Abstract
Detecting negation in biomedical texts entails the automatic identification of negation cues (e.g. “never”, “not”, “no longer”) as well as the scope of these cues. When medical concepts or terms are identified within the scope of a negation cue, their polarity is inferred as “negative”. All the other concepts or words receive a positive polarity. Correctly inferring the polarity is essential for patient cohort retrieval systems, as all inclusion criteria need to be automatically assigned positive polarity, whereas exclusion criteria should receive negative polarity. Motivated by the recent development of techniques using deep learning, we have experimented with a neural negation detection technique and compared it against an existing neural polarity recognition system, which were incorporated in a patient cohort system operating on clinical electroencephalography (EEG) reports. Our experiments indicate that the neural negation detection method produces better patient cohorts then the polarity recognition method.
Introduction
Clinical electroencephalography (EEG) is the most important investigation tool in the diagnosis and management of epilepsy. In addition, it is used to evaluate other types of brain disorders1, including encephalopathies, neurological infections, Creutzfelt-Jacob disease and other prion disorders, and even in the progression of Alzheimer’s disease. An EEG records the electrical activity along the scalp and measures spontaneous electrical activity of the brain. The signals measured along the scalp can be correlated with brain activity, which makes it a primary tool for diagnosis of brain-related illnesses2. However, the EEG signal is complex and inter-observer agreement for EEG interpretation is known to be moderate3. These interpretations of EEG recordings are documented in EEG reports. As more clinical EEG data becomes available, the interpretations of EEG signals can be improved by providing neurologists with results of search for patients that exhibit similar EEG characteristics. Recently, Goodwin & Harabagiu (2016)4 have described the MERCuRY (Multi-modal EncephalogRam patient Cohort discoveRY) system that relies on deep learning to represent the EEG signal and operates on a multi-modal EEG index resulting from the automatic processing of both the EEG signal and the EEG reports. The MERCuRY system allows neurologist to search a vast data archive of clinical EEG signals and EEG reports, enabling them to discover patient populations relevant to queries like Q1: “History of seizures and EEG with TIRDA without sharps, spikes, and electrographic seizures”.
The identification of relevant patient cohorts satisfying the characteristics expressed in queries such as Q1 relies on (1) the ability to automatically and accurately recognize in the query the inclusion and exclusion criteria; and (2) a relevance model capable to identify patients satisfying the inclusion criteria and not displaying any of the exclusion criteria. In Q1, the inclusion criteria are “history of seizures” and “EEG with temporal intermittent Delta activity (TIRDA)” while the exclusion criteria are “sharps, spikes and electrographic seizures”. The ability of automatically identifying the inclusion and exclusion criteria in the query is granted by the recognition of negation, leading to the inference of the polarity of query expressions. Expressions which are within the scope of negation are considered as having negative polarity, whereas the other query expressions are considered to have positive polarity. Alternatively, we can recognize polarity by taking advantage of the definitions used in the 2012 i2b2 challenge5 on evaluating temporal relations in medical text. In that challenge, each medical concept could have either a “positive” or a “negative” polarity, depending on the absence or presence of negation of its finding. Moreover, as implemented in the MERCuRY system, all the medical concepts from the EEG reports are indexed along with their polarity attribute, informing the relevance model. It is important to note that negation or polarity is recognized only in the queries or EEG reports, thus we ignore the multi-modal properties of MERCuRY system in this study, which tests the hypothesis that negation detection may have a greater impact on the quality of patient cohort retrieval than polarity detection methods. In Q1, when negation recognition is performed, all the terms identified as part of inclusion criteria received a polarity attribute of “positive”, whereas all the terms from expressions of exclusion criteria received a “negative” polarity attribute. Alternatively, when polarity recognition is performed, only the medical concepts from the inclusion criteria (e.g. “seizures”, “EEG” or “TIRDA”) receive a positive polarity, and similarly, only the concepts from the exclusion criteria receive a negative polarity (e.g. “sharps”, “spikes”, “electrographic seizures”). In this way, when searching for patient cohorts, both the inclusion and the exclusion criteria are taken into account based on the polarity attributes of the query terms or medical concepts, respectively, depending on whether negation detection or polarity recognition is considered. Moreover, the relevance model considers only patients having EEG reports with medical concepts having (a) “positive” polarity when those concepts/terms correspond to the inclusion criteria; and either (b) “negative” polarity for the concepts/terms corresponding to the exclusion criteria, or (c) concept/terms corresponding to the exclusion criteria are not mentioned in the EEG reports of the patients. An example of a relevant patient for the cohort described by Q1 is identified in the excerpts of Example 1, where expressions corresponding to inclusion criteria are bolded, and are inferred to have positive polarity, whereas expressions corresponding to the exclusion criteria are identified with negative polarity, and are underlined:
EXAMPLE 1: [This is a 44-year-old man with 2 seizures ….Abnormal EEG due to intermittent left anterior temporal slowing as well as intermittent left temporal intermittent rhythmic delta activity, (TIRDA)….No sharps, spikes, and electrographic seizures were seen in this recording.]
However, we have found that the incorrect identification of the polarity of concepts both in the queries and in the EEG reports leads often to false positive or false negative results. Example 2 illustrates a false positive result for Q1, in which one of the inclusion criteria (“history of seizures”) is incorrectly inferred to be met, while one of the exclusion criteria (“spikes”) is incorrectly inferred to be met. This is because although the concept “seizures” should receive a positive polarity, the expression “history of seizures” from Q1 is not recognized, and thus shouldn’t be inferred to meet the inclusion criterion. Moreover, the concept “spikes” is incorrectly recognized as having negative polarity, even if it is preceded by the contrastive “but”. As in the previous example, terms from the inclusion criteria are bolded, whereas terms corresponding to the exclusion criteria are underlined, when identified with negative polarity:
EXAMPLE 2: [A 41-year-old woman with a history of left MCA stroke with dilation of the left lateral ventricle admitted to evaluate for seizures….There is TIRDA noted in the left anterior temporal region…. no sharps but spikes on the left.]
As the query Q1 requires all inclusion and exclusion criteria to be met, example 2 illustrates a false positive relevant patient retrieved when polarity detection on medical concepts is used. Although the polarity inference of medical concepts has been considered in past i2b2 Challenges5,6 we found that the accuracy of 85% that was reported in previous work7 might not produce optimal results when included in a patient cohort retrieval. Therefore, we contemplated using negation detection methods, that operate on all the terms of the query and EEG reports and included it in the MERCuRY system, to enable comparisons of the patient cohorts.
Background
The ability to automatically identify patient cohorts satisfying a wide range of criteria – including clinical, demographic, and social information – has applications in numerous use cases, as pointed out in by Shivade et al.8 including (a) clinical trial recruitment; (b) outcome prediction; and (c) survival analysis. Although the identification of patient cohorts is a complex task, many systems aiming to resolve it automatically have used statistical techniques or machine learning methods taking advantage of natural language processing (NLP) of the clinical documents8. However, these systems cannot rank the identified patients based on the relevance of the patient to the cohort criteria. The MERCuRY system, presented in Goodwin & Harabagiu (2016)4 considers the problem of patient cohort identification as an Information Retrieval (IR) problem, adopting the same framework as the one used in the Medical Records track (TRECMed)9,10 of the annual Text REtrieval Conference (TREC) hosted by the National Institute for Standards and Technology (NIST). When patient cohort identification systems are presented with a query expressing the inclusion/exclusion criteria for a desired patient cohort, a ranked list of patients representing the cohort is produced where each patient may be associated with multiple medical records. Thus, identifying a ranked list of patients is equivalent to producing a ranked list of sets of medical records, each pertaining to a patient belonging to the cohort.
Detecting negation in biomedical texts, as reported in Morante et al.11 entails the automatic identification of negation cues (e.g. “never”, “not”, “no longer”) as well as the scope of these cues. When medical concepts or terms are identified within the scope of a negation cue, their polarity is inferred as “negative”. Early approaches to negation detection in clinical texts, such as NegEx12, relied on regular expressions and hand-crafted rules to capture lexical knowledge indicative of negated findings. Syntactic information in the form of (a) dependency parses, as reported in Sohn et al.13; (b) token chunks, as reported in Morante et al.11; or (c) predicate argument structures, as reported in Prabhakaran and Boguraev14, was also considered as informative for detecting the scope of negations. The advent of the BioScope corpus15, which has negation annotations in three genres: medical abstracts, scientific papers and clinical records, provided an impetus to multiple approaches using machine learning methods. These methods relied on lexical and syntactic features to inform Conditional Random Fields (CRF)16 classifiers, the k-nearest neighbors algorithm11, or support vector machines17 to detect negation cues and recognize their scope. More recently, several deep learning methods were designed for recognizing the negation scope18,19. A feed-forward neural network (NN) as well as a bi-directional Long-Short Term Memory (LSTM) NN were explored for recognizing the scope of negation18. The advantage of convolutional neural networks (CNNs), which have proven successful in a variety of natural language processing (NLP) tasks, was considered also for identifying the scope of negation in clinical texts19. However, both these methods using deep learning require prior knowledge of the negation cues, thus resolving only partially the problem of negation detection and polarity inference. In this paper we present a negation detection method makes use of a bi-directional LSTM, to jointly identify negation cues as well as their scope. Our method has the advantage of requiring no prior knowledge of the negation cues, enabling its incorporation in the MERCuRY system, thus allowing us to evaluate directly the impact of negation detection of a patient cohort retrieval system operating on EEG reports.
Data
The MERCuRY system was developed to identify patient cohorts from the big EEG data available from the Temple University Hospital (TUH) EEG Corpus20 (over 25,000 sessions and 15,000 patients collected over 12 years). This dataset is unique because, in addition to the raw signal information, physician’s EEG reports are provided for each EEG. Following the American Clinical Neurophysiology Society Guidelines for writing EEG reports21, the EEG reports from the TUH corpus start with a CLINICAL HISTORY of the patient, describing the patient’s age, gender, and relevant medical conditions at the time of the recording (e.g., “after cardiac arrest”) followed by a list of the medications which may influence the EEG. The INTRODUCTION section is the depiction of the techniques used for the EEG (e.g. “digital video EEG”, “using standard 10-20 system of electrode placement with 1 channel of EKG”), as well as the patient’s conditions prevalent at the time of the recording (e.g., fasting, sleep deprivation) and level of consciousness (e.g. “comatose”). The DESCRIPTION section is the mandatory part of the EEG report, and it provides a description of any notable epileptiform activity (e.g. “sharp wave”), patterns (e.g. “burst suppression pattern”) and events (“very quick jerks of the head”). In the IMPRESSION section, the physician states whether the EEG readings are normal or abnormal. If abnormal, then the contributing epileptiform phenomena are listed. The final section of the EEG report, the CLINICAL CORRELATIONS section explains what the EEG findings mean in terms of clinical interpretation22 (e.g. “very worrisome prognostic features”). Each EEG report in the TUH corpus is associated with the EEG signal recording it interprets.
Patient cohort retrieval from the TUH EEG corpus was performed based on 100 queries generated by 8 neurologists, selected when at least 3 of the 4 neurologists that reviewed each query decided that it is clinical relevant in their practice. Table 1 illustrates some of the queries we have evaluated, in addition to Q1 which was discussed previously.
Table 1:
Example queries used to evaluate the impact of negation on patient cohort retrieval
| Patient Cohort Description (Queries) |
|---|
| Patients under 18 years old with absence seizures |
| Patients without normal sleep architecture |
| Patients with disorganized and slow background, a clinical indication of altered mental status, and no epileptiform activity |
| Patients with EEG showing temporal slowing without epileptiform discharges |
| Patients with head rocking and no epileptiform activity |
| Patients with brain tumor and sharp waves without spike/polyspike and wave or spikes |
| Focal slowing and patients with migraine headache without a history of seizure or epilepsy |
Moreover, we considered the BioScope annotated corpus15 to test our deep learning method for detecting negation and compare it with previous methods. The BioScope corpus contains annotations for hedges and negations in sentences from the biomedical domain. For our evaluations, we were interested only in the negation annotations. Each negation annotation consists of (a) a cue and (b) its corresponding scope. Examples of negations cues which were annotated are “without”, “not”, and “no”. The scope of negation represents the contiguous text span associated with a negation cue, as shown in the example “Mildly hyperinflated lungs ([without]cue focal opacity)scope”. The BioScope corpus contains sentences from three sub-genres: (1) abstracts of biological papers; (2) full scientific papers, and (3) clinical radiology reports. Following the example of Qian et al19, we used only the abstracts of biological papers from the Bioscope corpus, which represents the largest sub-corpus of the Bioscope resource.
Methods
MERCuRY4 is a multi-modal patient cohort discovery system which allows neurologists to inspect the EEG records as well as the EEG signal recordings of patients deemed relevant to a query expressing inclusion and exclusion criteria through natural language. In order of evaluate the impact of negation detection on patient cohort retrieval operating on the TUH EEG corpus, we have considered only the EEG records and 100 queries that were generated by neurologists to express criteria for patient cohorts of clinical relevance in their practice. Many of the 25,000 EEG records from the TUH EEG corpus document findings of negative polarity, e.g. “no sharps or spikes” that need to be detected automatically. In addition, 17 of the 100 queries (hence 17%) exhibit negation that needs to be automatically detected, e.g. “without normal sleep architecture”. We considered a negation detection method that takes advantage of deep learning, relying on bi-directional LSTMs. We also considered a polarity detection methodology which we designed previously to annotate EEG reports with medical concepts and their attributes using a using a neural active learning framework, called the Multi-task Active Deep Learning (MTADL) and recently reported in Maldonado et al. 201723. Finally, we also considered several baselines for negation detection and select the best performing one for inclusion in MERCuRY. Formally, a negation detection methodology considers that in a natural language sentence, negation detection consists of (1) the identification of negation cues, e.g. “by no means”, “no longer”, “without”, “absence”; and (2) the recognition of the scope of each negation cue. The scope is defined by the words from the sentence which represent the negation instance, thus acquiring negative polarity. The MTDAL method does not discover negation cues or their scope, it simply assigns polarity values to medical concepts.
Negation Detection Method: Our deep learning architecture is based on a framework that casts the problem of negation detection as a sequence labeling problem. Each word from a sentence from the EEG reports or from a query is assigned a label l∈ {C, I, O} such that if it receives the label C, it means that the word belongs to a negation cue, if it receives a label O it means that the word is within the scope of the negation cues (i.e. words labeled with C), and if it receives a label of O it means that the word is outside of the scope of the negation cue. For example, the word sequence of Q1=[“History” “of” “seizures” “and” “EEG” “with” “TIRDA” “without” sharps” “,” “spikes” “,” “or” “electrographic” “seizures”] is labels with the sequence [O, O, O, O, O, O, O, C, I, I, I, I, I, I, I] because of the words preceding “without” are outside the scope of the negation of this cue, the word “without” is labeled as the only negation cue and all words following it are within its scope.
All the queries and the EEG reports were preprocessed before applying this method. First, we used the OpenNLP* sentence splitter on the EEG reports. Then, both on the queries and the EEG reports we applied the GENIA tokenizer24. This enabled us to use the Gensim implementation25 of the Word2vec26 model to generate embeddings for each token from every query and each EEG reports. As illustrated in Figure 1, the embeddings are provided as input to the bi-directional LSTM architecture which considers (1) the embedding representation of each token, ei, as well as (2) the embedding representation of tokens from both directions in the sentence. There are the two main challenges involved in negation detection: (1) a sentence or query can contain multiple negation cues; and (2) the negation scope of cues can extend in both directions. The most promising deep architecture for addressing these challenges is provided by bi-directional LSTMs. Bi-LSTM are sequential models that operate both in forward and backwards fashion; the backward pass is especially important in the case of negation scope detection, given that a token within the scope of a negation cue can appear in a string before the cue and it is therefore important that we see the latter first to classify the former. The forward pass is equally important, because of a token is within the scope of a negation cue, the next token may be in the scope as well.
Figure 1:
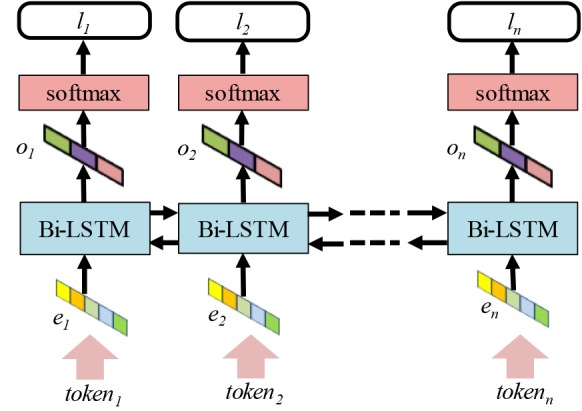
Bi-LSTM architecture for negation detection.
The output from the Bi-LSTM cells, oi, is a vector representing token ti and the rest of the tokens in the sentence. To determine the IOC label for token ti, the vector oi is passed through a softmax layer which produces a probability distribution over all IOC labels. This is accomplished by computing a 3-dimensional vector of probabilities, qi, such that qi1 is the probability of label I, qi2 is the probability of label O, and qi3 is the probability of label C. The predicted IOC label is then chosen as the label with the highest probability . Clearly, all tokens within the scope of a negation cue were considered to have negative polarity.
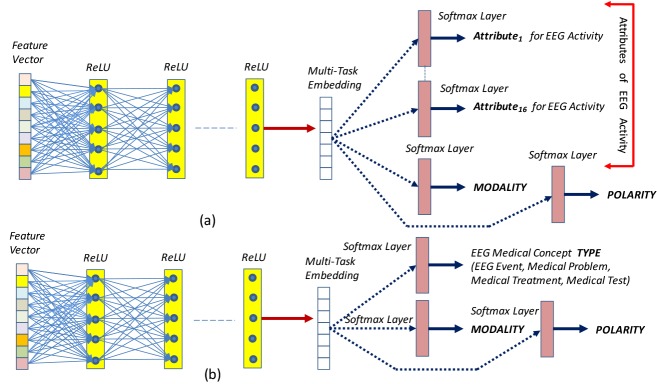
Polarity Detection Method: The polarity detection method was originally designed as a component of the Multitask Active Deep Learning (MTADL) paradigm23 aiming to perform concurrently multiple annotation tasks, corresponding to the automatic identification in EEG reports of (1) EEG activities and their attributes, (2) EEG events, (3) medical problems, (4) medical treatments and (5) medical tests mentioned in the narratives of the reports, along with their inferred forms of modality and polarity. When we considered the recognition of the modality and polarity, this method took advantage of the definitions used in the 2012 i2b2 challenge5 on evaluating temporal relations in medical text. In that challenge, modality was used to capture whether a medical event discerned from a medical record actually happens, is merely proposed, mentioned as conditional, or described as possible. In addition, each concept can have either a “positive” or a “negative” polarity, depending on absence or presence of negation of its findings. However, polarity was one of the possible 18 attributes that were discovered automatically, as illustrated in Figure 2(a). By leveraging the power of deep learning, this method used one multi-purpose, high-dimensional vector representation of medical concepts, or embedding, to determine each attribute simultaneously with the same deep learning network. Because EEG reports contain many mentions of EEG activities and EEG events, along with other medical concepts, such as treatments, medical problems, this method identified the polarity of EEG activities with one Deep Rectified Linear Network (DRLN) architecture, which also recognizes the other 17 attributes of EEG activities, whereas the polarity of the EEG events, medical treatments and tests is recognized with a second architecture, as illustrated in Figure 2(b). The 16 attributes of the EEG activities are identified in the EEG reports by feeding a multi-task embedding produced by passing the concept features through five fully connected Rectified Linear Unit (ReLU) layers. Then, a softmax layer learns the predicted value for attribute j of medical concept a. Let be the vector of probabilities produced by the softmax layer for attribute j of medical concept a. Note that polarity is one of these attributes. Each element of is defined as: with , the predicted attribute value .
Figure 2:
Deep Learning Architectures for Automatic Recognition of (a) attributes of EEG activities; (b) type for all the other medical concepts expressed in EEG reports; as well as the modality and polarity for all concepts.
Baseline Negation Detection detection methods: We also considered three baseline negation detectio methods. The first baseline negation detection method, Lingscope, reported by Agarwal et al16 detects the scope of negation. Lingscope trains a conditional random field (CRF) on the BioScope corpus. Its implementation is publicly available†. The second baseline also uses a CRF (“CRF baseline”) which we trained using the Scikit-learn27 CRF implementation with the same features employed in Lingscope. The third baseline method uses a bi-directional recurrent neural network (RNN) implemented using tensorflow28.
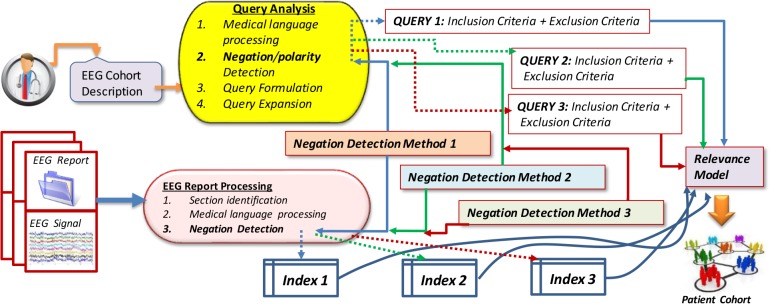
Incorporating Negation Detection and Polarity Detection Methods into MERCuRY: As originally reported in Goodwin & Harabagiu (2016)4, the MERCuRY system considered only the polarity of medical concepts, using it in (a) the query processing, (b) the generation of the index; and (c) the relevance models. The query analysis consists of (1) medical language processing, having the role of identifying medical concepts and terms used in the query; (2) negation or polarity detection, having the role of recognizing which concepts or terms correspond to inclusion criteria and which of them correspond to exclusion criteria; (3) query formulation, that generates a query that informs the relevance model; and (4) query expansion, which enhances the query, to produce improved results. When the negation detection methods are incorporated, we considered that not only the medical concepts, but all the terms from the scope of a negation receive a negative polarity, whereas all the other terms receive a positive polarity. If different negation/polarity detection methods are used, different queries are generated and expanded. For example, when processing the EEG cohort description Q1, the Lingscope method has identified the negation cue “without” and only the term “sharps” under its scope, leading to the consideration that the terms “history” “seizures”, “EEG”, “TIRDA”, “spikes”, and “electrographic” characterize the inclusion criteria, whereas the exclusion criteria are represented in the query only by the term “sharps”. The polarity detection method from MTADL similarly identified “sharps” as having negative polarity, but additionally identified the term “spikes” as having negative polarity, thus adding it correctly to the exclusion criteria representation. Finally, only the negation detection method recognized using bi-LSTMs correctly identified all the exclusion criteria. The decision of which negation method to incorporate in MERCuRY was based on an initial evaluation of several negation methods performed on a sub-set of EEG reports. We found that the bi-LSTM-based method and the Lingscope baseline performed the best, thus incorporated only these two negation detection methods in MERCuRY. In addition, we incorporated the polarity detection method from the MTDAL framework and used it in MERCuRY. Hence, as illustrated in Figure 3, the relevance model considered three different queries, generated by incorporating negation/polarity, when searching for the patient cohort against the indexed information.
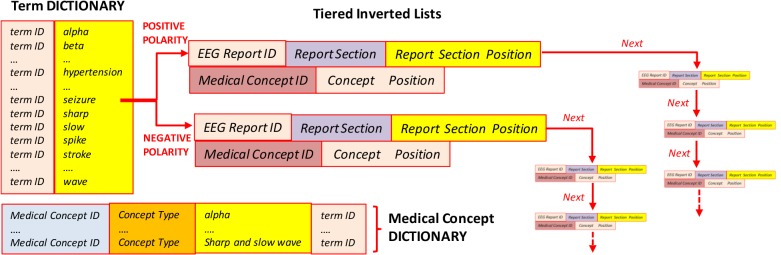
In order to produce the index of the patient cohort retrieval system, the EEG reports are also processed to (1) identify each section; (2) recognize both medical concepts and terms that need to be indexed; and (3) detect negation or polarity to inform the production of the posting files of each medical concept. As with the query processing, each negation/polarity detection method may produce different results, thus for each of them a separate index has been created. It is important to note that negation/polarity detection method i has informed the production of the query-i and index-i. This allows the relevance model to retrieve a patient cohort-i for each of the negation detection methods that we experimented with, as shown in Figure 3. Each of the three indexes illustrated in Figure 3 contains both a term dictionary and a medical concept dictionary, listing all the terms and medical concepts discerned from the EEG report. We considered five medical concept types: (1) EEG activities; (2) EEG events; (3) medical tests; (4) medical treatments (including medications); (5) medical problems. Because medical concepts often are multi-term expressions (e.g. “spike and slow waves”), the medical concept dictionary used term IDs to associate a concept with all terms expressing it (e.g. “spike and slow waves” is associated with the terms “spike”, “slow” and “wave”). When any of the two negation detection methods are used to build an index, they enable the organization of two different tiered inverted lists for each term from the dictionary. When the term is recognized to have positive polarity, because it was not within the scope of any negation cue, the positional tiered inverted list is recording (a) the EEG report ID where the term was found; (b) the section within the reports where the term was observed (c) its position in the section; and if the term was recognized as being part of a medical concept; (d) the medical concept ID and its position within the concept.
Figure 3:
Overview of the usage of negation/polarity detection methods in a patient cohort retrieval system.
The inverted list provides all such information for any time when the term is recognized with positive polarity in any EEG report, as illustrated in Figure 4. If a term is identified within the scope of a negation cue by the negation detection methods, it shall be linked to a similar positional tiered inverted list, which provides all the information for any time the term was recognized with negative polarity throughout the collection of EEG reports. It is important to note that the same term may have positive polarity sometimes and negative polarity other times: the role of the polarity-informed inverted lists is to organize information about the term by considering the value of its polarity. In contrast, when the polarity detection method is used, only the terms from the concept expression receive a positive/negative polarity, and thus only those terms have associated tiered lists.
Figure 4:
Index with (a) term dictionary and medical concept dictionary and (b) polarity-informed tiered positional inverted lists.
Finally, we used the same relevance models reported in Goodwin and Harabagiu, 20164. The relevance model assigns a score to an individual EEG report based on the BM25F ranking function29. BM25F measures the relevance of an EEG report based on the frequency of mentions of each inclusion criterion and the absence of each exclusion criterion. Moreover, BM25F is capable of adjusting the score for each criterion based on the tiers in the posting list: that is, a criterion mention is scored according to both the polarity and the section in the document.
Evaluation
We first evaluated the quality of the negation detection methods to select the best-performing methods that should be incorporated in the patient cohort retrieval system and to evaluate their impact on the results of the MERCuRY4 system. We considered (1) a set of EEG reports and (2) a set of patient cohort retrieval queries, annotated with negation cues and their spans. We also used the BioScope Abstracts corpus for evaluation of all these methods
Evaluation of Negation Detection Methods: As evaluation metrics we have used accuracy, defined as (#true positive+#true negatives) /(#tokens), precision, defined as (# true positives) / (# true positives+# false positives), recall defined as (#true positives) /(#true positives + #false negatives), and F1 score, defined as 2 × (precision × recall) / (precision + recall).
Evaluation on EEG reports: We evaluated the performance of each of the negation detection methods by performing 10-fold cross validation (8 folds for training, 1 fold validation, and 1 held-out fold as a test set) on the 169 EEG reports that we have annotated with (a) negation cues and (b) their scopes. The results shown in Table 2 indicate that the neural method using Bi-LSTMs demonstrated notably higher performance compared to the baselines. We tuned the hyperparameters of our bi-directional LSTM, bi-directional RNN, and CRF baseline by optimizing for the F1 score of the validation fold during cross validation.
Table 2:
Evaluation of negation scope detection on EEG reports
| Negation Method | Precision | Recall | F1 | Accuracy |
|---|---|---|---|---|
| CRF baseline | 83.72% | 72.23% | 77.40% | 97.59% |
| Bi-RNN baseline | 83.62% | 84.55% | 83.94% | 98.33% |
| Lingscope baseline | 80.57% | 80.06% | 80.04% | 97.68% |
| Neural Bi-LSTM | 85.27% | 86.31% | 85.61% | 98.34% |
Evaluation on queries deemed clinically relevant for retrieval from the EEG corpus: We also evaluated the performance of the negation detection methods on 100 annotated queries. Since the number of queries is too small to perform cross validation, we trained the neural models and the CRF baseline using the annotated EEG reports and then evaluated the trained models on all 100 queries. Table 3 shows the results. It is interesting to note that the accuracy of the Lingscope baseline is not notably different from the results of the neural Bi-LSTM method, probably because they were both applied on a very small set of queries. The precision of Lingscope however was higher, but the neural Bi-LSTM neural method produced better F1 score and recall.
Table 3:
Evaluation of negation scope detection on EEG queries
| Negation Method | Precision | Recall | F1 | Accuracy |
|---|---|---|---|---|
| CRF baseline | 69.13% | 68.87% | 68.88% | 94.91% |
| Bi-RNN baseline | 87.37% | 58.50% | 69.47% | 95.87% |
| Lingscope baseline | 95.38% | 77.50% | 85.51% | 97.82% |
| Neural Bi-LSTM | 89.33% | 83.75% | 86.45% | 97.83% |
Evaluation on the BioScope abstracts: Following the example of Qian et al19, we perform 10-fold cross validation on the BioScope Abstracts corpus. We compared the neural Bi-LSTM method against the reported results for Qian et al19 and Lingscope as shown in Table 4. Cleary, the of the neural Bi-LSTM method achieves the best results.
Table 4:
Evaluation of negation scope detection on BioScope Abstract
| Negation Method | Precision | Recall | F1 | Accuracy |
|---|---|---|---|---|
| Lingscope baseline | 84.74% | 84.07% | 84.37% | 94.61% |
| Bi-RNN baseline | 72.90% | 69.92% | 71.22% | 96.01% |
| Neural Bi-LSTM | 88.72% | 89.02% | 88.85% | 98.43% |
| Qian et al | 89.49% | 90.54% | 89.91% | Not Reported |
Evaluation of Patient Cohorts: We evaluated the impact of incorporating the Bi-LSTM and the Lingscope negation methods as well as the MTADL polarity detection method in the MERCuRY4 patient cohort retrieval system. To do this, we retrieved the ten most relevant EEG reports of patients for each query. Each patient was assessed to determine if they satisfied all inclusion and exclusion criteria by at least one of five researchers which consisted of a combination of neurologists and student researchers. The inter-judge agreement was 82%. To measure the impact of each negation method, we adopted standard measures for information retrieval effectiveness, where patients labeled as belonging to the cohort were considered relevant to the cohort query, and patients labelled as not belonging to the cohort were considered as non-relevant to the cohort query. Because our relevance assessments consider only a sample of patients retrieved for each query, we adopted the Mean Average Precision (MAP) as one of our measures. The MAP metric provides a single measurement of the quality of patients retrieved at each rank for a particular query. Formally, MAP is the mean of the average precision of each query. The average precision for a query is the average of the precision computed at each rank which has a relevant document. Additionally, we adopted the Mean Reciprocal Rank (MRR) which measures the average reciprocal rank of the first relevant patient. For example, if the first relevant patient is ranked 2nd on the MRR will be 1/2 = 0.5. If it is ranked third, the MRR will be 1/3 = 0.33. Lastly, we compute the “Precision at 10” metric (P@10), which measures the ratio of relevant patients retrieved in the first 10 ranks. Although less statistically meaningful, the precision is the easiest to interpret in terms of clinical application in that a 50% Precision at 10 indicates that half of the patients returned above rank 10 completely satisfy all criteria of the given cohort. Additionally, we report the improvement achieved when using the bi-LSTM neural method against the alternative negation/polarity detection methods. Table 5 lists the results and shows that the impact of the bi-LSTM based negation method is superior not only to Lingscope, but especially to the polarity detection method from MTDAL. To note that these results were obtained when the entire set of 100 queries were evaluated in the MERCuRY patient cohort retrieval system. We were also interested evaluate the impact of negation detection methods on patient cohorts when only queries that contained exclusion criteria were used. Table 6 lists the evaluation results for the patient cohorts obtained for 17 queries containing exclusion criteria. As shown, the neural Bi-LSTM method for negation detection method led to the best results for all three evaluation metrics. We also note that, as expected, the percent improvement was higher in this case than when all 100 queries were tried.
Table 5:
Quality of patient cohorts (All queries)
| Negation Method | MAP | MRR | P@10 |
|---|---|---|---|
| MTADL | 52.00% | 59.38% | 42.70% |
| Lingscope baseline | 55.40% | 62.61% | 45.70% |
| Neural Bi-LSTM | 56.89% | 64.36% | 46.40% |
| improvement over MTADL | 9.40% | 8.38% | 8.66% |
| improvement over Lingscope | 2.69% | 2.80% | 1.53% |
Table 6:
Quality of patient cohorts (Negation queries)
| Negation Method | MAP | MRR | P@10 |
|---|---|---|---|
| MTADL | 25.30% | 41.95% | 26.47% |
| Lingscope baseline | 39.82% | 55.78% | 38.82% |
| Neural Bi-LSTM | 43.00% | 60.99% | 42.35% |
| improvement over MTADL | 69.96% | 45.38% | 59.99% |
| improvement over Lingscope | 7.99% | 9.34% | 9.09% |
Discussion
In general, the negation detection method using bi-directional LSTMs performed well on EEG reports as indicated by the results reported in Table 2, especially when compared with the other three baselines. However, it also produced several errors. The first type of error was observed when automatically identifying negation cues. Sometimes, our Bi-LSTM method was not able to recognize some of the negation cues, especially those rarely occurring in the EEG corpus. For example, one negation cue that is seen rarely is “free” as in appears in the sentence “the patient has been seizure free for the past 6 months”. When a negation cue is not identified, as expected, its scope is also not recognized, which leads to incorrectly assigning positive polarity to all words that should be in its scope. In this way, “seizure” receives a positive polarity value, which may lead to the incorrect retrieval of a patient, when the exclusion criteria considered patients with no seizures. Conversely, some negation cues are observed quite frequently, like the cue “not”, leading our neural negation detection method to identify negative polarity when in fact it was not meant by the neurologist that wrote the EEG report. For example, in the sentence “Normal EEG does not exclude a diagnosis of epilepsy”, the words “diagnosis of epilepsy” receives a negative polarity, because it is in the negation scope of the cue “not”. But, in fact, the neurologist is not asserting such a fact. A hypothesis is made, asserting that the patient might have epilepsy since epilepsy cannot be ruled out by a normal EEG. Therefore, an automatic assertion detection method would select the value of hypothetical for the diagnosis, instead of absent, as inferred by a negative polarity. This suggests that performance of our negation detection method could be improved by incorporating information related to hedging, or assertion recognition.
A second source of errors generated by the bi-LSTM-based negation detection when operating on the EEG reports was observed in the quality of the spans of negation cues, even when cues where identified correctly. More specifically, the negation spans did not cover all the words correctly, either missing some words or expanding the span with words that should not be negated. For example, when the detected negation scope did not extend far enough, some words were left to acquire a positive polarity value. In the sentence “abnormal discharges: none”, the negation scope is underlined, incorrectly missing the word “abnormal”. When the negation detection methods operated on the query set, the results in Table 3 by themselves do not indicate any system as being clearly superior to all the others. This is due to the fact that the Lingscope system obtained results with superior precision, while the negation method using the bi-directional LSTMs produced results with better recall.
When evaluating the impact of the two negation detection methods and the polarity detection method incorporated in MERCuRY on the results of patient cohort retrieval, however, it is clear that the negation method employing the bi-directional LSTMs outperformed Lingscope in our experiments as demonstrated in Table 6. This difference in performance was the result of the Lingscope system producing results with inferior recall. For example, for the query Q1 “History of seizures and EEG with TIRDA without sharps, spikes, or electrographic seizures” Lingscope only identifies that “sharps” should have negative polarity even though the negation scope should extend all the way to the end of the query. In contrast, the bi-directional LSTM correctly identified all terms to the right of “without” as having negative polarity. This led to retrieving patients when using the Lingscope system that had “spikes” and “electrographic seizures”, which were exclusion criteria expressed in the query. Consequently, for many queries, when the Lingscope system was used for detecting negation, many irrelevant patients were retrieved. Finally, the notable increase in patient cohort quality when using the negation method based on bi-directional LSTMs indicates that this method is able to better identify inclusion and exclusion criteria in the queries and especially in the EEG reports. It is also important to note that this method has a simpler neural architecture than the MATDL method, which focused on the detection of polarity only for medical concepts used in EEG reports. However, it is obvious that there are still avenues for improvement which include detecting negation scopes more accurately, accounting for rare or frequent cues, and determining terms from the EEG reports which are not informative about the patients’ condition.
Conclusion
In this paper, we have shown that a bi-directional LSTM model for negation detection is a far better choice than a polarity detection method operating only on medical concepts from the query and the EEG reports when retrieving patient cohorts. Although this neural negation detection method uses a simpler architecture that the one used in the neural polarity recognition, reported in Maldonado et a. 201723, it provided superior results, which indicate improved patient cohort retrieval.
Acknowledgements
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under award number 1U01HG008468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
References
- 1.Smith SJM. EEG in the diagnosis, classification, and management of patients with epilepsy. J Neurol Neurosurg Psychiatry. 2005;76(suppl 2):ii2–7. doi: 10.1136/jnnp.2005.069245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatum WO., IV . Demos Medical Publishing; 2014. Handbook of EEG interpretation. [Google Scholar]
- 3.Beniczky S, Hirsch LJ, Kaplan PW, Pressler R, Bauer G, Aurlien H, et al. Unified EEG terminology and criteria for nonconvulsive status epilepticus. Epilepsia. 2013;54(s6):28–9. doi: 10.1111/epi.12270. [DOI] [PubMed] [Google Scholar]
- 4.Goodwin TR, Harabagiu SM. 2016. Multimodal Patient Cohort Identification from EEG Report and Signal Data. In: AMIA Annual Symposium Proceedings. American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- 5.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc JAMIA. 2013 Sep;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzuner O, South B, Shen S, Duvall S. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011 Jun 16;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts K, Rink B, Harabagiu SM. A flexible framework for recognizing events, temporal expressions, and temporal relations in clinical text. Journal of the American Medical Informatics Association. 2013 May 18;20(5):867–75. doi: 10.1136/amiajnl-2013-001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. JAMIA. 2014;21(2):221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voorhees EM, Hersh WR. 2012. Overview of the TREC 2012 Medical Records Track. In: TREC [Internet] [Google Scholar]
- 10.Voorhees EM. ACM; 2013. The trec medical records track. In: ACM-BCB; p. 239. [Google Scholar]
- 11.Morante R, Liekens A, Daelemans W. Learning the scope of negation in biomedical texts. Proceedings of the Conference on Empirical Methods in Natural Language Processing - EMNLP 08; 2008. pp. 265–274. [Google Scholar]
- 12.Chapman W, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 13.Sohn S, Wu S, Chute CG. 2012. Dependency Parser-based Negation Detection in Clinical Narratives. AMIA Summits on Translational Science Proceedings; pp. 1–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhakaran V, Boguraev B. Learning structures of negations from flat annotations. Conference on Lexical and Computational Semantics (*SEM 2015); 2015. pp. 71–81. [Google Scholar]
- 15.Vincze V, Szarvas G, Farkas R, Móra G, Csirik J. The BioScope corpus: biomedical texts annotated for uncertainty, negation and their scopes. BMC Bioinformatics. 2008;9(Suppl 11):S9. doi: 10.1186/1471-2105-9-S11-S9. doi:10.1186/1471-2105-9-S11-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal S, Yu H. Biomedical negation scope detection with conditional random fields. Journal of the American Medical Informatics Association : JAMIA. 2010;17(6):696–701. doi: 10.1136/jamia.2010.003228. doi:10.1136/jamia.2010.003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujikawa K, Seki K, Uehara K. 2012. Oct 29, A hybrid approach to finding negated and uncertain expressions in biomedical documents. Proceedings of the 2nd international workshop on Managing interoperability and compleXity in health systems - MIXHS 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fancellu F, Lopez A, Webber B. Neural networks for negation scope detection. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers); 2016. pp. 495–504. [Google Scholar]
- 19.Qian Z, Li P, Zhu Q, Zhou G, Luo Z, Luo W. Speculation and negation scope detection via convolutional neural networks. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing; 2016. pp. 815–825. [Google Scholar]
- 20.Harati A, Choi S-M, Tabrizi M, Obeid I, Picone J, Jacobson MP. IEEE; 2013 2013. The Temple University Hospital EEG Corpus. In: GlobalSIP. [Google Scholar]
- 21.Anonymous. Guideline 7: Guidelines for writing EEG reports. Am Electroencephalogr Soc. 2006;23(2):118. doi: 10.1097/00004691-200604000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan PW, Benbadis SR. 2013. Jan 1, How to write an EEG report: dos and don’ts. Neurology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldonado R, Goodwin TR, Harabagiu SM. 2017. Active Deep Learning-Based Annotation of Electroencephalography Reports for Cohort Identification. AMIA Summits on Translational Science Proceedings; pp. 229–238. [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuruoka Y, Tateishi Y, Kim J-D, Ohta T, McNaught J, Ananiadou S, et al. Springer; 2005. Developing a robust part-of-speech tagger for biomedical text. In: Panhellenic Conference on Informatics [Internet] [cited 2016 Sep 22]. p. 382–92. Available from: http://link.springer.com/chapter/10.1007/11573036_36. [Google Scholar]
- 25.Rehurek R, Sojka P. 2010. Software framework for topic modelling with large corpora. In Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks. [Google Scholar]
- 26.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. 2013. Distributed representations of words and phrases and their compositionality. In Advances in neural information processing systems; pp. 3111–3119. [Google Scholar]
- 27.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J. Scikit-learn: Machine learning in Python. Journal of machine learning research. 2011 Oct;12:2825–30. [Google Scholar]
- 28.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M. TensorFlow: A System for Large-Scale Machine Learning. In OSDI. 2016 Nov 2;Vol. 16:265–283. [Google Scholar]
- 29.Zaragoza H, Craswell N, Taylor MJ, Saria S, Robertson SE. Citeseer; 2004. Microsoft Cambridge at TREC 13: web and hard tracks. In: TREC; pp. 1–1. [Google Scholar]