Abstract
We propose a frame-based natural language processing (NLP) method that extracts cancer-related information from clinical narratives. We focus on three frames: cancer diagnosis, cancer therapeutic procedure, and tumor description. We utilize a deep learning-based approach, bidirectional Long Short-term Memory (LSTM) Conditional Random Field (CRF), which uses both character and word embeddings. The system consists of two constituent sequence classifiers: a frame identification (lexical unit) classifier and a frame element classifier. The classifier achieves an F1 of 93.70 for cancer diagnosis, 96.33 for therapeutic procedure, and 87.18 for tumor description. These represent improvements of 10.72, 0.85, and 8.04 over a baseline heuristic, respectively. Additionally, we demonstrate that the combination of both GloVe and MIMIC-III embeddings has the best representational effect. Overall, this study demonstrates the effectiveness of deep learning methods to extract frame semantic information from clinical narratives.
Introduction
The past decade has seen a massive increase in the amount of data stored in electronic health record (EHR). Patient data consists of both structured data and unstructured (free-text) data. Free-text data such as clinical notes, discharge summaries, lab reports, and pathology reports are documented in detail by healthcare professionals during the course of patient care. Compared to the structured data within the EHR, free-text data often conveys more granular and contextual information of clinical events, as well as enhancing communication between clinical teams. As such, extracting information from these document resources is valuable to supporting a multitude of aims, from clinical decision support and quality care improvement to the secondary use of clinical data for research, public health, and pharmacovigilance activities.
In this paper, we take a frame-based approach to extracting cancer-related information. Frames are schematic representations of situations based on the theory of frame semantics that involve a series of participants such as events, states, and relations1. In frame semantics, the words or phrases that trigger the frame are known as lexical units (LU), and the frame elements are described as the participants or roles of a frame that defines the characteristics or attributes associated with the lexical units. Using the cancer diagnosis frame as an example, the lexical units are different types of cancer (e.g., melanoma, carcinoma) and the frame elements are cancer-related information one would expect to see that describing a particular cancer, such as status, histological characteristics, and the anatomic location of a tumor. Our study utilizes a machine learning method for extracting the cancer-related information. Machine learning approaches have been widely adopted in clinical NLP clinical tasks such as concept recognition and relation extraction. Traditional machine learning and statistical approaches such as conditional random fields (CRF), support vector machines (SVM), or hidden Markov models (HMM) have been widely used for information extraction of rich free-text data in EHR2. However, these traditional machine learning methods require a set of manually hand-crafted features and often suffer in terms of performance. More recently, deep learning techniques have been successfully implemented in many domains due to its power of identifying hidden patterns in large amounts of data and consequently better performance than traditional machine learning3. There remains a huge potential for deep learning methods to extract information from clinical notes given the intricacy of free text. The most widely-used neural networks for concept recognition and relation extraction are convolutional neural networks (CNN), recurrent neural networks (RNN), and RNN variants such as bidirectional Long Short-Term Memory (Bi-LSTM).
The rest of this paper is organized as follows. First, we describe the relevant prior work, notably in cancer information extraction and deep learning for clinical NLP. Next, we outline our prior work in developing the dataset and its corresponding annotation process, including a detailed description of the cancer frames. Then, we detail the deep learning model used to extract the frames. After that, we describe our experiments and results. Finally, we conclude with a discussion, including the limitations of the method and directions for future work.
Background
1. Cancer-related information extraction
Many efforts have focused on extracting cancer-related information from clinical records. Similar studies to ours have applied either rule-based methods or traditional machine learning methods (or combination of both) to identify different varieties of cancer-related information. Taira et al.4 proposed an automatic frame-structured representation of radiology reports. Xu et al.5 explored the performance of an existing NLP system, MedLEE, in capturing narrative and tabular information related to cancer. Weegar and Dalianis6 developed a rule-based system for extracting cancer-related information from breast cancer pathology reports. McCowan et al. 7 adopted a support vector machine approach in order to extract TNM stages listed in cancer staging protocols with the Cancer Stage Interpretation System (CSIS). D’Avolio et al.8 applied a rule-based method including regular expressions to parse Gleason scores and TNM stages from prostate cancer post-operative pathology reports. Coden et al.9 proposed an integrated cancer disease knowledge representation model (CDKRM) in order to process cancer and its progression through another clinical NLP system, Medical Text Analysis System. Cheng et al.10 discerned tumor status, magnitude and significance from MRI brain reports using both SVMs and a rule-based system. Ou and Patrick11 applied a CRF-based system to extract melanoma-related entities including metastasis, site, and size from primary cutaneous melanoma pathology reports. Yala et al.12 demonstrated that using machine learning methods to extract breast cancer-related information from pathology reports is effective. Denny et al.13 conceived the use of clinical NLP to aid in cancer screening by identifying patients in need of a colonoscopy by extracting terms of interest from colon cancer testing reports through the Knowledge Map Concept Identifier (KMCI) system. Napolitano et al.14 defined a rule-based method to identify different formats of Gleason score, Clark level, and Breslow depth. Wilson et al.15 classified ancillary cancer history for mesothelioma patients by using two kinds of rule-based methods, Dynamic-Window and ConText, with the assistance of domain-specific lexicons. Martinez and Li16 used a traditional machine learning classification method to extract colorectal cancer diagnosis and staging information from pathology reports at the document level. Herkema et al.17 demonstrated that an NLP system can be tailored to aid in quality measurements for colonoscopy procedures. As we have listed here, it is widely acknowledged that different types of cancer information are overlapping to a large extent18-24. It is our hope that by focusing on a frame semantic approach, relevant and consistent information should be integrated together instead of relying on developing task-specific NLP systems.
2. Deep learning in biomedical language domain
There has also been a significant amount of attention paid by researchers to the use of deep learning for processing biomedical text, including not only clinical narratives, but scientific literature articles, drug labels, and other types of biomedical text. Chalapathy et al.25 proved the effectiveness of the Bi-LSTM-CRF model by experimenting on a set of public dataset, 2010 i2b2/VA challenge26, especially when using GloVe embeddings. Wu et al.27 demonstrated the increased performance of the RNN model by comparing a convolutional neural network (CNN) and an RNN on the 2010 i2b2/VA datasets. Further, Liu et al.28 proved that the RNN can achieve the state-of-the-art performances on i2b2 201027, i2b2 201229, and i2b2 201430 corpora. Habibi et al.31 compared traditional CRF and Bi-LSTM-CRF method with different embeddings on several biomedical corpora and drew the conclusion that deep learning methods with domain-specific embeddings can largely improve recall without significant cost of precision, thereby increasing overall model performance. Gridach et al.32 demonstrated the effectiveness of character-level embeddings with a Bi-LSTM-CRF. Jagannatha et al.33-34 experimented and assigned clinical-related labels to each word in clinical notes by using several variants of RNNs including Bi-LSTM, combinations of Bi-LSTM with CRF, Bi-LSTM CRF with pairwise modeling, and approximate Skip-chain CRF. They showed that variants of LSTM outperformed other methods especially in terms of attributes with intricate phrases like medication duration or frequency. Xu et al.35 applied Bi-LSTM-CRF to extract adverse drug reaction for the 2017 TAC track36 and achieved the best overall results in the challenge. Tao et al.37 used word embeddings trained on MIMIC-III38 as the features of a machine learning model to extract prescription information from clinical notes. Gehrmann et al.39 carefully compared outputs from an existing NLP pipeline, cTAKES, with CNN and concluded that the CNN is an alternative method in terms of patient phenotyping tasks and should be integrated into systems like cTAKES in order to improve predictive models. Luo et al.40-41 proposed a novel segment-level deep neural network model to classify relations in clinical notes, which is one of the early investigations in how to use neural networks for medical relation classification. Models only used word embedding that were trained on MIMIC-III for features. The author also showed the benefits of segment-level over sentence-level classification for relation extraction.
Method
1. Dataset and Annotation
The dataset and annotation processes are described in detail in Roberts et al42. Here, we describe the data in brief. We focus on frames based on typical cancer phenotype tasks involving the extraction of cancer diagnoses, cancer therapeutic procedures, and tumor descriptions. These correspond to three frames, respectively: CANCER_DIAGNOSIS, CANCER_THERAPEUTIC_PROCEDURE, and TUMOR_DESCRIPTION. An expert physician devised a list of lexical units (trigger phrases) and corresponding elements (attributes) for each frame. The elements were determined by an iterative process by which elements with sufficient frequency and importance were included in the schema. The final list is shown in Table 1. With the approval of UT Health Institutional Review Board, under protocol number HSC-SBMI-13-0549, we extracted cancer-related clinical notes from the UT Physicians data warehouse. Sentences containing candidate lexical units were extracted from the note. To avoid duplicate sentences and improve sentence diversity, the sentences were sorted by TF-IDF cosine distance. The sentences were manually de-identified and checked by an automatic de-identification system. The frame annotation was performed in Brat43 by two annotators and then reconciled by an expert in clinical NLP.
Table 1.
Frame Lexical Units and Elements
| Frame Lexical Units | Frame Element | Description |
|---|---|---|
| CANCER_MASTER_FRAME | ||
| CERTAINTY | DATETIME | POLARITY |
| Certainty/hedging of frame (e.g., possible, likely) | Temporal information for the frame | Existence/negation of frame (e.g., no, positive) |
| CANCER_DIAGNOSIS | ||
| adenocarcinoma, cancer, carcinoma, leukemia, lymphoma, malignancy, malignant, melanoma, myeloma, sarcoma | FAMILYHISTORY | Specifies a family member with the diagnosis |
| HISTOLOGY | Histological description | |
| LOCATION | Part of body associated with the cancer | |
| PATIENT | Reference to the patient (e.g., patient, female) | |
| QUANTITY | Some quantitative measure of the cancer | |
| STATUS | Diagnostic status (e.g., history, ongoing) | |
| CANCER_THERAPEUTIC_PROCEDURE | ||
| colectomy, hysterectomy, lymphadenectomy, mastectomy, palliative, pancreatectomy, prostatectomy, radiation, whipple | AGENT | Agent performing the procedure (e.g., surgeon) |
| COMPLICATION | Unexpected, undesirable outcome of procedure | |
| EXTENT | Extent of the procedure (e.g., partial) | |
| LOCATION | Part of body procedure targets | |
| PATIENT | Reference to the patient (e.g., patient, female) | |
| RESULT | Result of the procedure (e.g., successful, negative) | |
| STATUS | Procedure status (e.g., planned, postoperative) | |
| TUMOR_DESCRIPTION | ||
| lesion, mass, tumor | LOCATION | Part of body tumor is located in |
| MALIGNANCY | Whether the tumor is benign or malignant | |
| MARGINSTATUS | Description of tumor margin (e.g., superficial edge) | |
| METASTASIS | Whether the tumor has metastasized | |
| PATIENT | Reference to the patient (e.g., patient, female) | |
| QUANTITY | Some quantitative measure of the tumor | |
| RECURRENCE | Whether the tumor has recurred | |
| RESECTABILITY | Indicator of whether tumor is resectable | |
| MORPHOLOGY | Morphology of tumor | |
| SIZE | Diameter/volume of tumor, including unit (e.g., 3-4 mm) | |
| SIZETREND | Trend in tumor size over time (e.g., increasing, | |
| STAGE | Stage number (e.g., stage IV) | |
| STATUS | Tumor status (e.g., present, active) | |
2. Model and Architecture
This section details the components of the neural network and as well as the embedding philosophy employed to improve extraction performance.
Bi-LSTM CRF Networks
Long Short-Term Memory (LSTM) network is an extension of recurrent neural networks (RNN) and are capable of coping with long term dependencies by minimizing vanishing gradient problems. Further, apart from the forward sequence representation (first word to last word in the sentence) that makes use of previous context, by concatenating a second LSTM network that processes the same sequence in reverse (last word to first) should capture both past and future information. This forward and backward LSTM was introduced as bidirectional LSTM44. However, in the decoding layer, the network still makes independent token-level labeling decisions, which is not well-suited for concept recognition or relation extraction since sequential information is an important factor in language. Therefore, to consider the correlations and jointly decode the best series, a linear-chain conditional random field (CRF) algorithm was applied in a Viterbi-style manner in the decoding layer instead of independent decoding. For the CRF layer, a state transition matrix score was calculated. The final score from the Bi-LSTM network along with the transition score was used to make a decision on sequence prediction. The overall neural network follows the structure proposed in Lample et al45.
Embedding
Medical text information is characterized by its complexity in vocabulary and richness in morphology. Therefore, using character embeddings to capture morphological information from complex medical terms can avoid out of vocabulary problems and compensate traditional word embeddings. We implemented a Bi-LSTM neural network and adjoined the output vector from both forward and backward sequences to obtain a vector of the final character representation. The initial character embedding was randomly assigned, but dynamically altered during training. Besides this character representation, we also applied pre-trained word embeddings, which have been shown to result in significant improvements in many NLP tasks. Due to the fact that word embeddings vary largely between general and specific domains, we performed experiments from different public resources. Thus, we make full use of contextual representation of each word by feeding into both character embeddings along with pre-trained word embeddings of the same word into the input layer of the neural network.
System Architecture and Pipeline
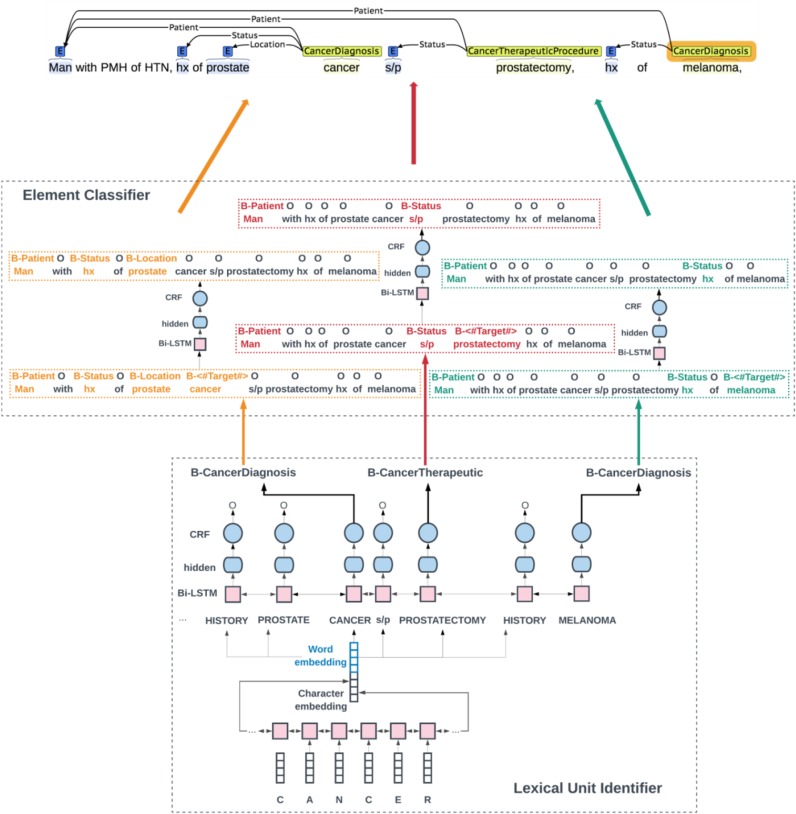
We propose a sequence labeling pipeline that extracts lexical units and elements in two steps. Figure 1 shows the main architecture of the system. The first step is a traditional concept recognition system that identifies all the frame-evoking lexical units in one sentence. As shown in Figure 1, the sentence has three lexical units predicted by the first system: cancer → B-CANCER_DIAGNOSIS, prostatectomy → B-CANCER_THERAPEUTIC_PROCEDURE, and melanoma → B-CANCER_DIAGNOSIS. Note that in the second step, during training we assign the lexical unit of interest in each sentence with a new tag called “B-<#Target#>” to make the system aware about which word is the lexical unit in the current sentence. Then the system splits the sentences into three samples and each sample has one lexical unit and its corresponding elements. For example, in sample 1, cancer was labeled as B-<#Target#> and the elements of this cancer: man, hx, prostate were labeled as B-PATIENT, B-STATUS, and B-LOCATION,respectively. Other lexical units and elements were not marked in this sample; thus, we emphasized the position information of the lexical unit to the element identifier system and avoid being confused with multiple candidate lexical units. Last, we merge the split sentences into the final output. In this way, we integrated cancer information into one large architecture. At run time, when a new sentence comes in, the first system would identify the lexical units in this sentence and transfer the identified lexical unit outputs into the input of the second system that could extract all the associated elements.
Figure 1.
Main Architecture of the System
3. Experiment and Evaluation
The annotated data was split into training, testing, and validation sets, with a ratio of 80 percent for training, 10 percent for testing and 10 percent for validation. The details of sample size information are shown in Table 2. The input embedding layer is concatenated with character representation with 100 dimensions and word representation. We implemented different word embedding methods: 300-dimesion GloVe embeddings, 100-dimension medical domain embeddings, and the combination of the two. The GloVe embeddings were pre-trained on 6 billion words from Wikipedia and web text46. For the medical domain embeddings, we adopted the pre-trained embeddings from a previous study47 , which was induced from the publicly available medical dataset MIMIC III. We also experimented by using GloVe and MIMIC-III embedding together with a dimensionality of 400.
Table 2.
Descriptive statistics of train, test, and dev set.
| Type | Train size | Test size | Dev size |
|---|---|---|---|
| Frame Lexical Units | 5696 | 759 | 708 |
| Frame Elements | 9108 | 1178 | 1092 |
During the training process, sentences and labels are split into two types of input data lists that contain all the necessary information. The first input list contains only lexical units with labels and are fed into the first system that can identify all lexical units in the sentence. The second input list consists of several labeled sentences and each sentence only has one lexical unit with its corresponding elements. All sentences in the second list are the input of the second system that extracts the elements of the target lexical units and classifies the relations between the lexical unit and candidate role in one step. During training, the gold standard lexical unit is fed into the second system. The final Bi-LSTM CRF neural network has one hidden layer that was implemented in Tensorflow48 on an NVidia Tesla GPU with the cuDNN library. We compare several combinations of parameters on the validation set and finally settle the following parameters: hidden unit dimension at 400, dropout probability at 0.5, learning rate at 0.0001, learning rate decay at 0.99, and Adam as the optimization algorithm. We also assign an embedding to provide target information for the second system. The target embedding for this information has 5 dimensions.
To understand how well the frame identification and frame element classification performs, we compare the model’s prediction with the annotations on the held-out test set. We calculate micro-averaged precision, recall, and F1-score for exact matching. In terms of the first step to extract lexical units, we compare the model with the baseline heuristic. The baseline simply identifies lexical unit that appears in the sentence as the frame-evoking lexical unit (i.e., a 100% recall baseline).
Results
Detailed performance comparison between the baseline heuristic and the proposed Bi-LSTM CRF method with different initialization is shown in Table 3. As a first note, the Bi-LSTM CRF method initialized with combination embeddings of GloVe and MIMIC-III outperforms all other embedding methods. Notably, the performance of GloVe embeddings have a slightly better result than MIMIC-III embeddings over the entire three frames.
Table 3.
Performance of System for Frame Identification
| metrics/th> | Method | CANCER_DIAGNOSIS | CANCER_THERAPEUTIC_PROCEDURE | TUMOR_DESCRIPTION |
|---|---|---|---|---|
| Precision (%) | Baseline | 70.91 | 91.35 | 65.48 |
| GloVe | 95.35 | 95.88 | 86.72 | |
| MIMIC-III | 93.65 | 94.40 | 79.87 | |
| GloVe+MIMIC-III | 92.70 | 94.40 | 78.81 | |
| Recall (%) | Baseline | 100 | 100 | 100 |
| GloVe | 91.99 | 95.88 | 86.05 | |
| MIMIC-III | 93.55 | 97.12 | 92.25 | |
| GloVe+MIMIC-III | 93.59 | 97.12 | 92.25 | |
| F1 (%) | Baseline | 82.98 | 95.48 | 79.14 |
| GloVe | 93.64 | 95.88 | 86.38 | |
| MIMIC-III | 93.60 | 95.74 | 85.61 | |
| GloVe+MIMIC-III | 93.70 | 96.33 | 87.18 |
The F1-scores of CANCER_DIAGNOSIS, CANCER_THERAPEUTIC_PROCEDURE, and TUMOR_DESCRIPTION are 93.70%, 96.33%, and 87.18% respectively. For CANCER_DIAGNOSIS, the performance of three embeddings are of about the same level (at around 93%), while the combined embeddings model shows an improvement of 10.72% F1 score over the baseline heuristic. CANCER_THERAPEUTIC_PROCEDURE already reaches a high baseline score of 95.48% and the combined embeddings improves on this a small amount. TUMOR_DESCRIPTION has a low baseline of 79.14% F1 and the combined embeddings improves 8.04% of F1 score over the baseline.
The element classifier is also measured in Precision, Recall and F1-score provided in Table 4&5. In terms of general performance results in Table 4, the combined embedding gets the best F1 score of 75.81% over two other embeddings. Similar with the first step, GloVe embeddings has a slightly better performance of 1.64% over MIMIC-III embeddings.
Table 4.
General Performance Evaluation of System for Element Classifier
| Embedding type | Accuracy(%) | Precision(%) | Recall(%) | F1(%) |
|---|---|---|---|---|
| GloVe | 94.73 | 77.39 | 73.83 | 75.57 |
| MIMIC-III | 93.99 | 70.54 | 77.66 | 73.93 |
| GloVe+MIMIC-III | 94.52 | 73.91 | 77.81 | 75.81 |
Table 5.
Cross-Frame Per Category Performance Evaluation of System for Element Classifier
| Precision (%) | Recall (%) | F1 (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| GloVe | MIMIC-III | GloVe +MIMIC-III | GloVe | MIMIC-III | GloVe +MIMIC-III | GloVe | MIMIC-III | GloVe +MIMIC-III | |
| Extent3 | 90.91 | 88.24 | 90.91 | 95.24 | 95.24 | 95.24 | 93.02 | 91.60 | 93.02 |
| Stage4 | 72.34 | 67.31 | 64.81 | 75.56 | 77.78 | 77.78 | 73.91 | 72.16 | 70.71 |
| Patient2,3,4 | 83.75 | 79.21 | 81.91 | 87.78 | 88.89 | 85.56 | 85.71 | 83.77 | 83.70 |
| Histology2 | 69.39 | 62.75 | 68.09 | 85.00 | 80.00 | 80.00 | 76.40 | 70.33 | 73.56 |
| Malignancy4 | 82.35 | 85.00 | 81.82 | 63.64 | 77.27 | 81.82 | 71.79 | 80.95 | 81.82 |
| Location2,3,4 | 79.69 | 73.55 | 79.13 | 76.40 | 78.76 | 80.53 | 78.01 | 76.07 | 79.82 |
| Family_history2 | 75.00 | 74.12 | 76.74 | 78.95 | 82.89 | 86.84 | 76.92 | 78.26 | 81.48 |
| Status2,3,4 | 78.76 | 71.64 | 71.60 | 73.86 | 79.67 | 76.35 | 76.23 | 75.44 | 73.90 |
| Size4 | 66.67 | 57.89 | 61.11 | 71.43 | 78.57 | 78.57 | 68.97 | 66.67 | 68.75 |
| Polarity1 | 70.73 | 54.90 | 65.91 | 69.05 | 66.67 | 69.05 | 69.88 | 60.22 | 67.44 |
| Resectability4 | 46.15 | 35.00 | 45.00 | 46.15 | 53.85 | 69.23 | 46.15 | 42.42 | 54.55 |
| Certainty1 | 59.65 | 45.56 | 48.89 | 40.96 | 49.40 | 53.01 | 48.57 | 47.4 | 50.87 |
| Datetime1 | 23.08 | 50.00 | 66.67 | 15.00 | 60.00 | 70.00 | 18.18 | 54.55 | 68.29 |
| Complication3 | 100.000 | 55.56 | 83.33 | 23.08 | 38.46 | 38.46 | 37.50 | 45.45 | 52.63 |
| Morphology4 | 30.00 | 50.00 | 28.57 | 33.33 | 33.33 | 22.22 | 31.58 | 40.00 | 25.00 |
| Sizetrend4 | 50.00 | 20.00 | 37.50 | 16.67 | 16.67 | 50.00 | 25.00 | 18.18 | 42.86 |
| Recurrence4 | 0 | 50.00 | 100.00 | 0 | 25.00 | 25.00 | 0 | 33.33 | 40.00 |
| Agent3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MarginStatus4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Quantity4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Result3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Superscripts indicate frame(s) associated with element (see Table 1 for more details):
CANCER_MASTER_FRAME
CANCER_MASTER_FRAME
CANCER_THERAPEUTIC_PROCEDURE
TUMOR_DESCRIPTION
We also measure the performance for each element category across three frames in Precision, Recall, and F1-score and compare the performance results of three models, differing only in word representations, shown in Table 5. The combination embeddings improves the performance in majority of categories (MALIGNANCY: 81.82%, FAMILYHISTORY: 81.48%, RESECTABILITY: 54.55%, CERTAINTY: 50.87%, DATETIME: 68.29%, COMPLICATION: 52.63%, SIZETREND: 42.86%, RECURRENCE: 40%). Among other categories, GloVe is next, followed by MIMIC-III. GloVe is pre-trained from news of general topics, which leads that it has much better representations for general categories including STAGE (73.91%), PATIENT (85.71%), STATUS (76.23%), SIZE (68.97%), POLARITY (69.88%). Note that the resource of MIMIC-III embedding is from ICU patient records, which presumably bring triple influence towards the representational effect. MIMIC-III outperforms in Morphology (40.00%). Generally speaking, three kinds of embeddings have slightly differences in performance for most categories (EXTENT: ~92%, STAGE: ~72%, PATIENT: ~84%, HISTOLOGY: ~73%, LOCATION: ~78%, FAMILYHISTORY: ~78%, STATUS: ~75%, SIZE: ~67%, POLARITY: ~65%, CERTAINTY: ~49%) except some categories (MALIGNANCY, RESECTABILITY, DATETIME, COMPLICATION, SIZETREND, RECURRENCE) where adding medical domain representation actually helps to improve the performance by over 1020%.
Discussion
Our study proposes a frame-based NLP system based on Bi-LSTM-CRF to extract important cancer-related information from clinical narratives. It achieves superior performance when compared to the baseline heuristics. The neural network is initialized with both character embeddings and word embeddings. We also apply three kinds of word embeddings and compare the performances. We find that the overall performance of combined GloVe and MIMIC-III embeddings generally outperform embeddings from only one dataset.
An inspection of the system errors reveals several patterns. One notable problem is that frame elements are often extracted for the wrong frame. For instance, excise is a RESECTABILITY element for TUMOR_DESCRIPTION frame but it is not an element in CANCER_DIAGNOSIS. This is a result of training a single model on all frames, and while the classifier is provided with information to indicate what the current frame is, this is not enough to remove all such errors. The alternative, training a different model for every frame, results in even more errors due to the significantly reduced data. Since the three frames share several frame elements, training together effectively increases the size of the training set, but causes the aforementioned types of errors. One solution to this would involve a multi-task learning approach, where all three frames are trained concurrently, and share parameters for part of the network, but also have corresponding task-specific aspects of the network as well.
As is typical with machine learning methods, both the frame identification and frame element classifiers the performances of different categories are variable due to the size of the training data. False negative samples probably result from the small size of the training set. For example, the performance of TUMOR_DESCRIPTION is poorer than that of other frames due to the smallest size in the training set. Similarly, the elements with lowest F1 score tend to be relatively rare in the training set (e.g., 6 AGENT, 18 MARGINSTATUS, 16 QUANTITY, 37 RESULT).
A limitation of this study is that we currently limit our evaluation to the gold standard for each process in the pipeline. However, instead of a multi-step evaluation and optimizing the pipeline in that manner, our future work will focus on applying joint models to share weights of both concept identification and relation extraction. This will achieve both steps at the same time, and should result in increased performance due to information sharing between the models. Prior work has explored end-to-end models for performing concept recognition and relation extraction jointly49-50, so this is a promising line of work for improving system performance.
Conclusion
In this paper, we design a frame-based NLP system based on a Bi-LSTM-CRF neural network to extract cancer-related information in clinical narratives. We also apply different pre-trained embeddings including character embeddings and word embeddings as input. This initial study achieved promising results, with frame identification F1 between 87.18% and 96.33% and element classification F1 up to 93.02%. Our primary goal is to test the feasibility of utilizing deep learning method to design a frame-based NLP system for extracting cancer frame elements from clinical notes. Our ultimate goal is to develop an integrated system that extracts all important cancer-related information from clinical narratives.
Acknowledgement
This work was supported by the U.S. National Library of Medicine, National Institutes of Health (NIH), under award R00 LM012104 as well as a UTHealth School of Biomedical Informatics doctoral fellowship.
References
- 1.Baker CF, Fillmore CJ, Lowe JB. editors. The Berkeley FrameNet project. Proceedings of the 17th international conference on Computational linguistics-Volume; Association for Computational Linguistics; 1998. [Google Scholar]
- 2.Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, Liu S, Zeng Y, Mehrabi S, Sohn S, Liu H. Journal of biomedical informatics. 2017. Nov 21, Clinical information extraction applications: A literature review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015 May;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 4.Taira RK, Soderland SG, Jakobovits RM. Automatic structuring of radiology free-text reports. Radiographics. 2001 Jan;21(1):237–45. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 5.Xu H, Anderson K, Grann VR, Friedman C. Studies in health technology and informatics. 2004. Facilitating cancer research using natural language processing of pathology reports; p. 1531. [PubMed] [Google Scholar]
- 6.Weegar R, Dalianis H. Proceedings of the Sixth International Workshop on Health Text Mining and Information Analysis. 2015. Creating a rule based system for text mining of Norwegian breast cancer pathology reports; pp. 73–78. [Google Scholar]
- 7.McCowan IA, Moore DC, Nguyen AN, Bowman RV, Clarke BE, Duhig EE, Fry MJ. Collection of cancer stage data by classifying free-text medical reports. Journal of the American Medical Informatics Association. 2007 Nov 1;14(6):736–45. doi: 10.1197/jamia.M2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Avolio LW, Litwin MS, Rogers Jr SO, Bui AA. Facilitating clinical outcomes assessment through the automated identification of quality measures for prostate cancer surgery. Journal of the American Medical Informatics Association. 2008 May;1, 15(3):341–8. doi: 10.1197/jamia.M2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coden A, Savova G, Sominsky I, Tanenblatt M, Masanz J, Schuler K, Cooper J, Guan W, De Groen PC. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. Journal of biomedical informatics. 2009 Oct 1;42(5):937–49. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Cheng LT, Zheng J, Savova GK, Erickson BJ. Discerning tumor status from unstructured MRI reports— completeness of information in existing reports and utility of automated natural language processing. Journal of digital imaging. 2010 Apr 1;23(2):119–32. doi: 10.1007/s10278-009-9215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou Y, Patrick J. Proceedings of the Seventh Australasian Workshop on Health Informatics and Knowledge Management-Volume. Australian Computer Society, Inc: 2014. Jan 20, Automatic population of structured reports from narrative pathology reports; pp. 153, 41–50. [Google Scholar]
- 12.Yala A, Barzilay R, Salama L, Griffin M, Sollender G, Bardia A, et al. Using machine learning to parse breast pathology reports. Breast cancer research and treatment. 2017;161(2):203–11. doi: 10.1007/s10549-016-4035-1. [DOI] [PubMed] [Google Scholar]
- 13.Denny JC, Choma NN, Peterson JF, Miller RA, Bastarache L, Li M, Peterson NB. Natural language processing improves identification of colorectal cancer testing in the electronic medical record. Medical Decision Making. 2012 Jan;32(1):188–97. doi: 10.1177/0272989X11400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Napolitano G, Fox C, Middleton R, Connolly D. Pattern-based information extraction from pathology reports for cancer registration. Cancer Causes & Control. 2010 Nov 1;21(11):1887–94. doi: 10.1007/s10552-010-9616-4. [DOI] [PubMed] [Google Scholar]
- 15.Wilson RA, Chapman WW, DeFries SJ, Becich MJ, Chapman BE. Journal of pathology informatics. 2010. Automated ancillary cancer history classification for mesothelioma patients from free-text clinical reports; p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez D, Li Y. Proceedings of the 20th ACM international conference on Information and knowledge management. ACM; 2011. Oct 24, Information extraction from pathology reports in a hospital setting; pp. 1877–1882. [Google Scholar]
- 17.Harkema H, Chapman WW, Saul M, Dellon ES, Schoen RE, Mehrotra A. Developing a natural language processing application for measuring the quality of colonoscopy procedures. Journal of the American Medical Informatics Association. 2011 Sep 21;18(Supplement_1):i150–6. doi: 10.1136/amiajnl-2011-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imler TD, Morea J, Kahi C, Imperiale TF. Natural language processing accurately categorizes findings from colonoscopy and pathology reports. Clinical Gastroenterology andHepatology. 2013 Jun 1;11(6):689–94. doi: 10.1016/j.cgh.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez D, Cavedon L, Pitson G. Louhi, The 4th International Workshop on Health Document Text Mining and Information Analysis. NICTA, Canberra, Australia: 2013. Feb 11, Stability of text mining techniques for identifying cancer staging. [Google Scholar]
- 20.Ping XO, Tseng YJ, Chung Y, Wu YL, Hsu CW, Yang PM, Huang GT, Lai F, Liang JD. Information extraction for tracking liver cancer patients’ statuses: from mixture of clinical narrative report types. TELEMEDICINE and e-HEALTH. 2013 Sep 1;19(9):704–10. doi: 10.1089/tmj.2012.0241. [DOI] [PubMed] [Google Scholar]
- 21.Vanderwende L, Xia F, Yetisgen-Yildiz M. Workshop on Events: Definition, Detection, Coreference, and Representation. 2013. Annotating Change of State for Clinical Events; pp. 47–51. [Google Scholar]
- 22.Ashish N, Dahm L, Boicey C. University of California, Irvine-Pathology Extraction Pipeline: the pathology extraction pipeline for information extraction from pathology reports. Health informatics journal. 2014 Dec;20(4):288–305. doi: 10.1177/1460458213494032. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Zhang W, Zeng Q, Li Z, Feng K, Liu L. Extracting important information from Chinese Operation Notes with natural language processing methods. Journal of biomedical informatics. 2014 Apr 1;48:130–6. doi: 10.1016/j.jbi.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Savova GK, Tseytlin E, Finan S, Castine M, Miller T, Medvedeva O, Harris D, Hochheiser H, Lin C, Chavan G, Jacobson RS. DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records. Cancer research. 2017 Nov 1;77(21):e115–8. doi: 10.1158/0008-5472.CAN-17-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalapathy R, Borzeshi EZ, Piccardi M. 2016. Nov 25, Bidirectional LSTM-CRF for clinical concept extraction. arXiv preprint arXiv:1611.08373. [Google Scholar]
- 26.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011 Jun 16;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Jiang M, Xun J, Zhi D, Xu H. AMIA Annual Symposium Proceedings. 2017. Clinical Named Entity Recognition Using Deep Learning Models. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Yang M, Wang X, Chen Q, Tang B, Wang Z, Xu H. Entity recognition from clinical texts via recurrent neural network. BMC medical informatics and decision making. 2017 Jul;17(2):67. doi: 10.1186/s12911-017-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. Journal of the American Medical Informatics Association. 2013 Apr 5;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stubbs A, Kotfila C, Uzuner Ö. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. Journal of biomedical informatics. 2015 Dec 1;58:S11–9. doi: 10.1016/j.jbi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habibi M, Weber L, Neves M, Wiegandt DL, Leser U. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics. 2017;33(14):i37–i48. doi: 10.1093/bioinformatics/btx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gridach M. Character-level neural network for biomedical named entity recognition. Journal of biomedical informatics. 2017;70:85–91. doi: 10.1016/j.jbi.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Jagannatha AN, Yu H. editors. Structured prediction models for RNN based sequence labeling in clinical text. Proceedings of the Conference on Empirical Methods in Natural Language Processing Conference on Empirical Methods in Natural Language Processing; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jagannatha AN, Yu H. editors. Bidirectional RNN for medical event detection in electronic health records. Proceedings of the conference Association for Computational Linguistics North American Chapter Meeting; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Lee H-J, Ji Z, Wang J, Wei Q, Xu H. UTH_CCB System for Adverse Drug Reaction Extraction from Drug Labels at TAC-ADR 2017. Proceedings of the Text Analysis Conference.2017. [Google Scholar]
- 36.Demner-Fushman D, Shooshan SE, Rodriguez L, Aronson AR, Lang F, Rogers W, Roberts K, Tonning J. A dataset of 200 structured product labels annotated for adverse drug reactions. Scientific data. 2018 Jan;30(5):180001. doi: 10.1038/sdata.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao C, Filannino M, Uzuner Ö. Prescription extraction using CRFs and word embeddings. Journal of biomedical informatics. 2017;72:60–6. doi: 10.1016/j.jbi.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson AE, Pollard TJ, Shen L, Li-wei HL, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Scientific data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gehrmann S, Dernoncourt F, Li Y, Carlson ET, Wu JT, Welt J, et al. Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PloS one. 2018;13(2):e0192360. doi: 10.1371/journal.pone.0192360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y. Recurrent neural networks for classifying relations in clinical notes. Journal of biomedical informatics. 2017;72:85–95. doi: 10.1016/j.jbi.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Y, Cheng Y, Uzuner Ö, Szolovits P, Starren J. Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. Journal of the American Medical Informatics Association. 2017;25(1):93–8. doi: 10.1093/jamia/ocx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts K, Si Y, Gandhi A, Bernstam E. A FrameNet for Cancer Information in Clinical Narratives: Schema and Annotation. Proceedings of the Language Resources and Evaluation Conference.2018. [Google Scholar]
- 43.Stenetorp P, Pyysalo S, Topic G, Ohta T, Ananiadou S, Tsujii JI. BRAT: a web-based tool for NLP-assisted text annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics; Association for Computational Linguistics; 2012. Apr 23, pp. 102–107. [Google Scholar]
- 44.Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM networks. InNeural Networks, 2005. IJCNN’05. Proceedings of the 2005 IEEE International Joint Conference; IEEE; 2005. Jul 31, pp. 2047–2052. [Google Scholar]
- 45.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. 2016. Neural architectures for named entity recognition. arXiv preprint arXiv:160301360. [Google Scholar]
- 46.Pennington J, Socher R, Manning C. editors. Glove: Global vectors for word representation. Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP).2014. [Google Scholar]
- 47.Roberts K. Proceedings of the Clinical Natural Language Processing Workshop (ClinicalNLP) 2016. Assessing the Corpus Size vs. Similarity Trade-off for Word Embeddings in Clinical NLP; pp. 54–63. [Google Scholar]
- 48.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M. TensorFlow: A System for Large-Scale Machine Learning. OSDI. 2016 Nov 2;Vol. 16:265–283. [Google Scholar]
- 49.Miwa M, Bansal M. 2016. End-to-end relation extraction using lstms on sequences and tree structures. arXiv preprint arXiv:160100770. [Google Scholar]
- 50.Li F, Zhang M, Fu G, Ji D. A neural joint model for entity and relation extraction from biomedical text. BMC bioinformatics. 2017;18(1):198. doi: 10.1186/s12859-017-1609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]