Abstract
Substance abuse carries many negative health consequences. Detailed information about patients’ substance abuse history is usually captured in free-text clinical notes. Automatic extraction of substance abuse information is vital to assess patients’ risk for developing certain diseases and adverse outcomes. We introduce a novel neural architecture to automatically extract substance abuse information. The model, which uses multi-task learning, outperformed previous work and several baselines created using discrete models. The classifier obtained 0.88-0.95 F1 for detecting substance abuse status (current, none, past, unknown) on a withheld test set. Other substance abuse entities (amount, frequency, exposure history, quit history, and type) were also extracted with high-performance. Our results demonstrate the feasibility of extracting substance abuse information with little annotated data. Additionally, we used the neural multi-task model to automatically annotate 59.7K notes from a different source. Manual review of a subset of these notes resulted 0.84-0.89 precision for substance abuse status.
Introduction
The negative impact of substance abuse (including alcohol, drug, and tobacco) on health is increasingly recognized as a key factor for morbidity and mortality.1-3 In clinical research, it has been established that 5-10% of cancers can be attributed to hereditary factors, while 90-95% have been found correlated with lifestyle and environmental factors, such as smoking and alcohol consumption.4 The consequences of illicit drug and prescribed opioid abuse are also widespread, causing permanent physical and emotional damage to users. In many cases, users die prematurely from drug overdoses or other drug-associated illnesses. Characterization of a patient’s substance abuse history can be used to assess risk of future negative health outcomes related to substance abuse. Clinical notes contain rich information detailing the history of substance abuse from caregivers’ perspective, beyond what is available from structured EHR databases and which can be used to quantify severity of abuse. To utilize the reported substance abuse information for the purposes of secondary use, automatic extraction approaches are needed to process free-text clinical notes. In this work, we extend prior work using machine learning to automatically extract substance abuse information from clinical text. We introduce and evaluate a neural information extraction (IE) model using a corpus of clinical notes annotated by Yetisgen and Vanderwende.5 The corpus was created using a publicly available data source (MTSamples) and annotated for alcohol, drug, and tobacco abuse information documented in social history sections of history and physical notes.
We create a neural multi-task model that outperformed Yetisgen and Vanderwende’s previous work and the strong baselines we created using discrete models. To assess the generalizability of the extraction model, we used the multi-task model to annotate 59.7K discharge summaries from the MIMIC-III corpus,6 and hand-scored the substance status predictions of a randomly selected subset of notes. The performance results are encouraging and demonstrate the feasibility and generalizability of our extraction approach. We make the annotated corpus, source code, and annotations for the MIMIC-III notes available online.7
Task
Details of substance abuse are often described in free-text clinical notes. The 2008 i2b2 smoking challenge leveraged de-identified discharge records annotated as “past smoker,” “current smoker,” “smoker,” “non-smoker,” and “unknown.”8 Since its release, many IE systems have been developed to automatically annotate clinical text with these smoking annotations.9-11 Presented methods included lexicon-based, rule-based, and supervised machine-learning models.
Melton et al. reviewed three widely used public health surveys, i.e. practitioners’ questions to measure behaviors that may be relevant to clinical care; they proposed an information model for survey items related to alcohol, drug, and tobacco use, as well as occupation.12-14 Dimensions included temporality, degree of exposure, and frequency. They also investigated longitudinal tobacco use information from clinical notes.15
In the present study, we used Yetisgen and Vanderwende’s annotated corpus to explore IE algorithms on multiple types of substance abuse in clinical notes.5 They followed Melton’s model14 in annotating substance use, because it reflects the practioners’ needs and insights into how these substances impact patient health. Most of the dimensions discussed in the Carter et al. study of drug use were also annotated.14 Chen et al.16 surveyed the use of free-text describing alcohol use, coding for “type,” “status,” “temporal,” and “amount,” which was also captured in Yetisgen and Vanderwende’s5 annotation. Wang et al. explored extracting similar dimensions using rule-based methods.17
In the Yetisgen and Vanderwende data, each category of substance abuse (alcohol, drug, and tobacco) is represented with seven entities: status, amount, frequency, exposure history, quit history, type, and method.5 Status is a sentence-level label with four classes: unknown, none, current, and past. The other annotations correspond to multi-word spans, which are referred to as “entities” although they are not always noun phrases. In this study, we explore the use of neural networks, since recent work on IE in other domains has shown that they give state-of-the-art performance. We did not extract method, because the number of occurrences is insufficient to adequately assess performance. The substance associated with an entity is implicit in the label, but we also separately detect whether or not each sentence has information related to substance use, referred to here as an “event.” Table 1 provides status label examples and span label examples for the other entities. The amount spans often have a similar format, ([quantity] [units]); however, the units generally vary by substance (e.g. “three packs” vs. “one beer”). The phrases used to describe frequency, exposure history, and quit history are similar for all substances.
Table 1.
Annotation examples (bold font indicates labeled span)
| Entity/label | Examples |
| status/tobacco-unknown | Currently employed. |
| status/tobacco-none | She does not smoke. |
| status/alcohol-current | He occasionally uses alcohol. |
| status/drug-past | He used cocaine in the past. |
| type | He occasionally has a beer. |
| amount | He smokes less than a pack of cigarettes daily. |
| frequency | She occasionally uses alcohol. |
| exposure history | ...smokes half a pack per day for 30 years. |
| quit history | The patient quit smoking 17 years ago. |
IE performance is evaluated as in prior studies.5 For each substance, status F1 scores are computed at the sentence level for each of the none, past, and current categories and the scores are combined with micro-averaging. The unknown category is our assumed default category for status, so we do not report its performance. Performance on labels involving multi-word spans is evaluated at the token-level, as in prior work on this corpus. Token-level (vs. span-level) evaluation is common in clinical IE tasks such as this, particularly when the dataset size is limited.
Related Work
Clinical IE tasks have been approached using rule-based and machine learning systems, and more recent work has utilized neural networks. Maldonado et al. automatically annotated electroencephalography (EEG) concepts and attributes using active learning in conjunction with a multi-task, deep neural network, including a long short-term memory (LSTM) network.18 Dernoncourt et al. detected protected health information (such as names, locations, dates, etc) in clinical notes using a bidirectional LSTM with character aware word embeddings.19 Other clinical natural language processing (NLP) tasks have also benefited from the use of neural approaches.20,21
There is a significant body of IE work outside of the clinical domain, including IE subtasks like named entity recognition (NER), slot filling, event extraction, and text classification. Several works have achieved high performance in IE tasks using the Conditional Random Field (CRF) model.22-24 More recent IE work utilized recurrent neural networks (RNN), including the LSTM network, which captures long-range word dependencies.25-27 A popular LSTM-based approach to sequence tagging IE tasks incorporates a CRF layer at the output of the LSTM.25,27,28 The inclusion of the CRF, allows the model to learn allowable transitions between labels and conditionally independent predictions. Convolutional neural networks (CNN) have also successfully been applied to IE tasks, including text classification, slot filling and event extraction.29-31 These methods were incorporated in the neural model developed in this study.
In multi-task modeling, a single model generates multiple outputs and/or leverages shared parameters across multiple data sets or tasks. Neural multi-task models have achieved state-of-the-art performance in a variety of NLP tasks, including IE.26,32-34 Neural multi-task modeling is also useful for applications in the medical domain outside of NLP. For example, Harutyunyan et al. used clinical time series data and neural multi-task modeling to predict in-hospital mortality, length of stay, phenotyping, and decompensation.35 Jaques et al. predicted health, stress, and happiness using a neural multi-task model and data from wearable sensors and smartphone logs.36
Attention mechanisms have been used in neural encoder-decoder models to identify and attend to the most relevant position within the input sequence.37 Attention has been extended to representation learning through self-attention.38 Self-attention learns a vector representation of target phenomena and calculates attention weights for an input sequence, to emphasize the elements within a sequence that are most salient to the task. The learned attention weights provide an interpretable result, which highlights the most predictive elements within the sequence.
Methods
This section describes the data that is the foundation for experimental work, the discrete models implemented as a baseline, and the new neural multi-task model. The training set and test set assignments from Yetisgen and Vanderwende’s previous work were unavailable. Consequently, we reimplemented discrete models, similar to Yetisgen and Vanderwende’s previous work, to facilitate direct comparison of the neural multi-task model with a discrete baseline. In the reimplementation of the discrete models, some additional features were explored.
In the discrete and neural multi-task modeling frameworks, a separate module was designed to determine whether or not a sentence is associated with any substance events (single binary detector), and the result was used to filter out sentences with no relevant information in both frameworks. Both frameworks also treat the span detection problems as sequence tagging, using a begin-inside-outside (BIO) approach to representing spans of different types.
Data
Yetisgen and Vanderwende annotated a corpus of 364 social history sections from 516 history and physical notes from MTSamples website (http://mtsamples.com).5 The annotated dataset is available for download at the UW-BioNLP website (http://depts.washington.edu/bionlp/index.html?corpora). Table 2 contains a summary of the entity frequencies. For multi-words span entities, these counts reflect the number of spans, not the number of associated tokens.
Table 2:
Annotation statistics
| Entity | Alcohol | Drug | Tobacco |
| status | 254 | 154 | 278 |
| type | 26 | 112 | 50 |
| method | 0 | 10 | 4 |
| amount | 69 | 25 | 78 |
| frequency | 65 | 6 | 59 |
| exposure history | 7 | 10 | 37 |
| quit history | 6 | 2 | 37 |
The MIMIC-III corpus6 discharge summaries (59.7K notes) and physician notes (142K notes) were used in unsupervised learning to pre-train word embeddings for the neural multi-task model. The discharge summaries were also used to evaluate the generalizability of the multi-task model and provide data with automatically detected labels. We experimented with using the entirety of these notes vs. only the “Social History” and “History of Present Illness” sections. Performance was similar, so the smaller subset was used. Note sections were identified using simple pattern matching. Some notes in the MIMIC-III corpus include extraneous line breaks within sentences. All of the lines within a given section were merged into a single line, and then a sentence boundary detector39 was used to parse the section into sentences. The extracted sections resulted in a corpus of 19M tokens from 113K notes for word embedding training.
Discrete Models
Events. A substance detection model was trained to predict the presence of any substance event (alcohol, drug, or tobacco) within a sentence. The predictions from the substance detection model were used as a first-stage mask, such that subsequent modeling only used sentences that were predicted to contain a substance event. The substance detection model was created using logistic regression (LR). The same substance detection model was used as the first-stage mask for both discrete and neural modeling approaches. Substance indicator models were also trained to predict each individual substance using LR. LR models used word n-gram features (unigram-trigram) and gazetteer features. The gazetteer features consisted of three binary features, indicating alcohol, drug, or tobacco. The gazetteer word lists were generated by searching WordNet for hyponyms of each substance, resulting in 324 alcohol, 271 drug, and 46 tobacco words (search terms included “tobacco,” “alcoholic_drink,” “sedative,” “narcotic,” and “controlled_substance”).
Status. The status was classified using separate Maximum Entropy (MaxEnt) models for each substance. The MaxEnt models used word n-gram features (unigram-trigram) and same gazetteer features as the LR models.
Sequence Tags. The amount, frequency, exposure history, quit history, and type entities were extracted using linear-chain CRF models.22 Features included word n-grams (unigram-trigram), part-of-speech (POS) tags, capitalization indicators (lowercase, uppercase, title case, and other), and string type indicators (punctuation, number, alphabetic, alphanumeric, and other). Amount, frequency, exposure history, and quit history entities were extracted using two approaches. In the first approach, a separate CRF model was trained for each substance to extract substance-specific amount, frequency, exposure history, and quit history entities. In the second approach, these labels were merged across all substances, and a single CRF model was created to extract these substance-independent entities. Then, the substance indicator models were used to associate a substance with each extracted entity using the heuristic that any predicted entity in the sentence is assigned to all substances detected for the sentence. In the Results section, the first approach is referred to as “CRF,” and the second, two-stage, approach is referred to as “CRF+LR.”
Neural Multi-task Model
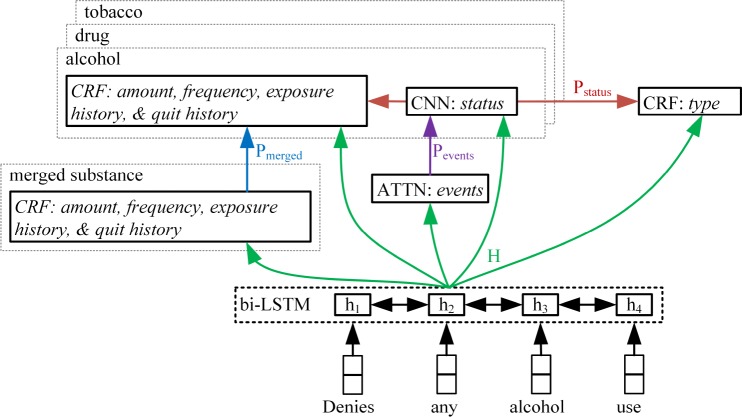
A single neural multi-task model was created to predict all entities for all substances, leveraging the shared information across entities and substances. Figure 1 is a diagram of the neural multi-task model. In the discrete modeling approaches, there is no shared learning between the different substances or entities. The multi-task approach is intended to take advantage of the similarities between substances and entities. The inputs to the model were pre-trained word embeddings, which fed into a bidirectional LSTM layer that used layer normalization.40 Word embeddings were pretrained using word2vec41 on the MIMIC-III data and held constant during the training of the multi-task model, due to the limited size of the annotated data. A 38K token vocabulary was chosen using tokens that occurred at least five times in the MIMIC-III subset, corresponding to a relatively low out-of-vocabulary rate of 2.3%. The forward and backward outputs states of the LSTM were concatenated resulting in n-by-2u matrix H, where u is the LSTM hidden size and n is the sequence length. H was used as features in downstream entity and substance-specific classifiers.
Figure 1.
Neural multi-task extraction model
Events. The same LR model as in the discrete model was used to filter out sentences with no events. Then, self-attention was used to estimate the probability of each substance event, automatically identifying word positions that best predict a given substance. Attention weights, Ai, were calculated as
| (1) |
where i denotes the substance, WS,i is a 1-by-2u learned vector, and Ai is a 1-by-n vector. The probability of a substance event was calculated using the weighted average of the hidden states, AiT, as
| (2) |
where WA,i is a 2-by-2u weight matrix and bA,i is a 2-by-1 bias vector. The probability of the substance events was concatenated to form a 3-dimensional vector (# substances) for use in status classification. The ground truth for learning Pevent was determined based on whether or not a given substance event occurred within the sentence. Error from the status classification was not back-propagated to Pevent.
Status. Status was predicted using separate, substance-specific classifiers. A CNN with max-pooling was used to create a low dimensional sentence representation for the target substance, xCNN,i, where i denotes the substance. The use of this CNN approach was motivated by the ability of CNNs to highlight salient information in the input sequence and preserve word order information. The CNNs used multiple filter widths, and multiple filters were created for each filter width. The substance probability vector, Pevents, was concatenated with xCNN,i to form a vector of size m-by-1 to predict status as
| (3) |
where Wstatus,i is a 4-by-m weight matrix and bstatus,i is a 4-by-1 bias vector(m = 3+#filters). Status probabilities, Pstatus,i, for each substance were concatenated to form 12-dimensional vector (4 labels × 3 substances), which was used as input features in the sequence tagging tasks. To prevent the sequence tagging tasks from negatively impacting the status classification, error from the sequence tagging tasks was not back-propagated to the status classifiers.
Sequence Tags. Linear-chain CRF models were used to extract sequence tags, because CRFs can learn and enforce allowable transitions between sequence labels (e.g. I-frequency label may follow a B-frequency label but not I-amount). The type entities for all substances were extracted using a single CRF, with input features H and Pstatus. The amount, frequency, exposure history, and quit history labels were merged across all three substances for training a substance-independent CRF that estimates probabilities, Pmerged, for these entities, with input features H and Pstatus. Then, substance-specific amount, frequency, exposure history, and quit history entities are extracted using separate, substance-specific CRFs, with input features H, Pstatus, and Pmerged.
Training
The annotated corpus was split into training and test sets using an 80%/20% split. All models were tuned using 5-fold cross validation (CV) on the training set. After determining the best configuration, models were retrained on the entirety of the training set. Only the highest performing discrete and multi-task models were applied to the withheld test set.
Discrete model tuning included the selection of feature types, including n-gram order, regularization type (L1 or L2), and regularization strength. Discrete models were created using Python scikit-learn.42 The best performing MaxEnt status models used the following features: unigrams for alcohol; unigrams and gazetteer for drug; and unigrams-bigrams and gazetteer for tobacco.
Neural multi-task model tuning included: selecting the word embedding training set, determining the connections within the multi-task graph, regularization strength, LSTM size, learning rate, number of epochs, CNN filter widths, number of CNN filters at each width, and layer normalization. The multi-task model was regularized using dropout in the LSTM and status CNN layers. The model was trained in multiple stages. In the first stage, the entire model was trained jointly to minimize average loss across all classifiers (tasks), updating all graph variables except for the pre-trained word embeddings. In the second stage, the learned parameters in the LSTM were frozen, and each set of entity-specific classifiers were trained jointly, with the other classifier variables fixed. The entity-specific classifiers were trained in the following order: 1) events, 2) status, 3) amount, frequency, exposure history, and quit history, and 4) type. The multi-task model configuration that achieved the best overall performance had the following configuration: H size = 200, H dropout = 20%, # joint epochs = 1000, # entity-specific epochs = 50, learning rate = 0.005, batch size = 20, CNN dropout = 40%, # CNN filters = 10, and CNN filter widths = [2, 3]. The neural multi-task model was created models using Google’s TensorFlow.43
All of the CV and test results presented reflect the end-to-end performance, i.e., any event detection errors impact the final results. Performance is presented in terms of the true positive count (TP), false negative count (FN), false positive count (FP), precision (P), recall (R) and F1 score (F1).
Results
The substance detector, which was used as a first-stage classifier, achieved a performance of F1=0.97 during CV and F1=0.98 on the test set. Tables 3 and 4 present the CV development and test set results, respectively, for the sentence-level status. Results are micro-averaged across labels.
Table 3:
Status CV development performance
| Substance | Model | TP | FN | FP | P | R | F1 |
| alcohol | MaxEnt | 168 | 32 | 17 | 0.91 | 0.84 | 0.87 |
| Multi | 178 | 22 | 23 | 0.89 | 0.89 | 0.89 | |
| drug | MaxEnt | 94 | 21 | 5 | 0.95 | 0.82 | 0.88 |
| Multi | 99 | 16 | 8 | 0.93 | 0.86 | 0.89 | |
| tobacco | MaxEnt | 172 | 42 | 33 | 0.84 | 0.80 | 0.82 |
| Multi | 185 | 29 | 33 | 0.85 | 0.86 | 0.86 |
Table 4:
Status test set performance
| Substance | Model | TP | FN | FP | P | R | F1 |
| alcohol | MaxEnt | 49 | 5 | 5 | 0.91 | 0.91 | 0.91 |
| Multi | 52 | 2 | 4 | 0.93 | 0.96 | 0.95 | |
| drug | MaxEnt | 20 | 6 | 3 | 0.87 | 0.77 | 0.82 |
| Multi | 24 | 2 | 1 | 0.96 | 0.92 | 0.94 | |
| tobacco | MaxEnt | 49 | 11 | 10 | 0.83 | 0.82 | 0.82 |
| Multi | 53 | 7 | 7 | 0.88 | 0.88 | 0.88 |
High performance was obtained for all substances in both data sets, with the neural model giving slightly better results in all cases. The multi-task model achieved higher performance on status extraction than Yetisgen and Vanderwende’s previous work previous work (F1 scores of 0.91 for alcohol, 0.86 for drug, and 0.80 for tobacco on the test set).5
Tables 5 and 6 present type, amount, frequency, exposure history, and quit history CV development and test set results, respectively. Results are presented for each entity, micro-averaged across substances. For amount, frequency, exposure history, and quit history extraction, the best performing CRF models used the following features: unigrams and gazetteer for alcohol; unigrams, POS, capitalization, string type, and gazetteer for drug; and unigrams and POS for tobacco. For the “CRF+LR” approach, the best performing CRF model used unigrams-trigrams, POS, capitalization, string type, and gazetteer features. The substance indicator models used in the “CRF+LR” approach achieved F1 performance of 0.95 for alcohol, 0.93 for drug, and 0.97 for tobacco during CV and F1 performance of 0.95 for alcohol, 0.88 for drug, and 0.95 for tobacco on the test set. The best-performing CRF model for type extraction used unigram and string type features. On the test set, the neural model produced similar or better results in all categories. The performance is robust in moving to the test set except for a small degradation for exposure history, which is from a drop in the drug subset where data is sparse. For amount, frequency, exposure history, and quit history, the multi-task model outperformed Yetisgen and Vanderwende’s previous work (F1 scores of 0.76 for amount, 0.75 for frequency, 0.63 for exposure history, and 0.75 for quit history)5 and both CRF baselines across each entity.
Table 5:
Entity CV development performance
| Entity | Model | TP | FN | FP | P | R | F1 |
| type | CRF | 119 | 53 | 5 | 0.96 | 0.69 | 0.80 |
| Multi | 123 | 49 | 8 | 0.94 | 0.72 | 0.81 | |
| CRF | 151 | 97 | 16 | 0.90 | 0.61 | 0.73 | |
| amount | CRF+LR | 168 | 80 | 90 | 0.65 | 0.68 | 0.66 |
| Multi | 180 | 68 | 39 | 0.82 | 0.73 | 0.77 | |
| CRF | 79 | 53 | 29 | 0.73 | 0.60 | 0.66 | |
| exposure hist. | CRF+LR | 89 | 43 | 23 | 0.79 | 0.67 | 0.73 |
| Multi | 85 | 47 | 17 | 0.83 | 0.64 | 0.73 | |
| CRF | 104 | 83 | 14 | 0.88 | 0.56 | 0.68 | |
| frequency | CRF+LR | 120 | 67 | 70 | 0.63 | 0.64 | 0.64 |
| Multi | 142 | 45 | 25 | 0.85 | 0.76 | 0.80 | |
| CRF | 59 | 55 | 6 | 0.91 | 0.52 | 0.66 | |
| quit history | CRF+LR | 82 | 32 | 19 | 0.81 | 0.72 | 0.76 |
| Multi | 65 | 49 | 23 | 0.74 | 0.57 | 0.64 |
Table 6:
Entity test set performance
| Entity | Model | TP | FN | FP | P | R | F1 |
| type | CRF | 31 | 3 | 1 | 0.97 | 0.91 | 0.94 |
| Multi | 31 | 3 | 2 | 0.94 | 0.91 | 0.93 | |
| CRF | 51 | 29 | 8 | 0.86 | 0.64 | 0.73 | |
| amount | CRF+LR | 61 | 19 | 22 | 0.73 | 0.76 | 0.75 |
| Multi | 65 | 15 | 12 | 0.84 | 0.81 | 0.83 | |
| CRF | 19 | 41 | 10 | 0.66 | 0.32 | 0.43 | |
| exposure hist. | CRF+LR | 31 | 29 | 27 | 0.53 | 0.52 | 0.53 |
| Multi | 37 | 23 | 12 | 0.76 | 0.62 | 0.68 | |
| CRF | 32 | 21 | 5 | 0.86 | 0.60 | 0.71 | |
| frequency | CRF+LR | 35 | 18 | 9 | 0.80 | 0.66 | 0.72 |
| Multi | 39 | 14 | 8 | 0.83 | 0.74 | 0.78 | |
| CRF | 18 | 9 | 12 | 0.60 | 0.67 | 0.63 | |
| quit history | CRF+LR | 18 | 9 | 4 | 0.82 | 0.67 | 0.73 |
| Multi | 21 | 6 | 4 | 0.84 | 0.78 | 0.81 |
Tables 7, 8, and 9 present the detailed status test set results for alcohol, drug, and tobacco, respectively. The none case is the dominant class for all substances. Again, the neural model gives the best result on the test set. For the cases where the difference between CV and test performance is greatest, the numbers of samples are small.
Table 7:
Alcohol status test set performance
| Label | Model | TP | FN | FP | P | R | F1 |
| current | MaxEnt | 17 | 3 | 2 | 0.89 | 0.85 | 0.87 |
| Multi | 20 | 0 | 2 | 0.91 | 1.00 | 0.95 | |
| none | MaxEnt | 30 | 1 | 3 | 0.91 | 0.97 | 0.94 |
| Multi | 30 | 1 | 1 | 0.97 | 0.97 | 0.97 | |
| past | MaxEnt | 2 | 1 | 0 | 1.00 | 0.67 | 0.80 |
| Multi | 2 | 1 | 1 | 0.67 | 0.67 | 0.67 |
Table 8:
Drug status test set performance
| Label | Model | TP | FN | FP | P | R | F1 |
| current | MaxEnt | 1 | 4 | 0 | 1.00 | 0.20 | 0.33 |
| Multi | 3 | 2 | 0 | 1.00 | 0.60 | 0.75 | |
| none | MaxEnt | 19 | 1 | 2 | 0.90 | 0.95 | 0.93 |
| Multi | 20 | 0 | 1 | 0.95 | 1.00 | 0.98 | |
| past | MaxEnt | 0 | 1 | 1 | 0.00 | 0.00 | 0.00 |
| Multi | 1 | 0 | 0 | 1.00 | 1.00 | 1.00 |
Table 9:
Tobacco status test set performance
| Label | Model | TP | FN | FP | P | R | F1 |
| current | MaxEnt | 8 | 3 | 1 | 0.89 | 0.73 | 0.80 |
| Multi | 8 | 3 | 1 | 0.89 | 0.73 | 0.80 | |
| none | MaxEnt | 34 | 3 | 6 | 0.85 | 0.92 | 0.88 |
| Multi | 35 | 2 | 2 | 0.95 | 0.95 | 0.95 | |
| past | MaxEnt | 7 | 5 | 3 | 0.70 | 0.58 | 0.64 |
| Multi | 10 | 2 | 4 | 0.71 | 0.83 | 0.77 |
MIMIC Annotation
The substance detection and multi-task models were used to predict substance events in the discharge summaries within the MIMIC-III corpus (59.7K notes). The resulting labeled texts are available online.7
The substance detection model predicted 40.3K MIMIC-III discharge summaries to have a substance event. Table 10 presents the entity occurrence counts from the unsupervised labeling of these notes.
Table 10:
MIMI-III annotation summary
| Entity | Alcohol | Drug | Tobacco |
| status | 44,536 | 20,725 | 45,244 |
| type | 4,756 | 14,509 | 11,443 |
| amount | 13,262 | 2,551 | 11,298 |
| frequency | 12,200 | 355 | 7,183 |
| exposure history | 770 | 396 | 4,639 |
| quit history | 176 | 72 | 10,241 |
Of the notes predicted to contain at least one substance event, 50 randomly sampled notes were hand-scored to evaluate the precision of status labels. Table 11 summarizes the manual review of 50 discharge summaries. The status classification precision was high for all substances (0.84-0.89).
Table 11.
MIMIC-III status prediction evaluation
| Substance | TP | FP | P |
| alcohol | 76 | 14 | 0.84 |
| drug | 80 | 10 | 0.89 |
| tobacco | 80 | 10 | 0.89 |
Conclusion
In summary, we implemented a state-of-the-art neural multi-task approach to the extraction of substance use information from clinical notes. The multi-task model outperformed Yetisgen and Vanderwende’s previous work and directly comparable discrete baselines on the test set for all entities, except type. For type, the multi-task model and the CRF baseline had similar performance. Excluding type, the performance gap between the multi-task model and the discrete baselines was larger on the test set than the training CV runs, suggesting the multi-task model generalized better to the test set. The improved extraction performance of the multi-task model will benefit downstream applications, including clinical decision support systems. Additionally, the multi-task modeling framework is well-suited to extending the current model to handle other types of information, such as socio-demographic, behavioral, and environmental exposure factors. A limited evaluation of the unsupervised labeling of the MIMIC-III discharge summaries indicated the multi-task model maintained high precision in the prediction of status.
This work is limited by the size and homogeneity of the annotated corpus, which may negatively impact generalizability of the extraction models to other clinical data sets. However, the initial results on the MIMIC-III data are encouraging. While semi-supervised learning techniques can be used to improve generalization,27,44,45 a larger annotated corpus with notes from different disciplines and institutions is required for assessing the generalizability of the extraction models.
References
- 1.Centers for Disease Control and Prevention. Annual smoking-attributable mortality, years of potential life lost, and productivity losses–United States, 1997-2001. Morb Mortal Wkly Rep. 2005;54:625–628. [PubMed] [Google Scholar]
- 2.World Health Organization. Global status report on alcohol and health, 2014; 2014. Available from: http://apps.who.int/iris/bitstream/10665/112736/1/9789240692763_eng.pdf.
- 3.Degenhardt L, Hall W. Extent of illicit drug use and dependence, and their contribution to the global burden of disease. The Lancet. 2012;379:55–70. doi: 10.1016/S0140-6736(11)61138-0. [DOI] [PubMed] [Google Scholar]
- 4.Anand P, Kunnumakara AB, Sundaram C, Harikumar KB, Tharakan ST, Lai OS, et al. Cancer is a preventable disease that requires major lifestyle changes. Pharmaceutical Research. 2008;25(9):2097–2116. doi: 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yetisgen M, Vanderwende L. 2017. Automatic identification of substance abuse from social history in clinical text. In: Artificial Intelligence in Medicine in Europe; pp. 171–181. [Google Scholar]
- 6.Johnson AE, Pollard TJ, Shen L, Lehman LwH, Feng M, Ghassemi M, et al. 2016. MIMIC-III, a freely accessible critical care database. Scientific Data; p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lybarger K. Clinical extraction; 2018. Available from: https://github.com/Lybarger/clinical_extraction.
- 8.Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.AM C. Five-way smoking status classification using text hot-spot identification and error-correcting output codes. J Am Med Inform Assoc. 2008;15(1):32–5. doi: 10.1197/jamia.M2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark C, Good K, Jezierny L, Macpherson M, Wilson B, Chajewska U. Identifying smokers with a medical extraction system. J Am Med Inform Assoc. 2008;15(1):36–39. doi: 10.1197/jamia.M2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonnagaddala J, Dai HJ, Ray P, Liaw ST. A preliminary study on automatic identification of patient smoking status in unstructured electronic health records. In: ACL-IJCNLP. 2015;vol. 2015:147–151. [Google Scholar]
- 12.Melton GB, Manaktala S, Sarkar IN, Chen ES. Social and behavioral history information in public health datasets. In: AMIA Annu Symp Proc. 2012;vol. 2012:625–34. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ES, Carter EW, Sarkar IN, Winden TJ, Melton GB. 2014. Examining the use, contents, and quality of free-text tobacco use documentation in the electronic health record. In: AMIA Annu Symp Proc; pp. 366–374. [PMC free article] [PubMed] [Google Scholar]
- 14.Carter EW, Sarkar IN, Melton GB, Chen ES. 2015. Representation of drug use in biomedical standards, clinical text, and research measures. In: AMIA Annu Symp Proc; pp. 376–385. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Chen ES, Pakhomov S, Lindemann E, Melton GB. 2016. Investigating longitudinal tobacco use information from social history and clinical notes in the electronic health record. In: AMIA Annu Symp Proc; pp. 1209–1218. [PMC free article] [PubMed] [Google Scholar]
- 16.Chen E, Garcia-Webb M. An analysis of free-text alcohol use documentation in the electronic health record. Applied Clinical Informatics. 2014;5(2):402–415. doi: 10.4338/ACI-2013-12-RA-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Chen ES, Pakhomov S, Arsoniadis E, Carter EW, Lindemann E. Automated extraction of substance use information from clinical texts. In: AMIA Annu Symp Proc. 2015;vol. 2015:2121–30. [PMC free article] [PubMed] [Google Scholar]
- 18.Maldonado R, Goodwin TR, Harabagiu SM. 2017. Active deep learning-based annotation of electroencephalography reports for cohort identification. In: AMIA Jt Summits Transl Sci Proc; p. 229. [PMC free article] [PubMed] [Google Scholar]
- 19.Dernoncourt F, Lee JY, Uzuner O, Szolovits P. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc. 2017;24(3):596–606. doi: 10.1093/jamia/ocw156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Zhang Y, Xu H. 2015. Clinical abbreviation disambiguation using neural word embeddings. In: Proceedings of BioNLP; pp. 171–176. [Google Scholar]
- 21.Jagannatha AN, Yu H. Bidirectional RNN for medical event detection in electronic health records. In: NAACL. 2016;vol. 2016:473–82. doi: 10.18653/v1/n16-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafferty J, McCallum A, Pereira FC. 2001. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In: Proc Int Conf Mach Learn; pp. 282–289. [Google Scholar]
- 23.Finkel JR, Grenager T, Manning C. 2005. Incorporating non-local information into information extraction systems by gibbs sampling. In: Proc Conf Assoc Comput Linguist Meet; pp. 363–370. [Google Scholar]
- 24.Peng F, McCallum A. Information extraction from research papers using conditional random fields. Inf Process Manag. 2006;42(4):963–979. [Google Scholar]
- 25.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. 2016. Neural architectures for named entity recognition. In: NAACL; pp. 260–270. [Google Scholar]
- 26.Augenstein I, Søgaard A. 2017. Multi-task learning of keyphrase boundary classification. In: Proc Conf Assoc Comput Linguist Meet; pp. 341–346. [Google Scholar]
- 27.Luan Y, Ostendorf M, Hajishirzi H. 2017. Scientific information extraction with semi-supervised neural tagging. In: Proc Conf Empir Methods Nat Lang Process; pp. 2641–2651. [Google Scholar]
- 28.Reimers N, Gurevych I. Optimal hyperparameters for deep LSTM-networks for sequence labeling tasks. arXiv preprint. 2017;1707:06799. [Google Scholar]
- 29.Johnson R, Zhang T. Effective use of word order for text categorization with convolutional neural networks. arXiv preprint. 2014;1412:1058. [Google Scholar]
- 30.Chen Y, Xu L, Liu K, Zeng D, Zhao J. 2015. Event extraction via dynamic multi-Pooling convolutional neural networks. In: Proc Conf Assoc Comput Linguist Meet; pp. 167–176. [Google Scholar]
- 31.Adel H, Roth B, Schutze H. 2016. Comparing convolutional neural networks to traditional models for slot filling. In: NAACL; pp. 828–838. [Google Scholar]
- 32.Collobert R, Weston J. 2008. A unified architecture for natural language processing: deep neural networks with multitask learning. In: Proc Int Conf Mach Learn; pp. 160–167. [Google Scholar]
- 33.Luan Y, Brockett C, Dolan B, Gao J, Galley M. Multi-task learning for speaker-role adaptation in neural conversation models. In: Int Joint Conf Natrual Lang Proc. 2017;vol. 1:605–614. [Google Scholar]
- 34.Jaech A, Heck LP, Ostendorf M. Domain adaptation of recurrent neural networks for natural language understanding. In: Interspeech. 2016;vol. 8:690–694. [Google Scholar]
- 35.Harutyunyan H, Khachatrian H, Kale DC, Galstyan A. Multitask learning and benchmarking with clinical time series data. arXiv preprint. 2017;1703:07771. doi: 10.1038/s41597-019-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaques N, Taylor S, Nosakhare E, Sano A, Picard R. 2016. Multi-task learning for predicting health, stress, and happiness. In: NIPS Workshop on Machine Learning for Healthcare. [Google Scholar]
- 37.Liu B, Lane I. Attention-based recurrent neural network models for joint intent detection and slot filling. In: Interspeech. 2016;vol. 8:685–689. [Google Scholar]
- 38.Lin Z, Feng M, Santos CNd, Yu M, Xiang B, Zhou B. A structured self-attentive sentence embedding. arXiv preprint. 2017;1703:03130. [Google Scholar]
- 39.Bird S, Loper E, Klein E, Inc OM. 2009. Natural language processing with Python. [Google Scholar]
- 40.Ba JL, Kiros JR, Hinton GE. Layer normalization. arXiv preprint. 2016;1607(06450) [Google Scholar]
- 41.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. arXiv preprint. 2013;1301:3781. [Google Scholar]
- 42.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O. Scikit-learn: machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 43.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C. TensorFlow: Large-scale machine learning on heterogeneous systems. 2015. Available from: https://www.tensorflow.org/.
- 44.Mohammad Aliannejadi SK, Masoud Kiaeeha, Ghidary SS. 2014. Graph-based semi-supervised conditional random fields for spoken language understanding using unaligned data. In: Australasian Lang Tech Assoc; pp. 98–103. [Google Scholar]
- 45.Yang Z, Cohen WW, Salakhutdinov R. Revisiting semi-supervised learning with graph embeddings. In: Proc Int Conf Mach Learn. 2016;vol. 48:40–48. [Google Scholar]