Abstract
The increasing adoption of electronic health record (EHR) systems and proliferation of clinical data offer unprecedented opportunities for cohort identification to accelerate patient recruitment. However, the effort required to translate trial eligibility criteria to the correct cohort identification queries for clinical investigators is substantial, at least in part due to the lack of clear definitions in both the free-text eligibility criteria and the data models used to structure the available data elements in target patient databases. We propose to adopt an ontology-driven data access approach that generates formal representations of the connections between the entities in eligibility criteria and the available data elements to (1) narrow the semantic gap between researchers’ cohort identification needs and the underlying database nuances, and (2) render the eligibility criteria computable. We implemented our approach based on an analysis of the eligibility criteria from 77 Hepatitis C trials. We found that 4 major types of data manipulation queries and 4 temporal patterns covered all eligibility criteria that were computable. We built a prototype system that helps researchers write computable eligibility criteria and execute them against clinical data in real-time to find potential trial cohorts.
Introduction
Low participation rates in clinical trials remain as a persistent barrier. To facilitate clinical trial recruitment planning, it is important to make eligibility criteria computable for automatic cohort identification. The increasing adoption of electronic health record (EHR) systems and proliferation of clinical data offer unprecedented opportunities for cohort identification to accelerate patient recruitment. Electronic screening for clinical studies has been shown to improve the efficiency of study recruitment1,2 and further ensure that all patients have the opportunity to be evaluated for participation3. The last few years have witnessed an increasing number of clinical research networks focused on building large collections of data from EHR and administrative claims to provide cohort discovery services. Two notable examples are the National Patient-Centered Clinical Research Network (PCORnet),4 funded by the Patient-Centered Outcomes Research Institute (PCORI), and the National Center for Advancing Translational Sciences (NCATS)’s Clinical and Translational Science Awards Accrual to Clinical Trials (CTSA ACT) initiative. In addition, a number of other national collaborative efforts are building tools, algorithms, and data models to support cohort discovery. For example, (1) the Informatics for Integrating Biology & the Bedside (i2b2) suite has a widely used cohort discovery tool;5 (2) the electronic MEdical Records and GEnomics (eMERGE) network funded by the National Human Genome Research Institute (NHGRI) has been building the Phenotype KnowledgeBase (PheKB),6 a repository of computable phenotypes for cohort identification; and (3) the stakeholders of the Observational Health Data Sciences and Informatics (OHDSI) consortium7 are developing open source analytical tools based on the Observational Medical Outcome Partnership (OMOP) Common Data Model (CDM), including CALYPSO–a study population evaluation tool8. However, these existing tools, algorithms, and data models have significant limitations: (1) computable phenotypes in PheKB were developed as specifications, and often without executable software code, requiring significant effort to implement; (2) i2b2 and computable phenotypes were not developed based on a universal CDM making them hard to use across different data sources and aggregate the results in a unified way; and (3) OHDSI tools, although based on the OMOP CDM, only support low-level Structure Query Language (SQL) queries rather than more advanced semantic queries (i.e., SPARQL queries), failing to address the gap between researchers’ mental models of what they want to query and the actual SQL queries they need to construct.
The effort required to translate clinical trial eligibility criteria to the correct cohort identification queries is substantial at least in part due to the lack of clear definitions (i.e., data semantics) of variables, measures, and constructs. For example, if a researcher is interested in identifying “hepatitis C (HCV) patients” in a clinical data warehouse, she will have to: (1) know not only multiple terminology standards but multiple versions of the same standard are used for coding diagnoses (i.e., the International Classification of Diseases, Ninth Revision/Tenth Revision, Clinical Modification, ICD-9/10-CM, and the Systematized Nomenclature of Medicine, Clinical Terms, SNOMED CT); (2) there are multiple ICD-9/10-CM and SNOMED CT codes for HCV (e.g., acute vs. chronic); (3) whether ICD, SNOMED or both standards are used in the target data warehouse, (4) these diagnosis codes have poor sensitivity and specificity for identifying HCV patients (which is why computable phenotypes were needed); (5) there is not a well-known computable phenotype for HCV; (6) decide whether developing a new HCV computable phenotype is necessary for her study; (7) (if not) need to conduct a literature review on what others used to identify HCV patients, and tailor or develop an algorithm to her needs; (8) translate the algorithm to queries in the specific low-level query language used by her data warehouse; and (9) understand the caveats and be able to interpret the results. The gap between researchers’ query intentions (research questions) and the actual constructed queries are widened when data are coming from disparate and heterogeneous sources (e.g., when a researcher wants to identify HCV patients from different EHR systems in a research network). The traditional approach of building federated databases using CDMs and common data elements (CDEs) is not necessarily the solution. Taking a reductionist approach and shoehorning data sources that are heterogeneous in nature into a CDM is not only labor-intensive and error-prone but also ill-fitted. Data integration is a daunting task with unique challenges because data from different sources can be heterogeneous in syntax (e.g., file formats), schema (e.g., data structures), and semantics (e.g., meanings or interpretations).
Thus, we propose to adopt a semantic-driven data access approach that generates a global conceptual representation of available “information” (including data and their relationships), via common controlled vocabularies or “ontologies”, to (1) narrow the semantic gap between researchers’ cohort identification needs and the underlying database nuances, and (2) render the eligibility criteria computable. To facilitate cohort discovery, a number of formal representations9 such as the Eligibility Criteria Representation Language (ECRL), the Eligibility Rule Grammar and Ontology (ERGO)10 and the eligibility criteria extraction and representation (EliXR)11 have been proposed to make eligibility criteria computable. Most existing studies have taken the approach to transform the human-readable free-text eligibility criteria into structured, computable formats, either manually or through developing natural language processing (NLP) tools11-13. Nevertheless, eligibility criteria are usually incomplete sentences, and some of which have compound semantics, making the NLP task challenging. Especially, eligibility criteria with temporal constraints pose technical challenges in formal representations. Even though these formal representations made eligibility criteria computable in theory, the heterogeneity of EHR data and the lacking capacity of NLP tools have limited the adoption of these approaches. Until recently, EliIE14 was developed to automatically structure and formalize free-text eligibility criteria following the OMOP CDM into computable SQL queries. Nevertheless, even though the evaluation results of EliIE were promising, the overall accuracy for its query formalization was still suboptimal (i.e., 0.71).14 Further, automated NLP tools might not be able to capture the subtle semantic gap between the free-text eligibility criteria and researchers’ mental models of the cohort identification queries. To the best of our knowledge, there is not an i2b2-like practical tool that allows researchers to construct computable eligibility criteria (CEC) using a formal representation (i.e., ontology), and enables the translation of the CEC into high-level semantic queries.
In this study, we first analyzed and categorized the eligibility criteria of 77 hepatitis C virus (HCV) clinical trials that we have conducted in the past decade. We then evaluated each inclusion and exclusion criterion against a large clinical data warehouse, the OneFlorida Data Trust—a centralized clinical data repository that contains 15 million patients’ linked EHR and claims data—to determine its computability. Based on the Ontop15 framework (i.e., an ontology-based data access framework over relational databases), we built the Ontology for Computable Eligibility Criteria of HCV (OCEC-HCV), created semantic mappings between the ontologically structured criteria to data elements in the OneFlorida Data Trust, and thus enabled high-level semantic queries (i.e., SPARQL) for cohort identification. To help researchers construct CEC, we created a prototype system with a user-friendly interactive CEC construction interface, capable of running CEC-based cohort identification queries against OneFlorida data in real-time.
Methods
Data sources
Hepatitis C virus clinical trials eligibility criteria. We obtained the free-text eligibility criteria from the ClinicalTrials.gov for 77 hepatitis C virus trials, primarily hepatitis C direct-acting antivirals (DAAs) trials, as part of our Hepatitis C Therapeutic Registry and Research Network (HCV-TARGET) effort16. ClinicalTrials.gov, created and maintained by the National Library of Medicine, is a clinical study registry in the United States. As of March 2018, over 267,636 research studies across all 50 states in the US as well as in 203 countries are registered on ClinicalTrials.gov. Study information is semi-structured in ClinicalTrials.gov: study descriptors such as study phase, intervention type, and locations are stored in structured fields, whereas eligibility criteria are largely free-text.
Real-world patient data from the OneFlorida Data Trust. In clinical trials, the target population represents the patients to whom the results of the clinical trials are intended to be applied. The study population represents the patients being sought as defined in the clinical trial eligibility criteria, which is a subset of the target population. We obtained individual-level patient data from the OneFlorida Data Trust—a centerpiece of the OneFlorida Clinical Research Consortium (OneFlorida CRC), one of the 13 Clinical Data Research Networks (CDRNs) as part of the PCORnet, funded by PCORI—as our target population. The Data Trust integrated various data sources from contributing organizations in the OneFlorida CRC including 9 unique clinical systems that provide care for approximately 48 percent of all Floridians through 4,100 physicians, 914 clinical practices, and 22 hospitals with a catchment area covering all 67 Florida counties. The Data Trust currently contains collated EHR, claims, and other data on a broad-based, unselected population of ~15 million people in Florida. The Data Trust follows the PCORnet CDM v3.1, that contains 15 domains in relational schemas, including patient demographics, enrollment status, death status, cause of death, vital signs (e.g., height, weight, and blood pressure), conditions (i.e., diagnosed and self-reported health conditions and diseases), encounters, diagnoses (i.e., results of diagnostic process and medical coding within healthcare delivery), procedures, prescribing (i.e., provider orders for medications), dispensing (i.e., outpatient pharmacy dispensing, such as filled prescriptions), and lab results. Since all the clinical trials we considered are specific to HCV intervention, we extracted both acute and chronic viral hepatitis C patients from OneFlorida as the target population using ICD-9/10-CM codes (i.e., ICD-9: 070.41, 070.51, 070.44, 070.54, 070.70, 070.71; ICD-10: B17.1, B17.10, B17.11, B18.2). We identified 40,029 HCV patients and extracted their data from 13 of the 15 PCORnet CDM domains, excluding the PCORNET_TRIAL and HARVEST tables. The extracted data were stored in a relational database (i.e., MySQL) without any manipulation.
A computable eligibility criteria framework for cohort identification through ontology-based data access
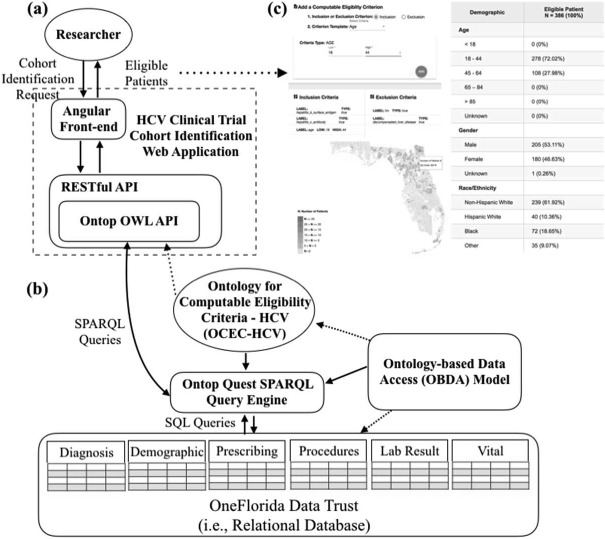
Our approach of making eligibility criteria computable is based on an ontology-based data access (OBDA) framework using Ontop15—a platform to query relational databases as virtual Resource Description Framework (RDF) graphs via SPARQL (SPARQL Protocol and RDF Query Language) queries. We also built a web-based front-end user interface to facilitate CEC construction and visualize the identified cohort. Figure 1 shows an overview of our CEC framework.
Figure 1.
An overview of the ontology-driven computable eligibility criteria framework for cohort identification
Our first step of making ontology-driven CEC was to construct the Ontology for Computable Eligibility Criteria -HCV (OCED-HCV) to represent the eligibility criteria commonly used in HCV trails. We then created semantic mapping axioms to link entities in OCED-HCV with the corresponding data constructs available in OneFlorida according to the PCORnet CDM. In Ontop, these semantic mapping axioms are stored in a OBDA model file (Figure 1.(b)). Given an ontological view of available data and the OBDA model, a CEC can be expressed in SPARQL queries and the Ontop’s Quest SPARQL query engine can translate the high-level SPARQL query over the ontology on-the-fly into a union of sub-queries over the patient data. The sub-queries are subject to the structure of source data and expressed in the low-level native query language of the source (i.e., SQL). The Ontop platform can efficiently aggregate the sub-query results as the response to the SPARQL query. In our current implementation, as shown in Figure 1 .(a), an researcher constructs her CEC (i.e., cohort identification requests) using our web application; and our system translates these CEC into SPARQL queries. Based on the query results (i.e., eligible patients), we present basic demographics of the eligible patients and plot their geographic locations at the zip code-level (Figure 1.(c)).
Constructing an Ontology for Computable Eligibility Criteria - HCV (OCEC-HCV)
Scope. The scope of OCEC-HCV was to provide researchers an efficient way to construct CEC for HCV trials. Further, as OneFlorida was our target database, the OCEC-HCV covered the necessary entities and their relationships to capture the data elements used in HCV trial eligibility criteria connecting to 6 domains (i.e., demographic, vital, diagnosis, procedure, prescribing, and lab result) of patient information in the PCORnet CDM v3.1. We used the Basic Formal Ontology (BFO) as the upper-level ontology, and imported the Time Event Ontology (TEO), Human Disease Ontology (DOID), Clinical Measurement Ontology (CMO), Units of Measurement Ontology, Drug Ontology (DrOn), Relations Ontology (RO), Ontology for Medical Related Social Entities (OMRSE), and Ontology for Biomedical Investigations (OBI) as the foundation of the OCEC-HCV as shown in Table 1.
Table 1.
The reference ontologies used to represent entities in HCV trial criteria based on the PCORnet CDM domain.
| PCORnet CDM domain | Reference ontology |
|---|---|
| Demographic (e.g., age, race, ethnicity) | OBI/OMRSE/OCEC-HCV |
| Vital (e.g., weight, height, body mass index) | CMO |
| Medication (e.g., ribavirin, telaprevir) | DrOn/OCEC-HCV |
| Lab Test (e.g., hepatitis C antibody test, HCV RNA test) | UO/OBI/CMO/OCEC-HCV |
| Diagnosis (e.g., HIV, hepatocellular carcinoma) | DOID |
| Procedure (e.g., liver transplant, antiretroviral therapy) | OCEC-HCV |
Approach. Using the BFO as the top-level ontology, the OCEC-HCV was first developed with a top-down approach, where we started by identifying candidate entities based on the eligibility criteria. A review of existing widely accepted ontologies was conducted, using the National Center for Biomedical Ontology (NCBO) BioPortal17 and Ontobee18, to find relevant entities that can be reused in OCEC-HCV. However, the top-down approach does not consider the data elements available in the patient database. Thus, we also took a bottom-up approach to examine the candidate entities that have been identified in the top-down approach and examined their corresponding data elements in the target patient database—OneFlorida—according to its data model (i.e., PCORnet CDM). The bottom-up approach can help us to determine what new entities and relations are needed to fully represent the eligibility criteria in HCV trials and refine the OCEC-HCV. More importantly, we considered the specific eligibility criteria use cases for cohort identification. For example, in HCV eligibility criteria, researchers often want to exclude patients with decompensated cirrhosis. However, decompensated cirrhosis is defined by the development of jaundice, ascites, variceal hemorrhage, or hepatic encephalopathy, including a group of diagnoses such as ‘chronic hepatitis C with hepatic coma’, ‘unspecified viral hepatitis C with hepatic coma’, and ‘other ascites’19. Thus, in OCEC-HCV, we declared these diagnoses as subclasses of the ‘decompensated cirrhosis’ diagnosis.
Implementation. We used Protégé 520 to construct the OCEC-HCV. We worked collaboratively, finalized the list of entities based on each one’s domain expertise (e.g., HCV clinical trials, clinical data warehouse, eligibility criteria, and ontology development), and defined the relationships among the entities according to the use cases.
Designing the semantic mapping axioms and developing semantic query scenarios
Semantic mapping axioms. In Ontop, an OBDA model with mapping axioms is used to specify how the data elements in the data source are linked to the entities in the ontology. We created three types of mapping axioms for classes, object properties, and data properties in OCEC-HCV. All mapping axioms in an Ontop’s OBDA model follow the same structure, including a mapping ID (i.e., a uniquely identifier for a mapping axiom), a source declaration (i.e., a SQL query to declare and retrieve the required data elements from the source database), and a target declaration (i.e., a RDF triple template to specify the corresponding entities in the ontology). The mapping process required a significant effort because all selected data elements in the source data and the relations among them need to be mapped with the classes and properties in the ontology accordingly. However, the semantic mappings in the OBDA model are highly reusable and extensible. To use our framework in a different disease domain (e.g., colorectal cancer trials), most of the classes and relationships (e.g., definitions of patient’s biological sex) in the ontology and the corresponding mappings (e.g., the connection between the concept biological sex class to the sex variable in PCORnet CDM) can be reused. Nevertheless, further adaption is also needed: 1) to tailor the ontology and corresponding mappings according to new eligibility criterion patterns in the new disease domain, and 2) to create new mappings when connect to a patient data source using different a data model (e.g., OMOP). Because of the flexibility in ontologically structured data, these extensions are straightforward. Further, the mappings between the ontology and a data source (i.e., the OBDA model) only need to be curated once for each CDM. With the OBDA model, we can then query relational source databases as virtual RDF graphs using SPARQL queries through the Ontop platform.
Semantic query scenarios. Through a thorough analysis of HCV eligibility criteria (see details in the Results section), we designed templates of SPARQL queries based on the eligibility criteria use cases. For each representative HCV eligibility criterion, the goal of its corresponding SPARQL queries is to return a list of patient identifiers that meet the criterion (whether it is an inclusion or exclusion criterion) from the source database. Therefore, based on the actual HCV trial eligibility criteria use cases, we categorized the needed SPARQL queries into four groups: (1) queries that examine patient characteristic variables directly without requiring any manipulation of the source data elements (e.g., criteria based on patient gender); (2) queries that need to handle the logic of how to process the raw data elements to produce the desired information (e.g., calculating a patient age from her date of birth); (3) queries that can leverage a semantic reasoner based on the knowledge encoded in the ontology (e.g., the knowledge of how the body mass index is calculated from height and weight was encoded in the OCEC-HCV); and (4) queries that correspond to eligibility criteria with temporal constraints. Note that these four types of SPARQL queries are not mutually exclusive. For example, temporal constraints can be applied on the other three types of queries. We will discuss these four groups of queries in detail in the Results section.
A prototype web-based HCV clinical trial Computable Eligibility Criteria Cohort Identification application
Equipped with the OCEC-HCV and the Ontop platform, we were able to query the HCV patient data we extracted from the OneFlorida Data Trust (i.e., considered as the target population) using semantic queries. We developed a prototype web-based application, the HCV clinical trial Computable Eligibility Criteria Cohort Identification (HCV-CECCI), to facilitate the constructions of CEC and to enable cohort identification with interactive feedback. As shown in Figure 1.(a), the web application consists of two parts: a user-friendly Angular front-end and a REpresentational state transfer (REST)-ful application programming interface (API) back-end built with the JAVA Spring framework. To interact with the Ontop platform, we integrated the Ontop Web Ontology Language (OWL) Java API into the backend of our web application following the best practices in developing RESTful API endpoints.
Results
Analysis and categorization of HCV clinical trial eligibility criteria
From a collection of 77 HCV trials, we extracted 1,043 eligibility criteria (i.e., 428 inclusion criteria and 615 exclusion criteria) from ClinicalTrails.gov. However, 10 of the 428 (2.33%) inclusion criteria contained negations. On average, each HCV study has 5.56 (2 to 17) inclusion criteria and 7.98 (2 to 25) exclusion criteria. We extracted the core elements of each inclusion/exclusion criterion and summarized these 1,043 eligibility criteria into 272 unique criterion patterns. For example, age-related criteria appeared 19 times in the 77 HCV trials with different age ranges. Thus, we consolidated these 19 age-related criteria into 1 age criterion pattern. Further, as many of the inclusion and exclusion criterion patterns were fundamentally similar (e.g., as shown in Table 2, gender can be either an inclusion or exclusion criterion), we did not distinguish between inclusion and exclusion criterion patterns. Note that many criteria were concerned with specific diagnoses, procedures, and medications; and even though they may function in the same way (e.g., a corresponding query will only need to change the ICD codes for two different diagnoses), we treated them as different criterion patterns. Nevertheless, we used the same query templates when we developed the SPARQL queries for these diagnosis/procedure/medication-related criteria. Table 2 shows the top 10 frequent criterion patterns, separated by inclusion vs. exclusion criteria.
Table 2.
Top 10 frequent criterion patterns used by the 77 HCV trials, separated by inclusion vs. exclusion.
| Rank | Inclusion Criterion Pattern | Study Coverage # of Studies (%) | Exclusion Criterion Pattern | Study Coverage # of Studies (%) |
|---|---|---|---|---|
| N = 77 | N = 77 | |||
| 1 | HCV genotype | 51 (66.23%) | Pregnancy | 39 (50.65%) |
| 2 | Gender | 36 (45.75%) | Decompensated cirrhosis | 34 (44.16%) |
| 3 | HCV RNA | 33 (42.86%) | HIVb | 31(40.26 %) |
| 4 | HCV treatment naive | 27 (35.06%) | Gender | 27 (37.66 %) |
| 5 | Cirrhosis of liver | 21 (27.27%) | Hepatitis Bc | 24 (31.69 %) |
| 6 | Age | 19 (24.68%) | Hepatocellular carcinoma | 22 (28.57 %) |
| 7 | PEGylated interferon | 13 (16.88%) | Chronic liver disease | 19 (24.68 %) |
| 8 | Chronic hepatitis C | 12 (15.58%) | Hepatitis B surface antigen testc | 18 (23.38 %) |
| 9 | ASTa level | 12 (15.58%) | HIV antibody test | 17 (22.08%) |
| 10 | Ribavirin | 11 (14.29%) | Chronic hepatitis C | 15 (19.48%) |
AST: aspartate aminotransferase, a blood test that checks for liver damage.
HIV: human immunodeficiency virus
We treated “Hepatitis B” and “Hepatitis B surface antigen test” as two different criterion patterns, even though “Hepatitis B surface antigen test” can be used to determine the status of “Hepatitis B”. However, some occurrences of the “Hepatitis B surface antigen test” criterion in the 77 trials were concerned with specific lab result values.
Table 3 shows the overall top 10 frequent inclusion/exclusion criterion patterns, along with the corresponding PCORnet CDM domains of the data elements that each criterion pattern intended to query and the value sets of these data elements for each criterion pattern. These top 10 criterion patterns appeared in 93.51% of the studies.
Table 3.
Top 10 criterion patterns and its corresponding data elements’ PCORnet CDM domains.
| Rank | Pattern of Inclusion/Exclusion Criterion | Study Coverage # of Studies (%) N = 77 | Value Sets | PCORnet CDM Domain |
|---|---|---|---|---|
| 1 | HCV genotype | 51 (66.23%) | LOINCa: 32286-7 | Lab result |
| 2 | Pregnancy | 40 (51.95%) | ICD-9-CMb: V22 ICD-10-CMc: Z34 | Diagnosis |
| 3 | Decompensated cirrhosis | 35 (45.45%) | ICD-9-CM: 070.44, 070.71, 348.3, 456.0, 456.1, 456.2, 572.2, 572.3, 572.4, 782.4, 789.59 ICD-10-CM: B18.2, B19.2, G93.40, 185.1, I85.00, K72.90, K72.91, K76.6, K76.7, R17, R18.8 | Diagnosis |
| 4 | HCV RNA | 34 (44.16%) | LOINC: 11011-4 | Lab result |
| 5 | Cirrhosis of liver | 32 (41.56%) | ICD-9-CM:571.2, 571.5, 571.6 ICD-10-CM: K70.3, K74.3, K74.4, K74.5, K74.60 | Diagnosis |
| 6 | HIV | 31 (40.26%) | ICD-9-CM: 042 ICD-10-CM: B20 | Diagnosis |
| 7 | HCV treatment naive | 27 (35.06%) | On HCV antiviral medications (can be identified by RxNORM or NDCd codes) or had HCV treatment procedures (can be identified by ICD-9/10 procedure codes or CPTe codes) such as liver transplant | Procedure/Medication |
| 8 | Hepatocellular carcinoma | 27 (35.06%) | ICD-9-CM: 155 ICD-10-CM: C22 | Diagnosis |
| 9 | Chronic hepatitis C | 22 (28.57%) | ICD-9-CM: 070.54, 070.44, 070.70 ICD-10-CM: B18.2 | Diagnosis |
| 10 | Age | 19 (24.68%) | 15-70 | Demographic |
LOINC: Logical Observation Identifiers Names and Codes
ICD-9-CM: The International Classification of Diseases, Ninth Revision, Clinical Modification
ICD-10-CM: The International Classification of Diseases, 10th Revision, Clinical Modification
NDC: National Drug Code
CPT: Current Procedural Terminology
However, not all eligibility criteria were computable against our patient database—OneFlorida. We found that 42 (15.44%) of the 272 unique patterns were not computable. The top 5 frequent criterion patterns that cannot be queried against the OneFlorida data were those that require: (1) information about patient consent (28 studies, 36.36%); (2) female with childbearing potential (20 studies, 25.97%); (3) information about previous studies (19 studies, 24.68%); (4) investigator’s judgement of the patient (15 studies, 19.48%); and (5) information about the stage of patient’s hepatocellular carcinoma (11 studies, 14.29%). In sum, there were two main reasons that these criteria were not computable: (1) the criterion asked for subjective information of the patient (e.g., patient’s consent or investigator’s judgement of patient’s health status); and (2) the data elements needed for the criteria were not presented in the OneFlorida data (e.g., tumor stage was not captured in the PCORnet CDM). We also found that many of the eligibility criteria contained temporal constraints. 74.03% of the 77 HCV trials had at least one criterion with temporal constraints. We summarized these temporal constraints into 4 temporal patterns, as shown in Table 4.
Table 4.
The temporal patterns appeared in HCV clinical trial eligibility criteria.
| Temporal Pattern | Example | Study Coverage # of Studies (%), N = 77 |
|---|---|---|
| Event X within t time unitsa before/after event Y | e.g., “Clinically significant gastrointestinal bleeding occurring <= 4 weeks prior to randomization” | 42 (54.55%) |
| Cumulative duration t time units of an event X | e.g., “Chronic HCV infection (>= 6 months)” | 25 (32.47%) |
| Event X occurred at least t time units before/after event Y | e.g., “Positive for anti-HCV antibody (Ab) or HCV RNA > 1,000 IU/mL at least 6 months before Screening” | 24 (31.69%) |
| Event X before/after Event Y (regardless of how long) | e.g., “A liver biopsy performed prior to the Baseline/Day 1 visit with evidence of chronic HCV infection” | 3 (3.90%) |
The time unit can be day, week, month, or year.
The Ontology for Computable Eligibility Criteria – HCV (OCEC-HCV) and query scenarios
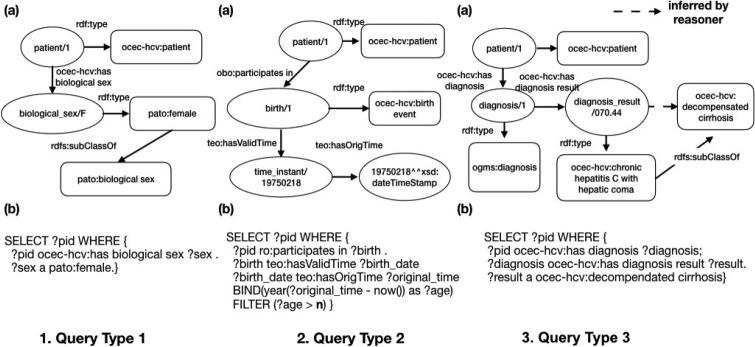
The OCEC-HCV was constructed as a formal representation of HCV trial eligibility criteria, but ultimately to support the translation of eligibility criteria into semantic (i.e., SPARQL) queries. Thus, our discussion focuses on how we implemented the OCEC-HCV to support the four types of semantic queries. Figure 2 shows three examples of the first three types of semantic queries, respectively.
Figure 2.
Examples for the first three types of semantic queries.
Query type 1: Queries that do not require any manipulation of the source data elements. For example, we used object property ‘ocec-hcv:has biological sex’ to link an instance of ‘ ocec-hcv:patient’ to its ‘ pato:biological sex’ (an instance of ‘pato:female’ in this case) as shown in Figure 2.1.(a). Then, we can directly use the SPARQL query listed in Figure 2.1.(b) to find all female patients through direct querying the demographic table in the source data. Note that in SPARQL syntax, query variables are prefixed with either “?” or “$”.
Query type 2: Queries that need to handle the logic of how to process the raw data elements to produce the desired information. For example, we wanted to identify eligible patients based on an age range (e.g., >= n years old). However, in OneFlorida, only patients’ birth dates were available in source data. Thus, we needed to calculate the patient’s age based on the date of birth as show in Figure 2.2.
Query type 3: Queries that can leverage a semantic reasoner based on the knowledge encoded in the ontology. As shown in Table 3, decompensated cirrhosis covered 45.45% of the studies, and is defined as a group of diagnoses. Thus, in OCEC-HCV, we declared these individual diagnoses as subclasses of the ‘decompensated cirrhosis’ diagnosis. To identify patients with decompensated cirrhosis, we can simply query the parent class, and the reasoner will automatically consider the subclasses of ‘ocec-hcv:decompensated cirrhosis’ as shown in Figure 2.3.
Query type 4: Queries that correspond to eligibility criteria with temporal constraints. Figure 3 shows an example criterion used to identify patients who have a history of alcohol abuse within n years before enrollment. The ‘input_date’ represents the time of enrollment. Note that both properties ‘ocec-hcv:has diagnosis result’ and ‘teo:hasValidTime” are defined as functional properties, ensuring that each instance of “ogms:diagnosis” class is linked to a specific diagnosis and the diagnosis happened at a specific time. We also used OWL restrictions on these two properties so that the instances linked with these properties were restricted to specific classes (i.e., owl:allValuesFrom indicates that the values of the property are all members of the class indicated by the owl:allValuesFrom class).
Figure 3.
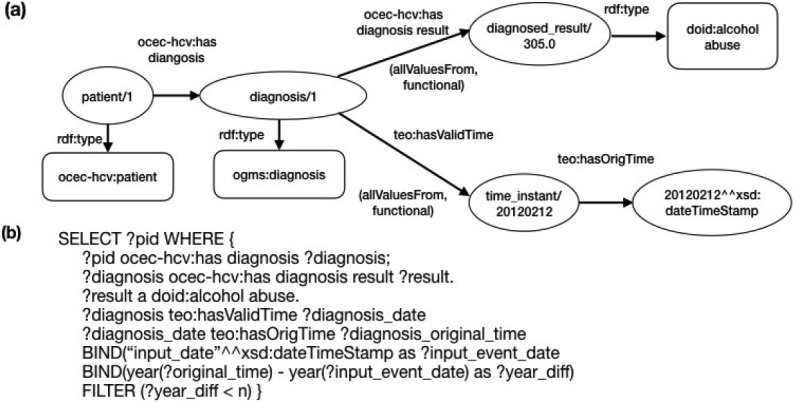
A query that identifies patients who have a history of alcohol abuse within n years before enrollment.
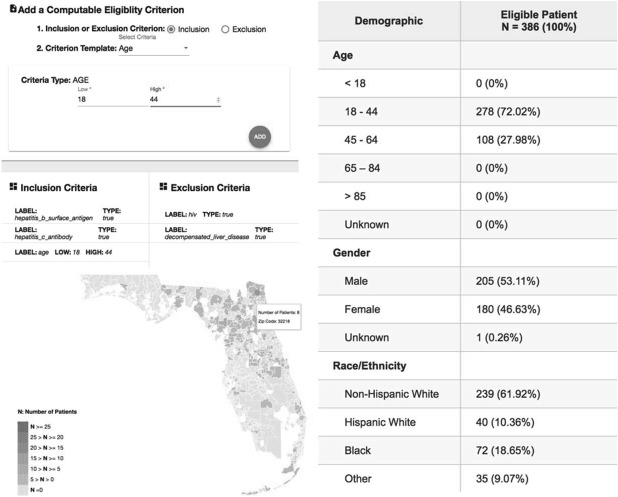
The HCV clinical trial Computable Eligibility Criteria Cohort Identification (HCV-CECCI) web application
We created a prototype web-based application, HCV-CECCI, to facilitate the construction of CEC and visualize the cohort identification results. Figure 4 shows a screenshot of the front-end interface. A researcher can easily add individual inclusion and exclusion criteria based on a list of CEC templates; and the system will return the basic demographics of the identified cohort along with an interactive map that shows the geographic distribution of the eligibility patients. Further, when the user hovers over a geographic region on the map, the system will show the demographics of eligible patients in that area. We also followed the anonymization strategy used in i2b2, masking the returned number of patients with (+- 5) to protect patient privacy.
Figure 4.
A screenshot of the HCV clinical trial Computable Eligibility Criteria Cohort Identification application.
Discussion and conclusion
Our experience in building the HCV-CECCI platform for HCV clinical trials has demonstrated the feasibility and benefits of making eligibility criteria computable with a formal representation (i.e., ontology). The use of an ontology can facilitate the construction of CEC for cohort identification in many ways and extend beyond traditional approaches. First, a shared, controlled vocabulary standardizes the definitions of the data elements and makes data understandable to both human (i.e., showing the preferred names for a class, and the synonyms and properties associated with it) and computers. Second, the OCEC-HCV linked the elements of the CEC to the underlying patient databases (i.e., OneFlorida in our case) at the semantic level. A high-level semantic query can leverage the vast amount of existing knowledge encoded in the ontologies. It is thus possible for researchers to use high-level concepts (e.g., ‘decompensated cirrhosis’) in their CEC without worrying about the technical details of the underlying queries (e.g., lookup for the right diagnosis codes, and write algorithms in the native database querying language). Third, explicitly modeling the semantic relationships among data elements makes domain and data assumptions clear and thus urges the researchers to validate their query intentions. Further, ontologies enable us to model the constrains of data elements using a formal and machine-readable language, which facilitates automatic validation and assurance of data quality. For example, the OneFlorida data have the raw height, weight as well as the calculated BMI for a patient. Figure 5 shows how we encoded the calculation process and implemented the constraints necessary for checking data consistency between the two different sources of the same data point.
Figure 5.
A representation of a patient’s height, weight, and body mass index measurements, their relationships, and the necessary constraints for data consistency checks.
Furthermore, it is clear that whether an eligibility criterion is computable or not depends on the data available in the target patient databases. For example, tumor information such as the stage of hepatocellular carcinoma, a commonly used criterion in HCV trials, typically does not exist in EHR data but does exist in sources such as tumor registries. Thus, it is necessary to have a flexible framework for integrating heterogeneous datasets to expand the capability of these computable eligibility criteria. The use of an ontology-driven data access model makes the integration of a new data source as simple as connecting the entities among the different data sources without the need to modify the underlying database structures and data models. Such an approach avoids the error-prone, and labor-intensive extract, transform, load (ETL) processes when transforming the source data into a CDM in typical data integration solutions. In addition to the benefits brought by the use of ontologies, we also built a user-friendly web-based system that can facilitate the construction of CEC. Our system can intuitively guide the users to make conscious choices when designing a trial’s eligibility criteria and give them not only real-time feedback of the eligible patients’ characteristics but also an interactive map that shows their geographic locations. With such a tool, investigators would be able to assess the feasibility of their studies and better plan the future recruitment efforts.
Our study is not without limitations. First, we used HCV diagnosis codes to generate the target population, which might not cover all types of HCV trials and all patients that we are interested in OneFlorida Data Trust. For example, HCV prevention and screening trials might not need participants with diagnosed HCV. Ideally, we shall use the entire OneFlorida Data Trust (an unselected population) as our target population. However, evaluating CEC against the entire OneFlorida Data Trust will be computationally expensive. Another source data related limitation is that the OneFlorida Data Trust only contains structured data (e.g., clinical notes). Thus, we built our platform with only structured data elements. However, unstructured data might be an important source of patients’ clinical information for evaluating patients’ eligibility. Third, the process of analyzing existing eligibility criteria to summarize criterion patterns was a manual process and both time- and labor-intensive. A few existing studies such as EliIE developed NLP tools to automatically transform free-text eligibility criteria into a structured representation. However, these methods are still premature and their accuracies were suboptimal. Further, to tease out the subtle ambiguities in free-text eligibility criteria, human judgments are inevitable. However, the number of unique criterion patterns is limited. Thus, it is possible to gradually expand the ontology and incorporate new patterns; and it may also be more desirable to have the correct representations. Fourth, a large percentage of eligibility criteria were concerned with whether an eligible patient has a particular disease or condition (e.g., HIV infected). We took a simplistic approach to map these entities to only diagnosis codes. However, it is well-known that identifying patients merely with diagnosis codes does not have high specificity and sensitivity. Thus, in the future, we will explore the potential to implement validated computable phenotyping algorithms in our CEC framework. Fifth, the system performance of using SPARQL queries against relational databases is also suboptimal. A typical SPARQL query in our system took about 0.4 to 78 seconds to run, depending on its complexity. Our future work will include tuning the performance of our query engines and exploring more capable SPARQL-to-SQL platforms other than Ontop. Our ultimate goal is to provide an easy-to-use computable eligibility criteria platform to facilitate clinical trial feasibility assessment and recruitment planning. Also, based on our existing computable eligibility criteria framework, we can build a more efficient screening tool for clinicians to identify eligible patients in clinical settings.
References
- 1.Thadani SR, Weng C, Bigger JT, Ennever JF, Wajngurt D. Electronic Screening Improves Efficiency in Clinical Trial Recruitment. J Am Med Inform Assoc. 2009;16(6):869–873. doi: 10.1197/jamia.M3119. doi:10.1197/jamia.M3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weng C, Batres C, Borda T, et al. A real-time screening alert improves patient recruitment efficiency. AMIA Annu Symp Proc AMIA Symp. 2011;2011:1489–1498. [PMC free article] [PubMed] [Google Scholar]
- 3.Penberthy L, Brown R, Puma F, Dahman B. Automated matching software for clinical trials eligibility: measuring efficiency and flexibility. Contemp Clin Trials. 2010;31(3):207–217. doi: 10.1016/j.cct.2010.03.005. doi: 10.1016/j.cct.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. J Am Med Inform Assoc JAMIA. 2014;21(4):576–577. doi: 10.1136/amiajnl-2014-002864. doi:10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc JAMIA. 2012;19(2):181–185. doi: 10.1136/amiajnl-2011-000492. doi:10.1136/amiajnl-2011-000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc JAMIA. 2016;23(6):1046–1052. doi: 10.1093/jamia/ocv202. doi:10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 8.OHDSI. Criteria Assessment Logic for Your Population Studies of Observations. http://www.ohdsi.org/web/calypso/.Published 2018. Accessed February 28, 2018.
- 9.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010;43(3):451–467. doi: 10.1016/j.jbi.2009.12.004. doi:10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu SW, Peleg M, Carini S, et al. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform. 2011;44(2):239–250. doi: 10.1016/j.jbi.2010.09.007. doi:10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc JAMIA. 2011;18(Suppl):6–124. doi: 10.1136/amiajnl-2011-000321. doi:10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel CO, Cimino JJ. AMIA Annu Symp Proc AMIA Symp. 2007. Oct, Semantic query generation from eligibility criteria in clinical trials; p. 1070. [PubMed] [Google Scholar]
- 13.Borlawsky T, Payne PRO. AMIA Annu Symp Proc AMIA Symp. 2007. Oct, Evaluating an NLP-based approach to modeling computable clinical trial eligibility criteria; p. 878. [PubMed] [Google Scholar]
- 14.Kang T, Zhang S, Tang Y, et al. EliIE: An open-source information extraction system for clinical trial eligibility criteria. J Am Med Inform Assoc JAMIA. 2017;24(6):1062–1071. doi: 10.1093/jamia/ocx019. doi:10.1093/jamia/ocx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvanese D, Cogrel B, Komla-Ebri S, et al. Ontop: Answering SPARQL queries over relational databases. Corcho Ó, ed. Semantic Web. 2016;8(3):471–487. doi:10.3233/SW-160217. [Google Scholar]
- 16.Mishra P, Florian J, Peter J, et al. Public-Private Partnership: Targeting Real-World Data for Hepatitis C Direct-Acting Antivirals. Gastroenterology. 2017;153(3):626–631. doi: 10.1053/j.gastro.2017.07.025. doi:10.1053/j.gastro.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 17.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37(Web Server issue):W170–173. doi: 10.1093/nar/gkp440. doi:10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong E, Xiang Z, Zhao B, et al. Ontobee: A linked ontology data server to support ontology term dereferencing, linkage, query and integration. Nucleic Acids Res. 2017;45(D1):D347–D352. doi: 10.1093/nar/gkw918. doi:10.1093/nar/gkw918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McAdam-Marx C, McGarry LJ, Hane CA, Biskupiak J, Deniz B, Brixner DI. All-cause and incremental per patient per year cost associated with chronic hepatitis C virus and associated liver complications in the United States: a managed care perspective. J Manag Care Pharm JMCP. 2011;17(7):531–546. doi: 10.18553/jmcp.2011.17.7.531. doi:10.18553/jmcp.2011.17.7.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanford Center for Biomedical Informatics Research. Protégé: A free, open-source ontology editor and framework for building intelligent systems. http://protege.stanford.edu/. Published 2016. Accessed March 7, 2018.