Abstract
HL7 Fast Healthcare Information Resources (FHIR) is rapidly becoming the de-facto standard for the exchange of clinical and healthcare related information. Major EHR vendors and healthcare providers are actively developing transformations between existing EHR databases and their corresponding FHIR representation. Many of these organizations are concurrently creating a second set of transformations from the same sources into integrated data repositories (IDRs). Considerable cost savings could be realized and overall quality could be improved were it possible to transformation primary FHIR EHR data directly into an IDR. We developed a FHIR to i2b2 transformation toolkit and evaluated the viability of such an approach.
Introduction
HL7 Fast Healthcare Information Resources (FHIR)1 is rapidly emerging as the de-facto standard for the interchange of healthcare related clinical information. EHR vendors and major healthcare providers are actively developing transformations between electronic health records (EHRs) and clinical data warehouses (CDW) to their corresponding FHIR representations2–7. At the same time, many of these organizations are creating another set of transformations from the same primary data onto Integrated Data Repositories (IDRs) for secondary use. While some of these organizations have created bespoke schemas tailored for the specific organization or institution8–11, others have chosen to collaboratively develop shared “integrative IDR schemas”12 such as the Informatics for Integrating Biology and Bedside (i2b2) star schema13, 14 and the Observational Medical Outcomes Partnership (OMOP) common data model(CDM)15. The emergence of the Shared Health Research Information Network (SHRINE)16 has led to the NIH NCATS project17, which has been developing the ACT network - “a nationwide network of sites that share EHR data”18. This community is in the process of developing of the ACT Common Data Model19 and the accompanying ACT SHRINE Query Ontology20.
We believe that significant benefit could be realized if these parallel efforts could be combined – if vendors and institutions could focus their resources on a single transformation between local data and their FHIR resource equivalents, while the FHIR and research communities produced a generalizable transformation between primary clinical data as represented in FHIR and a shared target IDR. We chose to focus our initial investigations on i2b2 because it used more of a “pure” Entity Attribute Value (EAV) model12 and, as such would be more amenable to a “metalevel” transformation, where transformation rules are specified between the model’s entities, attributes and values instead of what those elements represent: patient records, birthdates, genders, etc.. In addition, i2b2 based transformations had already been demonstrated from models closely related to FHIR including CCDA21,22, CDISC ODM23, OpenMRS24 and openEHR25 and from i2b2 to FHIR26. The Haarbrandt openEHR25 transformation is of particular interest as their approach is similar to the our own proposal. By specifying the transformation on the metamodel level, we produce a generic process that can represent any patient focused FHIR resource (e.g. Observation, DiagnosticReport, ImagingStudy, Careplan, RiskAssessment, DiagnosticReport, (genomics) Sequence, etc.) in a form amenable to secondary use. This approach allows the (necessarily) expensive and time consuming modeling effort to remain focused on primary clinical use cases, which then automatically can be made available for secondary use with only an incremental effort. Another approach that is potentially complimentary our proposal is a proposed implementation of i2b2 directly over a FHIR server27.
Material and methods
Materials
FHIR Specification The Fast Healthcare Interoperability Resources (FHIR)1 specification emerged in the 2012 timeframe as a response to the lack of adoption of the HL7 V3 specification. FHIR “…is a next generation standards framework created by HL7. FHIR combines the best features of HL7’s v2 , HL7 v3 and CDA product lines while leveraging the latest web standards and applying a tight focus on implementability”28. FHIR has developed a custom modeling language and methodology which is used by the FHIR community to define, as of the Standard for Trial Use 3 (STU3)29 release some 140 “Resource” definitions. Like many modeling environments, the models used in the tooling (i.e. the “metamodel”) are also represented in FHIR. FHIR resource definitions are represented as instances of the FHIR StructureDefinition resource, the model of which, in turn, is represented as an instance of itself.a FHIR initially defined two official data representation formats – XMLb and JSONc. The STU3 release proposed a third – the Resource Description Format (RDF)30.
FHIR RDF format The FHIR RDF interchange formatd specification states how FHIR instance data is to be represented in RDF as well as formally defining the complete set of RDF identifiers (URIs) used in this exchange, which turns out to be extremely useful. As Murphy noted in a 2011 presentation to the NCBO31, i2b2 has strong RDF underpinnings, and there is a strong similarity between the i2b2 concept_cd, modifier_cd, value pattern and the RDF subject predicate object equivalent – a fact that we were able to use to our advantage.
i2b2 i2b2 is an open-source clinical data analytics platform that provides a component-based architecture and a flexible analytical database design. The i2b2 repository provides an extensible framework allowing collaborative exchange of data including electronic health records, lab results, genetic and research data. The backend infrastructure is known as the “Hive”. The i2b2 data model employs the “star schema” dimensional analysis approach, with the observation_fact table at the center representing atomic assertions or “facts”, each of which, in turn, references elements in the accompanying dimension tables. The i2b2 dimension tables include the visit_dimension for information about encounters, the patient_dimension for baseline facts about the target patient, the provider_dimension for information about organizations and clinicians. The concept_dimension and modifier_dimension tables identify the particular “fact” itself (e.g. “patient age”, “systolic blood pressure”, “MCHV”). The i2b2 Hive is composed of six core cells – Project Management (PM), Data Repository (CRC), Ontology (Ont), Workplace (WORK), File Repository (FR), and Identity Management (IM)32. REST services implemented on top of each of these cells, allowing them to communicate with each other and external applications. The i2b2 softwaree comes pre-populated with a core ontology and sample data records.
FHIR sample datasets We used 2 sample datasets to evaluate the performed transformation. The first comes from SMART on FHIR, “a set of open specifications to integrate apps with Electronic Health Records, portals, Health Information Exchanges, and other Health IT systems”3. The SMART on FHIR platform provides the de-identified patients dataset for platform testing33. Our second dataset comes from the Synthea platform34, a synthetic patient population simulator which generates synthetic, realistic (but not real), patient data and associated health records for research and experiment usage35.
CTSA ACT ontology The CTSA ACT Network36 publishes an extensive i2b2 ontology to support shared demographics, diagnoses, laboratory tests, medications, procedures and visit details. We used Version 0.4 of this ontology, downloaded from the CTSA ACT Technology pagef to evaluate the generalizability of our transformations.
UNMC i2b2 metadata generator for SNOMED CT We used an unpublished tool developed by Jay Pedersen and Jim Campbell at the University of Nebraska Medical Center37 to transform subsets of SNOMED CT from the official RF2 distribution format into i2b2 Ontology. For the purposes of this experiment we transformed the SNOMED CT Allergic Condition (disorder) branch, consisting of 32,833 concepts from the January 2018 International Edition.
Methods
We developed two closely coupled software transformation tools. The first, loadfacts, transforms FHIR resource instances into their corresponding representation in the i2b2 CRC tables. The second, generate_i2b2 creates an i2b2 ontology hierarchy that reflects the FHIR resource model structure.
The loadfacts tool transforms FHIR resource instances represented in JSON or RDF into their i2b2 CRC table equivalents. FHIR patient references are recorded in the patient mapping table. The actual patient demographics in the FHIR Patient resource are recorded twice – once as a collection of individual facts in the observation_fact table and a second time as as the subset of facts that can be mapped to the patient_dimension tablea.
The provider_dimension table is intended to represent a hierarchy of organizations, practitioners and, (possibly) roles. We didn’t implement the FHIR to provider dimension transformation in this study, but anticipate that it could be used to represent a combination of the FHIR Practitioner, PractitionerRole and Organization resources.
The encounter_mapping and visit_dimension tables are intended to represent an aggregate “visit” or, according to the i2b2 web client dropdown, a “financial encounter”. In the long term, we would need to transform a combination of the FHIR Encounter and/or EpisodeOfCare resources to this table. In the short term, however, we found ourselves in need of one more “dimension”, and chose temporarily repurpose the visit_dimension for a different use. As noted by Haarbrandt25 and Husser12, the i2b2 model has a limited support for the hierarchical organization of information. The collection of facts for a given patient can be ordered by “event start date”b, “concept” and/or “financial encounter”. There is no obvious mechanism, however, to group information by “resource”, “order set” or other similar aggregation mechanisms. For this study, we need to show that both a white cell and a red cell count derive from the same specimen or the fact that a diastolic and systolic blood pressure are components of the same measurement session. The notion of Resource is integral to FHIR, meaning that we have to preserve and expose this organizational artifact in i2b2. While we believe that at least one more “dimension” will need to be added to the i2b2 model in the longer term, for purposes of this study, we use the i2b2 encounter_mapping as a proxy for a FHIR resource, with the encounter_ide column carrying the FHIR id component of the FHIR the resource and the encounter_ide_source the corresponding namespace. As an example, an instance of a FHIR Care-Plan resource with the URI http://example.org/fhir/CarePlan/e1172935 would have an encounter_ide of CarePlan/e1172935 and an encounter_ide_source of http://example.org/fhir/c.
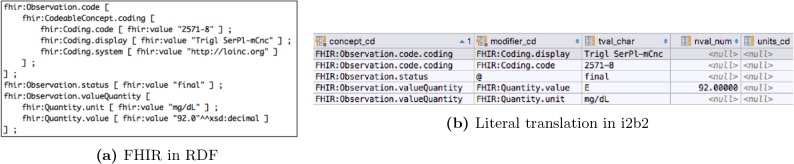
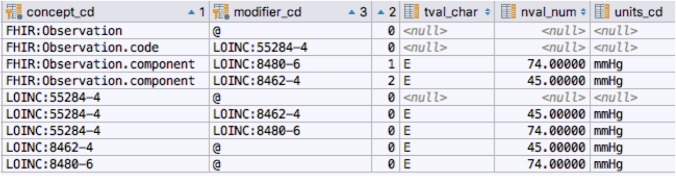
Literal mapping The FHIR RDF representation has already done the bulk of the work needed for the i2b2 loader. loadfacts creates an observation_fact concept_cd for each FHIR RDF value[x] predicate in the FHIR RDF representation. Figure 1 shows an fragment of an RDF FHIR Observation and its equivalent as observation_fact rows. The components of FHIR Quantity element are represented as i2b2 modifier codes. Some FHIR models include repeating groups. As an example, the Observation resource allows multiple component elements. i2b2 has the ability to represent one level of nesting through the instance_num attribute. Figure 2 shows how the i2b2 instance number (the third column on the right, labeled “2”) is used to represent the systolic and diastolic elements of a blood pressure observation.
Figure 1:
Literal transformation of FHIR RDF into i2b2
Figure 2:
Nested Observation components into i2b2
i2b2, however, only supports one level of repetition. FHIR Observation.component allows multiple occurrences of the ReferenceRange element within. Similarly, the AllergyIntolerance resource can include multiple reaction elements, each of which, in turn, can have multiple manifestation subcomponents. There is currently no way to represent these constructs in i2b2a
Secondary transformations Our goals in this study are twofold:
Determine whether it is possible to automatically transform a significant portion (ideally all) patient focused FHIR data into its i2b2 equivalent.
Determine whether it is possible to automatically enhance this transformation in a way that renders it (a) intuitive to an i2b2 user and (b) compatible with existing i2b2 ontologies such as CTSA ACT.
So far, all we have shown that goal (1) is achievable – by representing FHIR resources as EAV entries in the i2b2 tables. This step, when combined with the corresponding generate_i2b2 equivalent gives the end user the ability to query FHIR resources using the native FHIR Resource Model. We still need to speak to goal (2), however. i2b2 users, however, expect to ask about procedures, diagnoses, laboratory results etc. – not FHIR Observations, codes and quantity values. To meet these requirements, we need to augment the literal transformation by:
Representing “well known” FHIR coded concepts as i2b2 concept and modifier codes.
Identify implicit “tag/value” pairs in the FHIR information model and transform them to i2b2 code value entries.
Collapse the FHIR value[x] components into their i2b2 equivalents.
“Well Known” concept codes The established way of representing concept codes in the i2b2 space is the form of (Namespace):(code), where namespace represents the defining coding system. As an example, LOINC:2086-7 represents the HDL lipid test, ICD10:A05.1 botulism food poisoning, etc. We created a mapping from the FHIR Coding.system attribute to the i2b2 namespace equivalent. Every place a FHIR Coding or FHIR code element occurs we added an additional row with the actual code as the modifier code and, where nesting permitted, a second entry with with the code as the concept code.
Implicit tag/value pairs There are several places in the FHIR resource model where what is obviously intended to be a tag/value tuple is represented as sibling elements. The Observation resource, for example, uses the Observation.code to identify the observation and Observation.value[x] to record its value. In these situations we can combine the code and value into a single observation fact entry.
Collapse value components The i2b2 model supports a limited value representation. While it isn’t possible to represent more complex FHIR values like titers, ranges as observation entries, FHIR quantities, integers, strings, and dates can be collapsed into their i2b2 equivalents. One interesting outlier in this process are FHIR code values, where we have a choice of representing a code for, say an Observation.status as a string and using the FHIR metadata enumeration extension to allow the selection of possible values or to represent the code as a modifier. For the moment we do both.
Figure 3 shows how the LOINC codes for Blood Pressure, Systolic Blood Pressure and Diastolic Blood pressure have been added to the literal data shown earlier. These additions give us the ability to query by the entire observation, the observation code, the individual observation components or any combination thereof. In addition, the associated systolic and diastolic values have been mapped to their i2b2 equivalents. generate_i2b2 The loadfacts tool converts FHIR instance data into i2b2 observation fact and associated dimension entries. The job of the generate_i2b2 tool is to define a set of i2b2 ontology entries to expose and query the possible values. While loadfacts works with FHIR instance date, the generate_i2b2 module uses a subset of the FHIR Structure and Element definition resources, as represented in the FHIR Structure Vocabulary (FSV)a. The FSV specifies the name, type, domain and range of every element that can appear in a FHIR resource. generate_i2b2 transforms this information into a corresponding set of entries in the i2b2 ontology, concept_dimension and modifier_dimension tables. metadata_xml entries are added, where appropriate, to allow the specification of string, enumerated, numeric and date/time values where appropriate.
Figure 3:
Blood pressure observation concept and modifier codes
Results
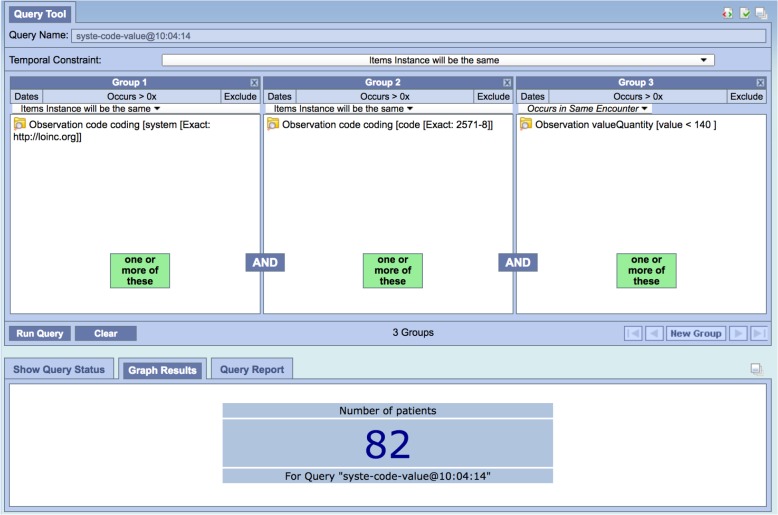
The literal mapping of the FHIR model made it possible for a FHIR expert to construct meaningful queries. Figure 4 shows a query for (FHIR) patients having one or more triglyceride results (LOINC 2571-8) whose values are less than 140 mg/dL. This query was run against our test target test and found 82 patients, as verified by accessing the source data directly. As mentioned earlier, this is not the sort of query that a researcher would want to use, as they would have to understand that the Observation resource carried laboratory results, that 2571-8 was the LOINC code for the triglycerides test, that http://loinc.org was the URI that FHIR used for LOINC, etc.
Figure 4:
Triglycerides < 140 using FHIR Resource model
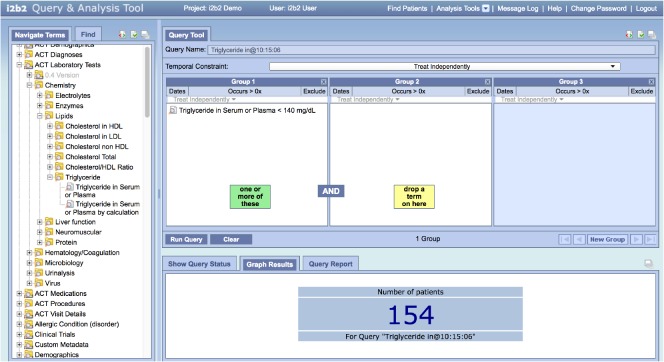
Figure 5 shows a similara query that utilizes secondary transformations described in the previous section. In this case we have used the ACT laboratory test ontology to select the test code and value. One will note, however, that this query is not identical to the previous query. It returned 154 patients vs. the earlier 82. The first query depended on FHIR Observation codes, something not present in the sample data that came with the i2b2 distribution. To match the first query exactly, we have to qualify this query with a requirement that the result is FHIR Observation as shown in Figure 6.
Figure 5:
Triglycerides < 140 using ACT Ontology
Figure 6:
FHIR Observation Triglycerides < 140 using ACT Ontology
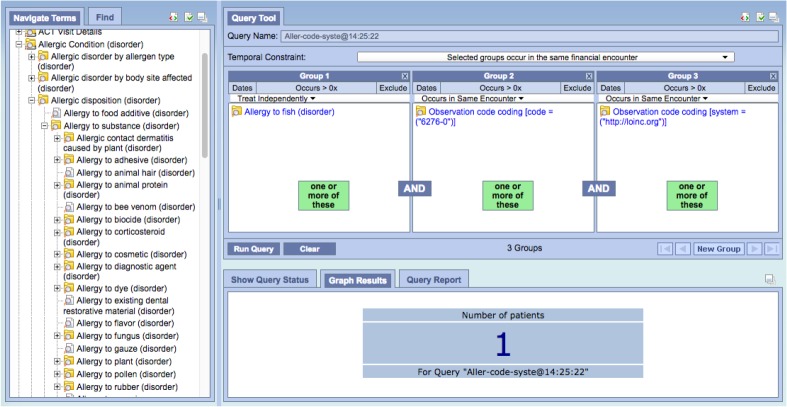
This leads to an interesting question: should we even load the “native” FHIR model or could we restrict the output to only the secondary transformations? We would argue that there is far more potentially relevant information in the FHIR models than are necessarily exposed in the accompanying ontologies. As an example, one might note that FHIR Observations have a status property that indicates whether the observation is preliminary or final, which leads to the question, “Have we been including preliminary observations in our queries?” While the long term answer is to expose this detail as a loader option, in the shorter term we can use a simple i2b2 query to count the number of patients having a status not equal to “final”. The other benefit of having the native FHIR ontology is that one can still construct queries before the “official” ontological infrastructure is in place. Figure 7 shows an example of such a query. In it we have asked for all patients diagnosed with fish allergy that also have taken an immunoglobulin E test for wheat antibodies. Note that i2b2 does not currently support an allergic reaction model. We were able to take advantage of the FHIR AllergyIntolerance resource which coded the allergies in SNOMED CT. We used the SNOMED CT Allergy ontology generated by the UNMC tool to provide a code selection list. Also note that V0.4 of the ACT ontology doesn’t carry the LOINC codes for IgE tests. In our case we added the LOINC code as a literal (“6276-0”). Obviously this would need to be an ontology entry in the longer term, but it serves to demonstrate the usefulness of the FHIR information in the absence of supporting ontologies
Figure 7:
AllergicReaction with wheat IgE test
Discussion
We set out to determine whether it would be possible to directly transform primary EHR data from standardized FHIR resources into an common IDR. We believe that we have been able to demonstrate that this is indeed possible. We have transformed FHIR sample data from a number of sources and have been able to construct meaningful queries against it. This process has exposed a myriad of things that still need to happen before day to day use of FHIR in i2b2 can be realized:
Additional i2b2 hierarchical grouping: We need to create a mechanism to represent arbitrary groups of information, as exemplified by the notion of “Resource”. This aspect will require careful planning, as FHIR has additional clustering levels such as Bundle and various mechanisms to incorporate cross resource references.
Multiple repeating group nesting: This is closely related to the previous item – we need a way to represent nested repeating values.
Alignment of FHIR profiles with i2b2 Ontology: At the moment, there is nothing in the FHIR core model that requires allergies reactions to be recorded using SNOMED CT or observation codes in LOINC. If we continue on this path, the i2b2 community will have to become an active participant in the FHIR modeling effort in order to be sure that the i2b2 ontologies align with those used in FHIR itself. In addition, a set of FHIR profiles will need to be identified that are considerably more deterministic than the core FHIR resource models.
Patient identifying information: FHIR resources may contain all sorts of identifying information in the form of comments, location information, names, etc. This issue will need to address and the appropriate filters and obfuscation mechanisms put into place before FHIR based i2b2 information could be shared beyond IRB protected environments.
FHIR value sets: A significant portion of the coded information in the FHIR models use FHIR specific coding systems. Observation.status, as described earlier is just one example. The tooling will need to be extended to represent these value sets as useful i2b2 ontologies.
Usability and performance: We have shown specific cases where FHIR sample data can be meaningfully queried in the i2b2 environment. From a performance perspective, however, sample queries directly against the FHIR model took between 3x and 4x times as long (~5.5 seconds for the FHIR approach vs. ~2.2 for the native). An obvious next step would be to construct some real world use cases and evaluate the usability, accuracy and performance features of this approach. We are guardedly optimistic from the performance perspective, as similar models such as Haarbrandt25 have already been shown to be acceptable.
Conclusion
We have demonstrated that it is possible to transform primary FHIR EHR data into an i2b2 IDR and that the resultant data can be represented and queried in a fashion that makes sense to the clinical researcher. Being able to do this means that it may no longer be necessary to maintain two separate modeling communities, one (FHIR) focused on the representation and exchange of primary EHR data and a second (ACT? NFACTS?) on secondary IDR information. In addition, it may be possible for individual vendors and organizations to focus exclusively on the transformation of bespoke clinical data into its FHIR equivalent and for the research community to develop a single transformation process from FHIR to its secondary IDR form.
Acknowledgements
This study is supported in part by NIH grants U01 HG009450 and U01 CA180940
The authors thank Jay Pedersen and Jim Campbell from University of Nebraska Medical Center for the i2b2 SNOMED CT metadata builder.
The loadfacts and generate_i2b2 toolkits can be found at https://github.com/BD2KOnFHIR/i2FHIRb2.
(All UR’s last referenced March 5, 2018 unless otherwise noted)
Footnotes
It has been noted that the patient_dimension table is redundant — any fact that can be recorded in this table can equally be represented as an observation fact. As there doesn’t appear to be widespread agreement on which to use, we currently load both forms. It should also be noted, however, that mapping from the FHIR Patient resource to the patient_dimension table is a non-trivial exercise.
As noted by Haarbrandt, the definition of “event start date” is not always obvious
Note that, as with the patient identifier, resource identifiers can be encrypted to prevent patient identification.
While this is a serious limitation, it should be noted that it only applies in the case where a repeating list of components occurs within another repeating list. In particular this situation does not affect repeating lists of data types (e.g. FHIR Codings within FHIR CodeableConcepts)
The equivalent query will be presented shortly.
References
- 1.FHIR® Release 3 (STU3); Available from: http://hl7.org/fhir/
- 2.Simplifier.net. 2018. Available from: https://simplifier.net.
- 3.SMART: Tech Stack for Health Apps. 2018. Available from: http://docs.smarthealthit.org/
- 4.Epic Systems Corporation: OpenEpic. 2018. Available from: https://open.epic.com/
- 5.Bresnick J. 2015. May, Epic, IBM Watson Embrace FHIR for Healthcare Big Data Analytics. HealthIT Analytics. [Google Scholar]
- 6.HAPI-FHIR: fhir made simple; Available from: http://hapifhir.io/
- 7.Cerner: Leverage the power of the HL7® FHIR®® standard in your SMART app; Available from: http://fhir.cerner.com/
- 8.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010;17(2):131–135. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE-An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009 Nov;2009:391–395. [PMC free article] [PubMed] [Google Scholar]
- 10.Wilcox AB, Vawdrey DK, Chen YH, Forman B, Hripcsak G. The evolving use of a clinical data repository: facilitating data access within an electronic medical record. AMIA Annu Symp Proc. 2009 Nov;2009:701–705. [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J. The DEDUCE Guided Query tool: providing simplified access to clinical data for research and quality improvement. J Biomed Inform. 2011 Apr;44(2):266–276. doi: 10.1016/j.jbi.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huser V, Cimino JJ. Desiderata for healthcare integrated data repositories based on architectural comparison of three public repositories. AMIA Annu Symp Proc. 2013;2013:648–656. [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, et al. 2007. Oct, Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc; pp. 548–552. [PMC free article] [PubMed] [Google Scholar]
- 14.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc. 2012;19(2):181–185. doi: 10.1136/amiajnl-2011-000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stang PE, Ryan PB, Racoosin JA, Overhage JM, Hartzema AG, Reich C, et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Ann Intern Med. 2010 Nov;153(9):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 16.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009;16(5):624–630. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinstein YR, McInnes P. NIH/NCATS/GRDR® Common Data Elements: A leading force for standardized data collection. Contemp Clin Trials. 2015 May;42:78–80. doi: 10.1016/j.cct.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ACT Network; Available from: http://www.act-network.org/
- 19.ACT Common Data Model v1.3.docx. https://ncatswiki.dbmi.pitt.edu/acts/attachment/wiki/DataHarmonization/ACT/20Common/20Data/20Model/20v1.3.docx.
- 20.ACT SHRINE Query Ontology v1.3. 2018. https://ncatswiki.dbmi.pitt.edu/acts/raw-attachment/wiki/DataHarmonization/ACT/20SHRINE/20Query/20Ontology/20v1.3.docx.
- 21.Klann JG, Mendis M, Phillips LC, Goodson AP, Rocha BH, Goldberg HS, et al. Taking advantage of continuity of care documents to populate a research repository. J Am Med Inform Assoc. 2015 Mar;22(2):370–379. doi: 10.1136/amiajnl-2014-003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majeed RW, Rohrig R. Automated realtime data import for the i2b2 clinical data warehouse: introducing the HL7 ETL cell. Stud Health Technol Inform. 2012;180:270–274. [PubMed] [Google Scholar]
- 23.Bauer CR, Ganslandt T, Baum B, Christoph J, Engel I, Lobe M, et al. Integrated Data Repository Toolkit (IDRT). A Suite of Programs to Facilitate Health Analytics on Heterogeneous Medical Data. Methods Inf Med. 2016;55(2):125–135. doi: 10.3414/ME15-01-0082. [DOI] [PubMed] [Google Scholar]
- 24.Fomunyam T, Symonds J, Lorenz S. Designing a Public Health Software Framework: Porting OpenMRS Data to i2b2; Available from: https://wiki.openmrs.org/display/docs/I2B2+Export+Module.
- 25.Haarbrandt B, Tute E, Marschollek M. Automated population of an i2b2 clinical data warehouse from an openEHR-based data repository. J Biomed Inform. 2016 Oct;63:277–294. doi: 10.1016/j.jbi.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Boussadi A, Zapletal E. A Fast Healthcare Interoperability Resources (FHIR) layer implemented over i2b2. BMC Med Inform Decis Mak. 2017 Aug;17(1):120. doi: 10.1186/s12911-017-0513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paris N, Mendis M, Daniel C, Murphy S, Tannier X, Zweigenbaum P. i2b2 implemented over SMART-on-FHIR. 2018;2017:369–378. [PMC free article] [PubMed] [Google Scholar]
- 28.Introducing HL7 FHIR. 2018. Available from: https://hl7.org/fhir/summary.html.
- 29.FHIR® Release 3 (STU); Available from: http://hl7.org/fhir/STU3/index.html.
- 30.Schreiber G, Raimond Y. editors. RDF 1.1 Primer. W3C Working Group Note 24 June 2014. 2014. Jun, Available from: https://www.w3.org/TR/rdf11-primer/
- 31.Murphy SN, Phillips L. i2b2 – NCBO Collaboration to Provide i2b2 Ontology Services; Available from: https://www.i2b2.org/events/slides/i2b2_OntologyTalk_20110629_Murphy.pdf.
- 32.Informatics for integrating biology and the bedside (i2b2); 2018. Available from: http://www.i2b2.org. [DOI] [PMC free article] [PubMed]
- 33.Core SMART Patients. Secondary Core SMART Patients. Available from: http://docs.smarthealthit.org/data/dstu2-sandbox-data.html.
- 34.SYNTHEATM: Synthetic Patient Generation; 2018. Available from: https://synthetichealth.github.io/synthea/
- 35.Post AR, Pai AK, Willard R, May BJ, West AC, Agravat S, et al. Metadata-driven Clinical Data Loading into i2b2 for Clinical and Translational Science Institutes. AMIA Jt Summits Transl Sci Proc. 2016;2016:184–193. [PMC free article] [PubMed] [Google Scholar]
- 36.CTSActs Act Network Home Page; 2018. Available from: https://www.act-network.org/
- 37.Campbell JR, Campbell WS, Hickman H, Pedersen J, McClay J. Employing complex polyhierarchical ontologies and promoting interoperability of i2b2 data systems. AMIA Annu Symp Proc. 2015;2015:359–365. [PMC free article] [PubMed] [Google Scholar]