Abstract
Prescription information is an important component of electronic health records (EHRs). This information contains detailed medication instructions that are crucial for patients’ well-being and is often detailed in the narrative portions of EHRs. As a result, narratives of EHRs need to be processed with natural language processing (NLP) methods that can extract medication and prescription information from free text. However, automatic methods for medication and prescription extraction from narratives face two major challenges: (1) dictionaries can fall short even when identifying well-defined and syntactically consistent categories of medication entities, (2) some categories of medication entities are sparse, and at the same time lexically (and syntactically) diverse. In this paper, we describe FABLE, a system for automatically extracting prescription information from discharge summaries. FABLE utilizes unannotated data to enhance annotated training data: it performs semi-supervised extraction of medication information using pseudo-labels with Conditional Random Fields (CRFs) to improve its understanding of incomplete, sparse, and diverse medication entities. When evaluated against the official benchmark set from the 2009 i2b2 Shared Task and Workshop on Medication Extraction, FABLE achieves a horizontal phrase-level F1-measure of 0.878, giving state-of-the-art performance and significantly improving on nearly all entity categories.
1. Introduction
Electronic health records (EHR) contain significant amounts of factual information that is crucial to the overall well-being of the patients. Within this information, prescription information is an important component that contains detailed medication instructions. However, significant amounts of medication information are captured in narrative free-text portions of EHRs and are unavailable to systems that rely on structured knowledge representations for processing and retrieval. Automatic extraction of prescription information from free-text narratives can benefit medical studies and daily administrative tasks that need to work with structured data. For example, hospitals can build relational databases that link medication entries to patients’ medical histories so that the physicians can quickly skim through these records on future visits, especially under time constraints. Also, healthcare researchers can use extracted medication entities to build systems that predict future health conditions or incorporate them into decision support systems that can help diagnose rare diseases.
Our goal, in this paper, is to present a system for extracting prescription information from narrative discharge summaries. Given free-text narratives of discharge summaries, our objective is to identify medication names, along with their dosages, modes (routes), frequencies, durations, and reasons. We refer to these categories of information as medication entities. We then link these medication entities with their associated medication names to generate medication entries. These entries constitute prescription information. For example, given the following excerpts:
Take Lopressor 25 mg by mouth twice a day for angina.
The patient was prescribed with Celebrex 200 mg p.o. bid for 3 days for pain relief.
Subcutaneous heparin 5000 units x3 daily to reduce the risk of blood clots.
our system generates the structured information in Table 1.
Table 1.
Medication entries for the textual excerpts above. Columns show the entities constituting each entry.
| Drug Name | Dosage | Mode | Frequency | Duration | Reason | |
|---|---|---|---|---|---|---|
| Entry for excerpt 1 | Lopressor | 25 mg | by mouth | twice a day | N/A | angina |
| Entry for excerpt 2 | Celebrex | 200 mg | p.o. | bid | for 3 days | pain |
| Entry for excerpt 3 | Heparin | 5000 units | subcutaneous | x3 daily | N/A | blood clots |
Automatic extraction of medication entities and entries from narratives can be challenging. This is because only some of the medication entity types are well-defined lexically, syntactically, and semantically. When expressed in free-text form, categories such as durations and reasons tend to be lexically and syntactically diverse, and at the same time sparsely represented in the data. Furthermore, even in the case of well-defined medication entities, such as medication names, dictionaries can fall short of identifying all medication names, their generics, and group names. This limits the use of dictionaries and at the same time poses challenges for supervised machine learning approaches to medication extraction. As a result, we need methods that can handle sparsely represented, and at the same time lexically and syntactically sparse medication entity categories. In this paper, we describe FABLE, a system for automatically extracting medication entries from electronic health records. FABLE stands for Factual Analysis in Biomedical Language Extraction. FABLE employs a semi-supervised machine learning approach based on Conditional Random Fields (CRFs) in order to capitalize on unlabeled (but more abundantly available) data to enhance recognition of sparsely represented, lexically and syntactically diverse medication entities while at the same time compensating for incomplete dictionaries of even the well-defined categories.
2. Related Work
2.1. Prescription Extraction
Early prescription extraction systems relied mostly on rules. One of the earliest medication extraction systems, MedLEE1 was built with the purpose of extracting, structuring, and encoding clinical information within free-text patient reports. Similar to MedLEE, MetaMap2 extracts medication information by querying the Unified Medical Language System (UMLS) Metathesaurus. These systems take advantage of external resources (e.g., professional gazetteers, clinical databases) and combine them with pattern-matching rules to identify medication entities from unstructured free texts. They solely focus on recognizing clinically-relevant entities without extracting the relations among them.
The 2009 i2b2 Shared Task and Workshop on Medication Extraction3 vitalized the research in prescription extraction. The workshop focused on the extraction of medication names and medication entities that are related to prescription information from medical discharge summaries. The workshop provided 1,243 records, 251 of which were annotated for the gold standard. Most teams in this workshop tackled the medication entity and entry extraction task using rule-based approaches. However, the highest performing team4 hybridized machine learning classifiers and handwritten rules, achieving a horizontal phrase-level F1-measure of 0.857, when evaluated on the official benchmark set. This system performed particularly well on medication names, dosages, modes, and frequencies. However, the best performance on durations and reasons was achieved by a rule-based system5. The rules written for these medication entities focused on the linguistic signals found in sample duration and reason phrases and achieved phrase-level F1-measures of 0.525 and 0.459, respectively.
2.2. Semi-Supervised NER
One reason for the limited performance in medication extraction is the limited amount of annotated training data. Additionally, sparsity of some medication entity categories (such as medication names, reasons, and durations) that exhibit high lexical variation presents a challenge for supervised machine learning systems.
The preparation for annotated data is labor-intensive and time-consuming, limiting the amount of available training data for supervised learning. However, availability of unannotated data sets opens up the possibility of combining annotated data with unannotated data for training semi-supervised models. One common semi-supervised method automatically annotates unlabeled data and iteratively introduces these automatic annotations into the training set using a confidence-rated classifier, for a fixed number of iterations or until performance converges. We refer to this method as bootstrapping with unlabeled data. Outside of the medical domain, several studies6-10 utilized this method with successful results, applying the same high threshold for semi-supervised learning of all entities in the task.
During the 2010 i2b2/VA Shared-Task and Workshop on Medical Concept Extraction (where the medical concepts consisted of problems, lab tests, and treatments)11, the highest performing system used machine learning classifiers with rule-based components. They trained a semi-Markov model using passive-aggressive online updates. They then added semi-supervised training by bootstrapping with unlabeled data to exploit more entities that can be used to train models on relation extraction for an improved performance12.
In our study, we experiment with semi-supervised learning using a sequence labelling classifier, aimed at improving overall performance on prescription extraction, with particular focus on the sparsest and (lexically and syntactically) most diverse medication entities: medication names, durations, and reasons. We hypothesize that a confidence-based bootstrapping approach needs to be tailored to fit the characteristics of individual entity categories. Sparse and diverse entity categories may not produce as high confidence as better defined and better represented entity categories; so we identify optimal confidence scores per entity category rather than per task. We refer to these per-entity confidence rates as “conditional confidence rates.”
3. Data
We used the dataset provided by the 2009 i2b2 Shared Task and Workshop on Medication Extraction. This dataset contains a total of 1,243 de-identified discharge summaries. From these summaries, two subsets have been manually annotated: (1) 145 records annotated by Patrick et al. from the University of Sydney4. We refer to this subset as the Sydney set and use it for training our supervised models. (2) 251 records annotated, under the guidance of shared task organizers, during the shared task by the participants3. This set was the official benchmark set of the shared task and is referred to as the i2b2 set. We use the i2b2 set for final system evaluation. There is no overlap between the Sydney and the i2b2 sets. The remaining 846 unannotated documents contribute to semi-supervised learning. We refer to these unannotated records as the unlabeled set. See Table 2 for the per-category statistics in each subset.
Table 2.
Number of entities per category and entries in the Sydney, i2b2, and unlabeled subsets.
| Subset | Record | Token | Medication | Dosage | Mode | Frequency | Duration | Reason | Entry |
|---|---|---|---|---|---|---|---|---|---|
| Sydney | 145 | 250,436 | 7,988 | 4,132 | 3,052 | 3,881 | 508 | 1,516 | 8,516 |
| i2b2 | 251 | 284,311 | 8,440 | 4,371 | 3,299 | 3,925 | 499 | 1,325 | 9,193 |
| Unlabeled | 846 | 925,703 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
4. Method
Our system, FABLE, automatically extracts medication entries from electronic health records. Extraction of medication entries requires first the identification of medication entities, and then the linkage of these entities to the medication that they describe so as to create entries. This means that poor performance on medication entity extraction dooms medication entry extraction from the outset. In other words, improved performance on medication entity extraction also enables improvement in medication entry extraction. Therefore, FABLE focuses on extraction of medication entities. It employs semi-supervised learning to capitalize on unlabeled data to enhance recognition of sparsely represented, lexically and syntactically diverse medication entities. The unlabeled data also help it compensate for potentially incomplete dictionaries.
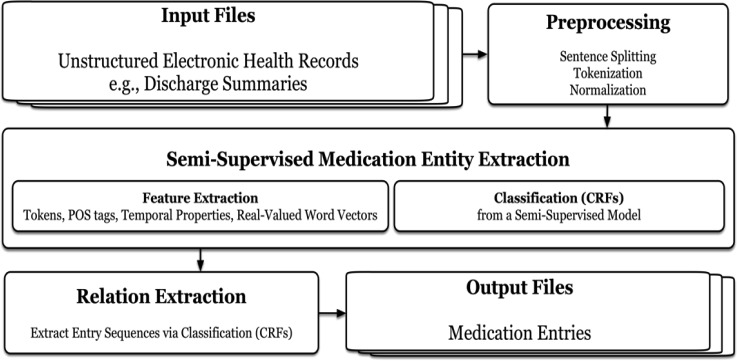
Figure 1 shows FABLE’s workflow. The workflow starts with pre-processing and feature extraction which help semi-supervised learning of entities. The entities then become input to a relation extractor which links together all entities related to the same medication. Final output is a prescription consisting of medication entries.
Figure 1.
FABLE’s workflow: a semi-supervised prescription extraction system
4.1. Pre-processing
We pre-processed our data as follows. We split the documents into sentences and into tokens by splitting at periods and whitespaces, while also maintaining the punctuation in acronyms, lists, and numbers. We lowercased all tokens and paired them with part-of-speech (POS) tags using Natural Language Toolkit (NLTK)13. Finally, we replaced numbers, including literal and numerical forms, with placeholders (e.g., 200 ml → DDD ml, Seven hours → D hours, the fifth day → the D day).
4.2. Feature Extraction
We extracted the following features from our data. Tokens and POS tags are considered the base features for our models. We then added six temporal binary features14 to represent medication entities that contain temporal expressions, such as durations and frequencies. These features indicate whether the current token is a temporal signal (e.g., “for” in for three weeks, “before” in “before admission”, “after” in “after 24 hours”), a time (e.g., 8am, 7pm), a temporal period (e.g., decades, weekends), a part of the day (e.g., morning, afternoon), a temporal reference (e.g., today, yesterday), and a number (e.g., 0.25, 700).
We also included word embeddings15 in the feature set to capture meaningful syntactic and semantic regularities based on statistical co-occurrences of the tokens. The extraction of word vectors is unsupervised. Real-valued word vectors in clinical NER were proven to be effective16 for significantly improved performance.
We extracted 100-dimensional real-valued word vectors using GloVe17 from a subset of MIMIC III18. This subset contains about 2 million clinical notes from about 46 thousand patients. We pre-processed this dataset using the same pre-processing steps described in Section 4.1 to create a training corpus that is compatible with our task. We used the vector from token <unknown> for words that did not exist in MIMIC III.
4.3. Semi-Supervised Models
Prior to the generation of semi-supervised models, we trained a base learner on the labeled data using the extracted features described in Section 4.2. We utilized CRFSuite19 with feature window size of ± 2 as the optimal setting for this task16. The base learner serves two purposes. First, it predicts medication entities from the unlabeled set in various settings. Second, it provides a baseline for our proposed system.
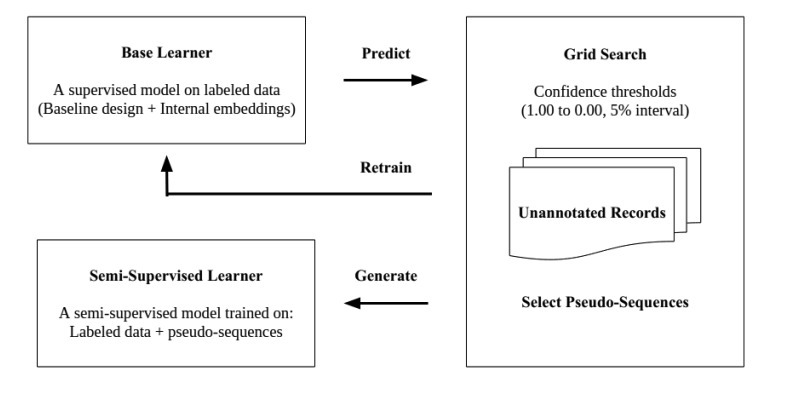
We experimented with semi-supervised learning for medication entity extraction using the base learner. We utilized the base learner trained on the labeled data to identify pseudo-sequences (i.e., medication entities) in the unlabeled data. When predicting pseudo-sequences, the base learner also assigns a confidence rate to each sequence (ranges from 0.00 to 1.00). Confidence rates indicate the level of trust the base learner has in predicted sequences. We used these confidence rates to expand the labeled data with only the high scoring pseudo-sequences (with respect to a threshold) and retrained the base learner. We repeated this process until no predicted pseudo-sequences passed the threshold or until performance converged. Figure 2 shows the workflow of semi-supervised approach.
Figure 2.
Generating semi-supervised models using grid search for medication entity extraction
The literature20 suggests picking a confidence threshold of 0.90 or 0.95 for semi-supervised learning. In such a case, the classifier only utilizes high-confidence sequences from the unannotated records. Such a high threshold would minimize noise that can come from incorrect pseudo-sequences. However, setting a high confidence threshold also potentially excludes some correctly predicted pseudo-sequences from the training model. In our case, this can be problematic: we are predicting multiple entity categories, some of which are easier than others. A high threshold would therefore favor the easier categories and filter out some potentially useful but low confidence samples from the harder categories. The intuition is that the confidence rates of the sequences that come from low performing entity categories (e.g., durations and reasons) can be lower than others.
In our study, we expand the literature’s suggestion by performing grid search on the full range [0,1] of confidence rates with 0.05 increments, with the aim of selecting the optimal threshold for each entity type. We collect five-fold cross-validated extraction results from 20 different confidence thresholds. For each threshold, for each fold, we train the base learner on four folds and predict pseudo-sequences on the unlabeled set. We add the sequences that score higher than the threshold into the labeled dataset and retrain the model. We then use this resulting model to extract pseudo-sequences from the unlabeled set again and retrain the model on the labeled data set expanded with new sequences that score higher than the threshold. We repeat these steps iteratively until no sequence passes the threshold or performance converges. We evaluate the resulting model on the held-out fold. We repeat this process five times, once per fold, and compute average cross-validation performance per entity type per threshold. We identify the highest performing threshold for each entity category. If we observe the same performance on multiple thresholds, we select the highest threshold. The rationale is to filter out pseudo-sequences that potentially have low quality, even if they do not negatively impact overall performance. We refer to the identified optimal per entity category thresholds as the conditional thresholds.
As a last step in medication entity extraction, we train an optimal semi-supervised model using the conditional thresholds. We report final results on the test set.
4.4. Relation Extraction
At the end of relation extraction, the extracted utterances are textual excerpts that contain medication entities. We then grouped medication entities inside each utterance into individual entries. We evaluated this approach using gold After the extraction of medication entities, we organize them into individual medication entries via relation extraction. We tackled this task as a sequence labeling problem using CRFs from the observation that medication names are mostly related to their neighboring entities. Therefore, instead of extracting binary relation pairs, we extracted each medication entry by predicting boundaries of utterances that contain related medication entities. We trained a model on tokens and the previously predicted medication entities to predict spans of utterances containing medication entries. We added to these the absolute distances between the current token and its nearest medication name (both to the left and right), and the collapsed word shape (e.g., “Advil” → “Aa”, “40ml” → “0a”). Figure 3 shows an example utterance containing a medication entry. All underlined medication entities are related to “Aspirin”.
Figure 3.
Example medication entry utterance
At the end of relation extraction, the extracted utterances are textual excerpts that contain medication entities. We then grouped medication entities inside each utterance into individual entries. We evaluated this approach using gold standard medication entities in the training data and achieved a horizontal phrase-level F1-measure of 0.89316. Final results are reported on the test set.
4.5. Evaluation Metrics and Significance Tests
The results from each experiment on medication entity extraction were evaluated using phrase-level F1-measure (i.e., exact match21). Final output of FABLE was evaluated using the official evaluation metrics from 2009 i2b2 Shared Task and Workshop on Medication Extraction: horizontal phrase-level F1-measure. This metric evaluates the final output at the medication entry level. Therefore, every entity in the entry needs to match the gold standard. In other words, horizontal phrase-level F1-measure provides a compound metric that simultaneously evaluates medication entity extraction and relation extraction.
We use the z-test on two proportions to test statistical significance of our performance gains. We use a z-score of 1.96. This corresponds to a = 0.05.
5. Results and Discussion
We first evaluated the performance of the base learner on the extraction of medication entities by five-fold cross validating on the Sydney set. Table 3 shows phrase-level F1-measure on each medication entity category.
Table 3.
Cross-validated phrase-level F1-measure of the base learner
| Medication | Dosage | Mode | Frequency | Duration | Reason |
|---|---|---|---|---|---|
| 0.885 | 0.918 | 0.933 | 0.931 | 0.623 | 0.407 |
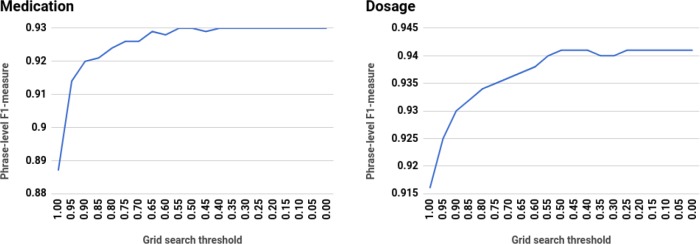
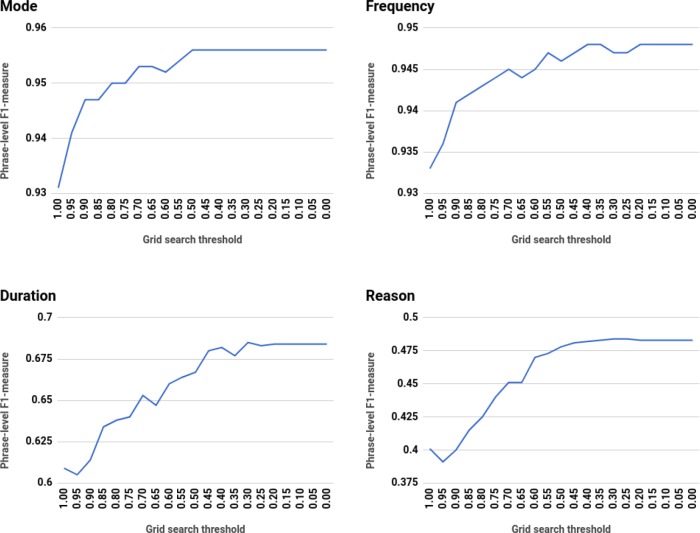
Given base learner results, we evaluated the performance gain after the introduction of unlabeled data using grid search. Figure 4 shows average five-fold cross-validated phrase-level F1-measure on each medication entity category at each confidence threshold. Overall, we achieve statistically significant performance improvements in prescription extraction task when we include pseudo-sequences in the labeled data. However, we found that most of the entity categories reach top performance when the threshold is set to 0.55 or below (see Table 4). This is particularly prominent for durations and reasons, when the pseudo-sequences still positively contribute to the model with threshold set to 0.30.
Figure 4.
Average cross-validated phrase-level F1-measure from grid search with unlabeled set
Table 4.
Conditional thresholds for medication entities
| Medication | Dosage | Mode | Frequency | Duration | Reason |
|---|---|---|---|---|---|
| 0.55 | 0.50 | 0.50 | 0.40 | 0.30 | 0.30 |
Figure 4 also shows that despite minor fluctuations, the extraction performance converges during grid search. Furthermore, the extraction performance remains identical when we set the threshold to 0.20 or under. We manually analyzed predictions from the base learner to understand this phenomenon. We found that there were only five pseudo-sequences that scored below 0.20 (three medication names, one dosage, and one reason). Such a small number of pseudo-sequences could not have a significant effect on the classifier.
Given these findings, for each entity category, we only included the sequences from the unlabeled set with confidence rates higher than the conditional threshold for that entity category (see Table 4). We retrained the base learner with the addition of these selected pseudo-sequences to generate semi-supervised models.
Table 5 shows five-fold cross-validated performance of the conditional threshold model (semi-supervised) against the base learner (fully supervised) results from Table 3 (repeated here for convenience). Results show significant performance improvement on all entity categories.
Table 5.
Average cross-validated phrase-level F1-measure of supervised and semi-supervised methods
| Approach | Medication | Dosage | Mode | Frequency | Duration | Reason |
|---|---|---|---|---|---|---|
| Supervised | 0.885 | 0.918 | 0.933 | 0.931 | 0.623 | 0.407 |
| Semi-Supervised | 0.930 | 0.941 | 0.955 | 0.947 | 0.685 | 0.481 |
* Bold numbers indicate statistical significance with respect to other results in the same column. We use a z-score of 1.96, which corresponds to α = 0.05.
To conclude our experiments, we evaluated FABLE on the official benchmark set from the 2009 i2b2 Shared Task and Workshop on Medication Extraction (i.e., i2b2 set). We trained the base learner on the Sydney set and used this supervised model to predict the unlabeled set with conditional thresholds. We then retrained the base learner with the addition of the pseudo-sequences from the unlabeled set and predicted new pseudo-sequences, further expanding the labeled set with pseudo-sequences that passed the conditional threshold. We repeated this process until no pseudo-sequences passed the threshold or performance converged. We then used the final semi-supervised model to predict the i2b2 set. Medication entries were then generated from these predicted entities, using the relation extraction module described in Section 4.4. Table 6 shows phrase-level F1-measure on the medication entities as well as horizontal score (last column on the right) on the predicted medication entries using various methods. In comparison to the fully-supervised baseline, as well as the semi-supervised models that used a fixed 0.95 or 0.90 threshold (as suggested by literature), the results from semi-supervised model that utilized conditional thresholds (last row) were significantly higher on medication entries and on all entity categories. Our method also performed significantly higher than the top system4 from 2009 i2b2 Challenge on medication entries and on nearly all entity categories.
Table 6.
Phrase-level F1-measure and horizontal score on the i2b2 set
| Approach | Medication | Dosage | Mode | Frequency | Duration | Reason | Entry |
|---|---|---|---|---|---|---|---|
| Patrick et al.4 | 0.884 | 0.893 | 0.909 | 0.902 | 0.446 | 0.444 | 0.864 |
| Supervised | 0.857 | 0.860 | 0.885 | 0.893 | 0.473 | 0.320 | 0.832 |
| 0.95 threshold | 0.866 | 0.873 | 0.896 | 0.890 | 0.500 | 0.341 | 0.841 |
| 0.90 threshold | 0.869 | 0.874 | 0.896 | 0.890 | 0.511 | 0.346 | 0.843 |
| Conditional | 0.907 | 0.896 | 0.919 | 0.914 | 0.597 | 0.422 | 0.878 |
* Bold numbers indicate statistical significance with respect to other results in the same column. We use a z-score of 1.96, which corresponds to α = 0.05.
We investigated the reason behind performance gains over the fixed thresholds of 0.95 and 0.90. We calculated the min, max, and average confidence rates as well as standard deviations of confidence rates on the correctly-predicted sequences per entity category from the base learner in a five-fold cross-validation setup (see Table 7). We found that confidence rates ranged from 1 to 0.2 for all entity categories. But the average confidence rates were lower for the entity categories that performed poorly in the base learner, i.e., durations and reasons. This confirmed our hypothesis that higher thresholds can be too conservative in their choice of pseudo-sequences and miss learning opportunities. In other words, models that use high thresholds (e.g., 0.95 or 0.90) exclude most of the duration and reason sequences from the semi-supervised model. Conditional confidence rates solve this problem and achieve significant gains.
Table 7.
Confidence rates for medication entities
| Stat. | Medication | Dosage | Mode | Frequency | Duration | Reason |
|---|---|---|---|---|---|---|
| Average | 0.913 | 0.961 | 0.951 | 0.940 | 0.805 | 0.808 |
| Maximum | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Minimum | 0.271 | 0.261 | 0.244 | 0.278 | 0.278 | 0.336 |
| Std. Deviation | 0.125 | 0.088 | 0.095 | 0.106 | 0.106 | 0.156 |
Overall, our system improves upon the state of the art in extraction of medication entries. We manually analyzed extraction results from the semi-supervised approach with conditional thresholds. The optimal model resolved 2,306 prediction errors when compared against the fully-supervised baseline. We randomly checked 20% of these instances (corresponding to 461 instances). We found that semi-supervised approach with conditional thresholds recognized more entities that contain non-specific tokens (e.g., “mildly” in “mildly tachycardic”, “persistent” in “persistent hypoglycemia”, “her” in “her arthritis”). These tokens have weak relationships with the overall semantics of the entity categories because they frequently appear in phrases that are not medication entities. Furthermore, with the help of pseudo-sequences, our semi-supervised approach recognized 790 more hapax legomena, i.e., entities that appear only once in the data, when compared to the baseline. These entities constitute 34.26% of the prediction errors that were resolved by semi-supervised approach. Table 8 shows three example entries from our system’s final output.
Table 8.
Medication entries from the system’s final output using the i2b2 set
| Drug Name | Dosage | Mode | Frequency | Duration | Reason | |
|---|---|---|---|---|---|---|
| Entry 1 | nitroglycerin | 1 tab | sl | x3 doses prn | N/A | chest pain |
| Entry 2 | coumadin | 1 mg | p.o. | q. h. s. | N/A | N/A |
| Entry 3 | glucophage | 1000 mg | p.o. | t.i.d. | x 7 days | N/A |
Despite its gains, the semi-supervised model with conditional thresholds still makes mistakes. For example, similarly to other models, it still has a hard time recognizing entities that occur only once in the corpus. Most of these entities come from medication names and show high morphological and lexical variation. Word embeddings and pseudo-sequences are beneficial when extracting medication names but extraction of these entity categories still needs more work. Also, our system has difficulty distinguishing generic entities. For example, some medication names refer to groups of medications (e.g., “home medication”, “meds”, “antibiotics”, “supplements”). These entities can be ambiguous and appear as section headings (which do not belong in a prescription and should not be extracted) or as a reference to groups of medications that the patient is taking (which should be extracted). Our system has a hard time distinguishing the two.
6. Conclusion
In this paper, we presented FABLE, a system to automatically extract prescription information (i.e., medication entries) from discharge summaries. FABLE utilizes a semi-supervised machine learning approach with CRFs and conditional confidence thresholds that are tuned to individual entity categories. The proposed system significantly improves the extraction performance on all medication entity categories. When evaluated against the official benchmark set from the 2009 i2b2 Shared Task and Workshop on Medication Extraction, FABLE achieves a horizontal phrase-level F1-measure of 0.878, giving state-of-the-art performance16.
References
- 1.Friedman C. American Medical Informatics Association. A broad-coverage natural language processing system. In Proceedings of the AMIA Symposium 2000 (p. 270) [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010 May 1;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association. 2010 Sep 1;17(5):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patrick J, Li M. In Proceedings of the Australasian Language Technology Association Workshop. 2009. A cascade approach to extracting medication events; pp. 99–103. [Google Scholar]
- 5.Spasić I, Sarafraz F, Keane JA, Nenadic G. Medication information extraction with linguistic pattern matching and semantic rules. Journal of the American Medical Informatics Association. 2010 Sep 1;17(5):532–5. doi: 10.1136/jamia.2010.003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson A, Betteridge J, Wang R. C, Hruschka E. R, Jr, Mitchell T. M. Coupled Semi-supervised Learning for Information Extraction. In Proceedings of the Third ACM International Conference on Web Search and Data Mining; New York, NY, USA: ACM; (2010). pp. 101–110. [DOI] [Google Scholar]
- 7.Erkan G, Ozgur A, Radev DR. Semi-supervised classification for extracting protein interaction sentences using dependency parsing. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL).2007. [Google Scholar]
- 8.Li X, Wang YY, Acero A. Extracting structured information from user queries with semi-supervised conditional random fields. In Proceedings of the 32nd international ACM SIGIR conference on Research and development in information retrieval; ACM; 2009. Jul 19, pp. 572–579. [Google Scholar]
- 9.Thenmalar S, Balaji J, Geetha TV. 2015. Nov 21, Semi-supervised bootstrapping approach for named entity recognition. arXiv preprint arXiv:1511.06833. [Google Scholar]
- 10.Wu T, Pottenger WM. A semi-supervised active learning algorithm for information extraction from textual data. Journal of the Association for Information Science and Technology. 2005 Feb 1;56(3):258–71. [Google Scholar]
- 11.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011 Jun 16;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. Journal of the American Medical Informatics Association. 2011 May 12;18(5):557–62. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird S. In Proceedings of the COLING/ACL on Interactive presentation sessions. Association for Computational Linguistics; 2006. Jul 17, NLTK: The Natural Language Toolkit; pp. 69–72. [Google Scholar]
- 14.Filannino M, Nenadic G. Temporal expression extraction with extensive feature type selection and a posteriori label adjustment. Data & Knowledge Engineering. 2015 Nov 1;100:19–33. [Google Scholar]
- 15.Mikolov T, Chen K, Corrado G, Dean J. 2013. Jan 16, Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781. [Google Scholar]
- 16.Tao C, Filannino M, Uzuner Ö. Prescription extraction using CRFs and word embeddings. Journal of Biomedical Informatics. 2017;72:60–6. doi: 10.1016/j.jbi.2017.07.002. doi:10.1016/j.jbi.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennington J, Socher R, Manning CD. Glove: Global Vectors for Word Representation. In EMNLP 2014 Oct 25. Vol. 14:1532–43. [Google Scholar]
- 18.Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Scientific data. 2016;3 doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okazaki N. CRFsuite: a fast implementation of conditional random fields (CRFs) 2007. [Google Scholar]
- 20.Abney S. Semi-supervised learning for computational linguistics. CRC Press; 2007. Sep 17, [Google Scholar]
- 21.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011 Jun 16;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]