more usable EMR would act like an intelligent agent and provide concise, relevant, and context sensitive data. An EMR can identify and provide relevant data, if information seeking behaviors can be predicted accurately. This paper describes a method for predicting clinician information seeking behaviors and a preliminary evaluation of the method.
Background
Clinician information seeking behavior varies by context. Context includes (1) EMR user type—a clinician, a nurse and a physician assistants have different information seeking behaviors6; (2) clinical task—a clinician has different information seeking behavior when performing differential diagnosis than when performing medication reconciliation; and (3) patient case—the same clinician when performing the same clinical task for different patients may have different information seeking behavior that are driven by differences in diagnoses.
Current EMRs support data and functionality needs across the spectrum of user-task-case contexts, and this broad support comes at a cost of decreased usability in any specific context. Customization by vendors or by local EMR technical teams improves EMR usability, but these are both difficult and inflexible. To improve EMR usability across many specific contexts, Covvey et al.7 proposed a systematic approach to “defining, eliciting, and specifying the structure and the information content of the electronic health record” for all unique contexts.
Anticipating information seeking behavior for every EMR user-task-case context and meeting those needs is very challenging. It has been done for some specific contexts (e.g., intensive care unit (ICU) clinicians determining diagnosis and treatment for newly admitted patients8) and the knowledge gained has been applied to create novel EMR interfaces9. A structured approach for eliciting data needs (i.e., questionnaires10 and observations11) and a manual approach for then tailoring the EMR interface to meet those needs might be feasible for a specific user type and context. However, this approach is time consuming, and it would be very expensive to replicate it for every user type and context. The approach also ignores the wide variation in patient cases within a given context. It is not feasible to elicit and manually tailor an EMR interface to all the possible user types, contexts, and patient cases. Is there a feasible way to automatically model data needs for a given user-task-case context? If so, such a model could be applied to anticipate data that will be sought and adapt the EMR interface to facilitate clinician access to that data.
We are developing a data-driven approach to predict and highlight data in the EMR that are most likely to be sought in a user-task-case context. The method relies on a combination of data of past patient cases that are readily available in the EMR, information seeking behavior data that we elicit from clinicians, and on construction of machine learning models that can be applied to the current patient to identify relevant patient data. Our ultimate goal is to apply such models to dynamically adapt the EMR interface to highlight context-relevant patient data12 We call such a system a Learning Electronic Medical Record (LEMR) system. In this paper, we describe the ICU EMR data, the information seeking behavior data that were elicited from clinicians, the machine learning models that we developed, and preliminary evaluation of these models.
Methods
Our goal is to build models that use patient data that are recorded in the EMR to predict which data items would be sought by clinicians and, therefore, should be highlighted in a future patient case. The predictors are all the data items in the patient record and the targets are items that might be highlighted, with one distinct model for each item. In the rest of this section, we describe our methods for collecting clinician information seeking behavior (including a LEMR interface, a set of de-identified patient cases, clinician participants, a procedure for reviewing patient cases, and data collected) and for training machine learning models of clinician information seeking behaviors (including a description of data representation and data preprocessing, machine learning algorithms, and learning rate calculations).
Methods for collecting clinician information seeking behavior
LEMR interface
A LEMR interface was developed using Bitnami Django stack (RRID:SCR_012855)*, Bootstrap CSS (RRID: SCR 016096), and High Charts (RRID: SCR 016095). The interface consists of three vertical columns: the left column displays temporal charts of vital sign measurements, ventilator settings, intake and output, and medication administrations; the middle column contains similar temporal displays of laboratory test results; and the right column displays free-text notes, reports, and procedures. The interface tracks what data are displayed on screen and provides a selector for participants to select which data are relevant. Figure 1 shows a screenshot of the interface. The source code for the interface is available at https://github.com/aik77/LEMRinterface.
Figure 1.
Screenshot of the LEMR interface used to display patient data and to capture selections made by participants. The check box on the top left of a data item becomes checked when the participant clicks antwhere the area associated with the item.
Selection of patient cases
A set of ICU patients was selected who (1) were admitted between June 2010 and May 2012 and (2) had a diagnosis of either acute kidney failure (AKF; ICD-9 584.9 or 584.5; 93 cases) or acute respiratory failure (ARF; ICD-9 518.81; 85 cases). EMR data were extracted from a research database13 and a clinical data warehouse.14 The data were de-identified (DE-ID Data Corp, RRID:SCR_008668) to create a limited data set that included dates and times related to the events.
Clinician participants, procedure for reviewing patient cases and data collection
The recruited participants included ICU fellows and attending clinicians from the Department of Critical Care Medicine at the University of Pittsburgh. Each participant was compensated $100 per hour of participation. Data collection occurred in a meeting room where a participant sat in front of a laptop computer that was pre-loaded with 30 patient cases. The first four cases were common to all participants (these are called burn-in cases) and the remaining 26 cases were different for each participant. Each participant reviewed and annotated as many cases as they could during one to two sessions that lasted a total of four to six hours.
The participants reviewed and annotated the cases using the interface shown in Figure 1. The case review procedure included the following tasks (see Figure 2).
Figure 2.
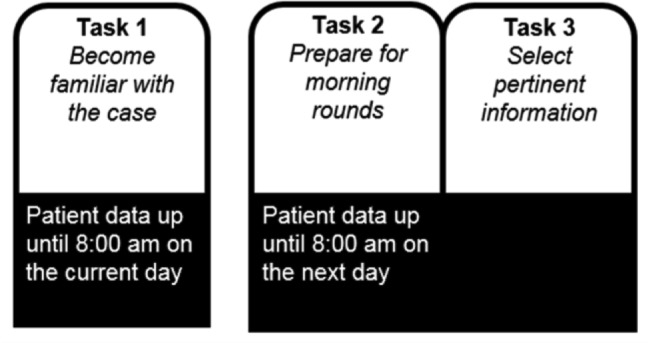
Case review procedure showing the three tasks that are performed for each patient case.
Task 1: For this task a random day between day two of admission to the ICU and the day before discharge from the ICU was selected as the patient’s current day. All available patient data up until 8:00 am on the current day was displayed to the participant. Structured data were shown in graphical time series plots and free-text notes were shown in a separate area on the interface. The participant was instructed to “use the available information to become familiar with the patient case as if they are one of your own patients.” After becoming familiar with the case, the participant clicked on a button to proceed to Task 2.
Task 2: An additional day (from 8:00 am on the current day to 8:00 am on the next day) of the patient’s data was added to the interface. The participant was prompted with “24-hours have passed” and directed to “use the available information to prepare to present the case during morning rounds.” After preparation was complete, the participant clicked on a button to advance to Task 3.
Task 3: On the interface, each available data item (e.g., glucose levels, insulin dosage regimen) was accompanied with a check box, and clicking on the area associated with data toggled the check box. The participant was directed to “select the information you consider pertinent when preparing to present this case at morning rounds.” The participant selected relevant data items by toggling the accompanying check box to the checked state.
Information seeking behavior was collected during Task 3. Selected targets were assigned the value yes, and variables that were available but were not selected were assigned the value no.
Agreement among participants
To determine agreement among participants, we calculated intraclass correlation coefficient (ICC).15 ICC ranges from 0 to 1, where values less than 0.50, between 0.50 and 0.75, and between 0.75 and 0.90 are indicative of poor, moderate, and good reliability, respectively.12 ICC was computed on the first four (burn-in) cases based on a single rater, absolute-agreement, two-way mixed-effects model (R Project for Statistical Computing, RRID:SCR_001905; CRAN, RRID:SCR_003005; psych package, ICC3 method). Only four cases were reviewed by all participants because we wanted to maximize the total number of reviewed cases. To increase the power of the ICC calculation, we aggregate across all targets and compute a single ICC score. While the time to task completion for the burn-in cases is longer because users are not yet familiar with the interface, the targets selected by the participants are presumed to be less affected by burn-in because data sought by clinicians is likely to be the same regardless of the time it takes them to navigate the interface and complete the tasks.
Methods for training machine learning models
Before training models of clinician information seeking behavior, the data was preprocessed into a representation that is suitable for machine learning. We applied and evaluated three different machine learning algorithms. Additionally, we calculated the learning rates for the best performing models and used them to estimate the sample sizes needed to train them.
Data representation
For each patient case, we created a training data sample that is comprised of a vector of values for predictor variables and is augmented with values for target variables. A predictor variable is any patient data item and includes observations, measurements, and actions that are recorded in the EMR. Examples of predictor variables include demographics, diagnosis, vital sign measurements, ventilator settings, intake and output, laboratory test results and medication administrations. For example, diagnosis = diabetes mellitus denotes that the predictor variable diagnosis has the value diabetes mellitus in a particular patient, and glucose = 80, 90, 100 mg/dL denotes that the predictor variable glucose consists of a series of glucose levels over a period of time. A target variable (or simply target) is any patient data item that a clinician can potentially seek as relevant for a specific task in a specific patient case. For example, glucose target is a target that denotes if glucose levels are sought by a clinician. A target is a binary variable that is assigned the value yes if it is available for the patient case under consideration and a clinician would seek it for the given task and is assigned the value no if it is available but a clinician would not seek it. If the target is not measured in the patient’s record then it is not defined for that patient. For example, a clinician who is preparing to present at morning rounds a patient who has diabetes mellitus and is on insulin may seek the patient’s glucose levels and insulin dosing regimen; thus, in this example glucose target = yes and insulin target = yes. Consider a different patient who has kidney failure and glucose levels are measured but are not sought by a clinician; in this example glucose target = no and insulin target is not defined because it was not measured. The target data are not directly available in the EMR, but rather, are a function of how the EMR is used. Therefore, we developed a LEMR interface and a procedure for obtaining them from clinicians in a laboratory setting.
Data preprocessing
The predictor variables include simple atemporal variables, such as diagnosis and demographics, and complex variables that represent multivariate time series for laboratory test results, medication administrations, and vital sign measurements. We expand each complex variable into a fixed set of features as described below.
Expansion of predictor variables. While some of the predictor variables are atemporal and their values for a patient do not change during the ICU stay, most of the variables are temporal and consist of a time-stamped sequence of values.
For each atemporal variable, such as diagnosis and demographics, we generate a single feature that is assigned a single value for a patient for the duration of ICU stay (e.g., diagnosis = diabetes mellitus).
For each medication variable, we generate several features to summarize the temporal aspects. For example, for a time-stamped sequence of administrations of insulin, we generate 4 features that include 1) an indicator of whether the patient is currently prescribed insulin, 2) the time since its first administration to the current time, 3) the time since its most recent administration to the current time, and 4) its dose at the most recent administration.
For each laboratory test result, we generate a rich set of features. For example, for a time-stamped sequence of glucose measurements, we generate 36 features that include the first glucose value during the ICU stay, the most recent value, the highest and lowest values until current time, the difference between the most recent two values, and so on.
For each vital sign, we generate a set of features similar to a laboratory test result.
The participants were represented by a set of 11 binary features. If a participant reviewed a case then the value of the corresponding participant feature for the patient case was assigned the value 1 and the remaining 10 features were assigned the value 0.
These variables form the basis of a vector of predictor values for each patient case. The vector of values summarizes the clinical evolution of the patient’s condition from the time of admission to the ICU to the current day.
Target variables. Targets are binary variables indicating whether associated patient data items were sought as relevant or not (e.g., glucose target is yes if glucose levels were sought by a clinician).
In the data representation that we have described, the temporal aspects of time series data are implicitly summarized in the vector of predictor values; this has the advantage that standard machine learning methods can be applied.
Missing values. Missing values were imputed by two different methods. In the first method, they were imputed with the median. In the second method, they were imputed via linear regression and logistic regression for continuous and discrete predictive features, respectively. The regression model’s predictive features were all available features except for the feature being imputed. Both imputation methods were applied, creating two distinct data sets (a median imputed data set and a regression imputed data set). We train models on each data set separately and compare performance.
Feature selection. Feature selection was necessary to reduce the dimensionality of the data and was performed in two steps. In the first step, for each set of features derived from a predictor variable (e.g., features such as the most recent value, the slope between the two most recent values, etc.), we assessed if the set is predictive of the target by itself using cross-validating models. Every set of features with an area under the Receiver Operator Characteristic curve (AUROC) of less than 0.6 was removed. In the second step, the features that remained after the first step were reduced further using recursive feature elimination and cross-validation (RFECV in scikit-learn, RRID:SCR_002577). The final set of features was used for model construction. Feature selection is target specific, so it was done separately for each target variable model.
Machine learning algorithms
Three different machine learning algorithms were applied: lasso logistic regression (LR),16 support vector classifier (SV),17 and random forest classifier (RF).18 Models were constructed using these algorithms using leave-one-out cross-fold validation. The imputation and variable selection steps were performed within the cross folds. Results are reported as AUROC with 95% confidence intervals estimated by bootstrapping. The scikit-learn19 (RRID:SCR_002577) implementation of each algorithm was used.
Learning rate calculations
Since acquiring target values is expensive, we wanted to measure the learning rate of models to assess the number of training cases needed to obtain optimal model performance. To calculate the learning rate of models, we trained each model with varying numbers of training cases. Each model was trained with 25, 50, 75, and 100 percent of its respective training set, and the resulting AUROCs are reported using box and whisker plots.
Results
Patient cases, participants and agreement
A total of 178 patient cases were reviewed and annotated by the clinician participants between August and October of 2017. Of these patient cases, 52% had AKF, the average age was 60, and the median ICU day at the time of target selection was 7. These numbers do not include the four burn-in cases that were annotated by all participants.
Eleven clinician participants reviewed the cases. Nine were fellows, two were faculty attendings, seven were male, the average years of experience since graduating from medical school was 5.5 years, and the average years of ICU experience was 1.8 years.
The aggregate ICC score of the 11 participants’ selections on the first four burn-in cases is 0.40 (95% confidence interval 0.36 - 0.45).
Description of data set
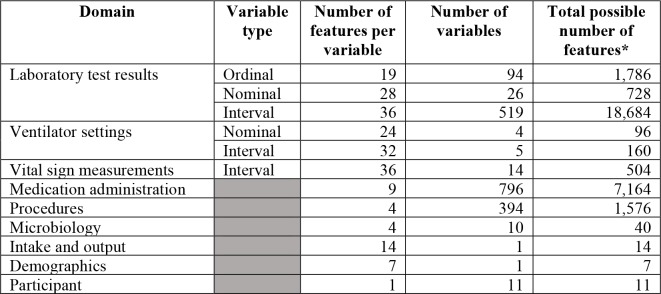
The data set was assembled from 178 patient cases and 1,875 variables from 9 domains; the details on the number of variables in each domain are given in Table 1. Each complex variable was expanded to several features as described under preprocessing in the Methods section. The total number of features in the final data set was 6,935 after expansion. The final data matrix consisted of 178 rows (one row for each patient case), 6,935 predictor columns (one column for each predictor feature), and 80 target columns (one column for each target that had been assigned the value yes in 10 patient cases or more). Missingness in the data set was 41%. Feature selection resulted in 6,935 predictor features being reduced to an average of 88 predictor features.
Table 1.
Predictor variables and the number of features constructed from them.
|
The actual number of features is smaller because many variables (e.g., rare laboratory tests) were not measured in this cohort of patient cases.
Model performance
We constructed models to predict 80 distinct targets. The 80 targets were chosen such that there were adequate cases to train on: in at least 20 cases (of 178 cases) the target was available and in at least 10 cases where the target was available it was assigned the value yes. In Table 2, due to space limitations, we report the performance of 46 of the 80 models where each of the 46 models had at least 100 cases where the target was available; the remaining 34 models were for targets that were available for less than 100 cases. Column 3 of Table 2 shows for each target the number of cases it was selected and the total number of cases in which it was available. Of the 46 models in the table, 14 had an AUROC of at least 0.7. Thirteen of the 34 models not shown in the table had an AUROC of at least 0.7.
Table 2.
Performance of models that predict clinician information seeking behavior. To reduce clutter the word “target” is not included in the names of the target variables. Domain: L = laboratory test, V = vital sign, M = medication, and S = ventilator setting. Results are reported as AUROC with 95% confidence intervals. For each target variable, the AUROC of the best performing model is bolded.
| Target variable | Domain | Selected/ available | Logistic regression median impute | Random forests median impute | Random forests regression impute |
|---|---|---|---|---|---|
| red blood cells | L | 24/165 | 0.68 (0.55, 0.80) | 0.94 (0.87, 0.99) | 0.94 (0.89, 0.98) |
| ventilator status | S | 20/131 | 0.54 (0.42, 0.67) | 0.72 (0.60, 0.83) | 0.83 (0.72, 0.92) |
| pH | L | 46/137 | 0.57 (0.49, 0.66) | 0.77 (0.70, 0.84) | 0.71 (0.64, 0.79) |
| bicarbonate (blood gases) | L | 11/108 | 0.63 (0.49, 0.77) | 0.75 (0.62, 0.86) | 0.61 (0.47, 0.73) |
| oxygen saturation | V | 103/177 | 0.64 (0.57, 0.71) | 0.74 (0.68, 0.80) | 0.61 (0.53, 0.68) |
| anion gap | L | 19/118 | 0.45 (0.34, 0.58) | 0.69 (0.58, 0.80) | 0.74 (0.63, 0.84) |
| prothrombin time | L | 21/125 | 0.64 (0.52, 0.76) | 0.73 (0.60, 0.86) | 0.73 (0.61, 0.84) |
| bilirubin total | L | 39/110 | 0.62 (0.53, 0.70) | 0.73 (0.64, 0.81) | 0.73 (0.64, 0.81) |
| chloride | L | 106/178 | 0.63 (0.56, 0.71) | 0.71 (0.64, 0.78) | 0.67 (0.60, 0.74) |
| lactate | L | 50/117 | 0.70 (0.61, 0.78) | 0.71 (0.63, 0.79) | 0.71 (0.63, 0.79) |
| glucose | L | 114/175 | 0.71 (0.64, 0.78) | 0.64 (0.57, 0.72) | 0.67 (0.59, 0.74) |
| potassium chloride | M | 28/136 | 0.52 (0.41, 0.63) | 0.71 (0.62, 0.79) | 0.56 (0.46, 0.66) |
| heparin | M | 41/102 | 0.71 (0.62, 0.80) | 0.58 (0.48, 0.67) | 0.50 (0.40, 0.60) |
| alkaline phosphatase | L | 16/109 | 0.53 (0.39, 0.67) | 0.71 (0.61, 0.79) | 0.57 (0.44, 0.70) |
| fraction of inspired O2 | S | 99/151 | 0.50 (0.41, 0.59) | 0.69 (0.62, 0.77) | 0.68 (0.61, 0.75) |
| central venous pressure | V | 31/111 | 0.62 (0.52, 0.72) | 0.68 (0.58, 0.77) | 0.69 (0.59, 0.78) |
| magnesium | L | 74/173 | 0.58 (0.51, 0.65) | 0.69 (0.62, 0.75) | 0.62 (0.55, 0.69) |
| blood urea nitrogen | L | 114/177 | 0.62 (0.54, 0.70) | 0.68 (0.60, 0.75) | 0.60 (0.53, 0.68) |
| respiratory rate | V | 121/178 | 0.58 (0.51, 0.66) | 0.56 (0.48, 0.64) | 0.68 (0.61, 0.75) |
| calcium | L | 41/163 | 0.65 (0.57, 0.73) | 0.68 (0.59, 0.76) | 0.67 (0.59, 0.75) |
| partial thromboplastin time | L | 21/108 | 0.68 (0.55, 0.79) | 0.64 (0.52, 0.77) | 0.64 (0.55, 0.73) |
| hematocrit | L | 11/166 | 0.61 (0.35, 0.85) | 0.59 (0.40, 0.76) | 0.68 (0.46, 0.88) |
| neutrophils | L | 31/156 | 0.49 (0.38, 0.59) | 0.67 (0.57, 0.78) | 0.65 (0.56, 0.74) |
| partial pressure of CO2 | L | 31/138 | 0.65 (0.55, 0.74) | 0.67 (0.58, 0.75) | 0.62 (0.53, 0.72) |
| temperature | V | 144/178 | 0.51 (0.42, 0.61) | 0.61 (0.52, 0.71) | 0.67 (0.58, 0.75) |
| ventilator mode | S | 72/148 | 0.66 (0.59, 0.74) | 0.67 (0.59, 0.74) | 0.66 (0.59, 0.73) |
| intake and output | V | 81/178 | 0.64 (0.57, 0.71) | 0.66 (0.59, 0.73) | 0.55 (0.47, 0.62) |
| blood pressure | V | 151/178 | 0.47 (0.36, 0.58) | 0.65 (0.56, 0.74) | 0.53 (0.43, 0.63) |
| INR | L | 74/125 | 0.65 (0.57, 0.73) | 0.59 (0.51, 0.67) | 0.63 (0.54, 0.71) |
| creatinine | L | 132/177 | 0.62 (0.55, 0.71) | 0.64 (0.56, 0.72) | 0.65 (0.57, 0.73) |
| aspartate aminotransferase | L | 28/113 | 0.65 (0.54, 0.75) | 0.64 (0.55, 0.74) | 0.52 (0.41, 0.63) |
| alanine aminotransferase | L | 26/111 | 0.57 (0.46, 0.68) | 0.65 (0.56, 0.74) | 0.50 (0.40, 0.60) |
| platelets | L | 125/166 | 0.65 (0.57, 0.73) | 0.58 (0.50, 0.66) | 0.56 (0.48, 0.64) |
| phosphate | L | 69/170 | 0.53 (0.45, 0.61) | 0.65 (0.58, 0.72) | 0.56 (0.48, 0.63) |
| potassium | L | 121/178 | 0.64 (0.57, 0.71) | 0.43 (0.36, 0.50) | 0.56 (0.48, 0.65) |
| white blood cells | L | 141/166 | 0.60 (0.50, 0.69) | 0.63 (0.55, 0.72) | 0.64 (0.57, 0.72) |
| sodium | L | 128/178 | 0.47 (0.39, 0.55) | 0.64 (0.56, 0.72) | 0.58 (0.50, 0.66) |
| bicarbonate (chemistry) | L | 104/178 | 0.58 (0.51, 0.65) | 0.64 (0.57, 0.71) | 0.62 (0.54, 0.69) |
| albumin | L | 21/114 | 0.56 (0.44, 0.68) | 0.56 (0.45, 0.67) | 0.64 (0.53, 0.76) |
| partial pressure of O2 | L | 30/137 | 0.62 (0.52, 0.71) | 0.64 (0.54, 0.74) | 0.64 (0.55, 0.72) |
| hemoglobin | L | 130/166 | 0.63 (0.55, 0.72) | 0.59 (0.51, 0.67) | 0.59 (0.51, 0.67) |
| heart rate | V | 152/178 | 0.48 (0.39, 0.57) | 0.62 (0.52, 0.73) | 0.48 (0.38, 0.58) |
| ionized calcium | L | 30/132 | 0.62 (0.52, 0.71) | 0.59 (0.50, 0.68) | 0.55 (0.45, 0.65) |
| glomerular filtration rate | L | 19/166 | 0.47 (0.36, 0.58) | 0.61 (0.50, 0.72) | 0.60 (0.48, 0.72) |
| ventilator tube status | S | 45/130 | 0.45 (0.36, 0.55) | 0.60 (0.50, 0.69) | 0.44 (0.35, 0.52) |
| sodium chloride 0.9% | M | 69/154 | 0.56 (0.49, 0.63) | 0.49 (0.42, 0.57) | 0.48 (0.40, 0.56) |
| Average | 0.59 | 0.65 | 0.62 | ||
| Average of best (bolded AUROCs) | 0.68 | ||||
Learning rates
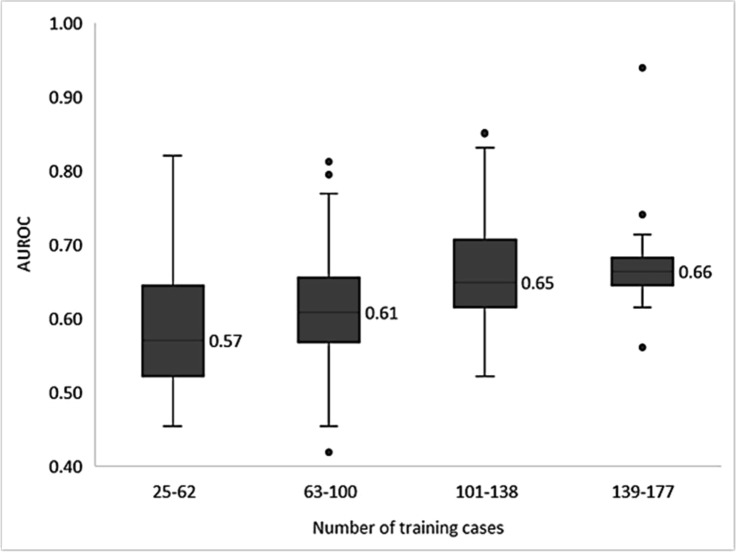
Learning rate calculations were performed by training all models in Table 2 at four training set sizes: 25, 50, 75, and 100 percent of each model’s respective training set. The median AUROCs for the varying training set sizes are shown in Figure 3. Overall, the median AUROC increases as the number of training cases increases.
Figure 3.
Learning rates of models trained on the different sized training sets. Each model is trained on 25, 50, 75, and 100 percent of relevent data sets. AUROCs were binned into four equal-interval by the number of training cases.
Discussion
We developed models to predict clinician EMR information seeking behavior from a set of ICU patient cases that were reviewed by clinicians who selected targets. The targets are variables such as a laboratory test value or a vital sign measurement that are available in the patient case of interest and are selected as relevant if the reviewing clinician considered them to be pertinent information when preparing to present that case for morning rounds. In obtaining data for training models, we focused on the context of clinicians (user), preparing for morning rounds (task), for patients with AKF and ARF (cases).
The ICC results showed poor agreement across participants regarding which patient data were relevant when rounding for a patient case. Some of the variation may be due to our intentional choice of a broad task that is subject to interpretation. We tried to standardize the task by clearly and consistently explaining the task to each participant. High variability between clinicians makes learning information seeking behavior more difficult. We plan to purse different directions that might lead to improved performance including (1) additional predictor variables (e.g., from natural language processing of free text), (2) more samples, (3) hierarchical modeling to leverage across contexts, from the most general to the most specialized, and (4) clinician-specific models that have the potential to overcome clinician variability as indicated by the low ICC values.
Sufficient sample sizes were available for building models to predict 80 distinct targets and, despite relatively small training sets, AUROC performance was greater than 0.70 for more than 30% of the models. These encouraging results are bolstered by the learning rate results. All but one model with at least 120 training samples had an AUROC greater than 0.60, and most models showed an upward trend in AUROC values as the number of training samples increased.
Superior performance with practically attainable numbers of training samples is important because manual collection of clinician information seeking behavior is laborious and time consuming, and thus, it is unlikely to scale well in obtaining training data for building predictive models across many different contexts. This limitation will be addressed in the future through the use of automatic methods of observing clinician information seeking behavior. For example, we could imitate how other researchers capture clinician behaviors without interrupting workflow,20 use EHR metadata, such as page visits and mouse clicks, or use newer technologies, such as eye-tracking.21
Open questions about the effectiveness of a LEMR system remain to be studied. We do not know the AUROC performance necessary for a clinician to trust models of their information seeking behavior. Future work is needed to determine the degree to which a LEMR system should favor precision (i.e., highlight only the patient data that are definitely relevant) versus recall (i.e., highlight any patient data that are potentially relevant); it could vary by the target and the context. Also, knowledge of clinical and cognitive-behavior can be leveraged to determine how and when highlighting patient data will be most effective.22
Conclusion
The results reported here contribute to the long-term development of a LEMR system that uses models to predict clinician information seeking behavior, dynamically adapts the LEMR interface to highlight relevant patient data, and thus provides concise, context sensitive data. We developed a data set from which we built models with good performance in predicting clinician information seeking behavior. As a next step, we plan to apply these models to a new set of patient cases and evaluate the quality and usefulness of the predictions in a new study. In the longer term, we plan to explore alternative methods of capturing targets that are more efficient and scale to larger data sets for building models. The LEMR system, we hope, will improve EMR usability by intelligently drawing the clinician’s attention to the right data, at the right time.
Acknowledgements
The research reported in this paper was supported by the National Library of Medicine of the National Institutes of Health under award numbers T15LM007059 and R01LM012095, and by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM088224. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
RRID’s are research resource identifiers. More information at https://scicrunch.org/resources
References
- 1.Friedberg MW, Chen PG, Van Busum KR, et al. Factors Affecting Physician Professional Satisfaction and Their Implications for Patient Care Health Systems and Health Policy. Rand Heal Q. 2014;3(4):1. [PMC free article] [PubMed] [Google Scholar]
- 2.Sinsky C, Colligan L, Li L, et al. Allocation of physician time in ambulatory practice: a time and motion study in 4 specialties. Ann Intern Med. 2016;165(11):753–760. doi: 10.7326/M16-0961. doi:10.7326/M16-0961. [DOI] [PubMed] [Google Scholar]
- 3.Street RL, Liu L, Farber NJ, et al. Keystrokes, mouse clicks and gazing at the computer: how physician interaction with the EHR affects patient participation. J Gen Intern Med. 2018;33(4):423–428. doi: 10.1007/s11606-017-4228-2. doi:10.1007/s11606-017-4228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marmor RA, Clay B, Millen M, Savides TJ, Longhurst CA. The impact of physician EHR usage on patient satisfaction. Appl Clin Inform. 2018;9(1):11–14. doi: 10.1055/s-0037-1620263. doi:10.1055/s-0037-1620263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Medical Association. Improving Care: Priorities to Improve Electronic Health Record Usability. 2014. https://www.aace.com/files/ehr-priorities.pdf. Accessed August 26, 2015.
- 6.Kannampallil TG, Jones LK, Patel VL, Buchman TG, Franklin A. Comparing the information seeking strategies of residents nurse practitioners and physician assistants in critical care settings. J Am Med Inform Assoc. 2014;21(e2) doi: 10.1136/amiajnl-2013-002615. doi:10.1136/amiajnl-2013-002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covvey HD, Zitner D, Berry DM, Cowan DD, Shepherd M. Proceedings 11th IEEEInt Requir Eng Conf 2003. 2003. Formal structure for specifying the content and quality of the electronic health record; pp. 162–168. [Google Scholar]
- 8.Pickering BW, Gajic O, Ahmed A, Herasevich V, Keegan MT. Data utilization for medical decision making at the time of patient admission to ICU. Crit Care Med. 2013;41(6):1502–1510. doi: 10.1097/CCM.0b013e318287f0c0. doi:10.1097/CCM.0b013e318287f0c0. [DOI] [PubMed] [Google Scholar]
- 9.Pickering BW, Herasevich V, Ahmed A, Gajic O. Novel representation of clinical information in the ICU. Appl Clin Inform. 2010;01(02):116–131. doi: 10.4338/ACI-2009-12-CR-0027. doi:10.4338/ACI-2009-12-CR-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolan ME, Cartin-Ceba R, Moreno-Franco P, Pickering B, Herasevich V. A multisite survey study of EMR review habits information needs and display preferences among medical ICU clinicians evaluating new patients background and significance. Appl Clin Inf. 2017;8:1197–1207. doi: 10.4338/ACI-2017-04-RA-0060. doi:10.4338/ACI-2017-04-RA-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolan M, Siwani R, Helmi H, Pickering B, Moreno-Franco P, Herasevich V. Health IT usability focus section: data use and navigation patterns among medical ICU clinicians during electronic chart review. Appl Clin Inform. 2017;08(04):1117–1126. doi: 10.4338/ACI-2017-06-RA-0110. doi:10.4338/ACI-2017-06-RA-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King AJ, Cooper GF, Hochheiser H, Clermont G, Visweswaran S. AMIA Annu Symp Proc. 2015. Development and preliminary evaluation of a prototype of a learning electronic medical record system; pp. 1967–1975. [PMC free article] [PubMed] [Google Scholar]
- 13.Visweswaran S, Mezger J, Clermont G, Hauskrecht M, Cooper GF. AMIA Annu Symp Proc. 2010. Identifying deviations from usual medical care using a statistical approach; pp. 827–831. [PMC free article] [PubMed] [Google Scholar]
- 14.Yount RJ, Vries JK, Councill CD. The Medical Archival System - an information-retrieval system based on distributed parallel processing. Inf Process Manag. 1991;27:379–389. [Google Scholar]
- 15.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15(2):155–163. doi: 10.1016/j.jcm.2016.02.012. doi:10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gortmaker SL, Hosmer DW, Lemeshow S. Applied logistic regression. Contemp Sociol. 1994;23(1):159. doi:10.2307/2074954. [Google Scholar]
- 17.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(3):27–1. doi:10.1145/1961189.1961199. [Google Scholar]
- 18.Breiman L. Random forests. Mach Learn. 2001;45:5–32. doi:10.1023/A:1010933404324. [Google Scholar]
- 19.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 20.Vankipuram A, Vankipuram M, Ghaemmaghami V, Patel VL. A mobile application to support collection and analytics of real-time critical care data. Comput Methods Programs Biomed. 2017;151:45–55. doi: 10.1016/j.cmpb.2017.08.014. doi:10.1016/j.cmpb.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 21.King AJ, Hochheiser H, Visweswaran S, Clermont G, Cooper GF. AMIA Jt Summits Transl Sci Proc. 2017. Eye-tracking for clinical decision support: a method to capture automatically what physicians are viewing in the EMR; pp. 512–521. [PMC free article] [PubMed] [Google Scholar]
- 22.Medlock S, Wyatt JC, Patel VL, Shortliffe EH, Abu-Hanna A. Modeling information flows in clinical decision support: key insights for enhancing system effectiveness. J Am Med Informatics Assoc. 2016;23(5):1001–1006. doi: 10.1093/jamia/ocv177. doi:10.1093/jamia/ocv177. [DOI] [PubMed] [Google Scholar]