Abstract
We present the outcome of an annotation effort targeting the content-sensitive segmentation of German clinical reports into sections. We recruited an annotation team of up to eight medical students to annotate a clinical text corpus on a sentence-by-sentence basis in four pre-annotation iterations and one final main annotation step. The annotation scheme we came up with adheres to categories developed for clinical documents in the HL7-CDA (Clinical Document Architecture) standard for section headings. Once the scheme became stable, we ran the main annotation campaign on the complete set of roughly 1,000 clinical documents. Due to its reliance on the CDA standard, the annotation scheme allows the integration of legacy and newly produced clinical documents within a common pipeline. We then made direct use of the annotations by training a baseline classifier to automatically identify sections in clinical reports.
Introduction
Clinical decision support has become increasingly dependent on hospital information systems (HIS), the Electronic Medical Record (EMR), in particular. The data contained in such repositories are either structured (e.g., measurements taken from all sorts of technical devices such as blood pressure, body weight, temperature, etc.) or unstructured (basically, all sorts of clinical reports and free-text notes). The added value of taking the latter data into account for clinical decision making, as well as for clinical and translational research, has repeatedly been shown. This holds true especially for large biomaterial collections (biobanks) where samples from selected phenotypes have to be collected and linked to clinical context information in order to support healthcare-integrated biobanking activities. However, automated workflows for the extraction and acquisition of high-quality clinical phenotype data, as a prerequisite for the proper selection of respective samples for high-quality biobanking, are still missing.
Since the extraction of information from unstructured documents cannot be reasonably delegated to humans (e.g., medical documentation officers) due to the enormous volume of already existing and the continuously growing number of clinical documents, the automatic analysis of clinical text by natural language processing (NLP) systems has become a viable alternative. While substantial progress has already been made, lots of problems remain because of the intrinsic complexity and non-canonicity of clinical language (e.g., abbreviations, acronyms, paragrammaticality, and the ambiguity resulting therefrom).
Some of these problems can already be resolved by properly contextualizing patient-relevant information in clinical reports. For instance, a phrase such as “Patient suffered from severe rash.” will be highly relevant for taking immediate actions if it occurs in the Admission Diagnosis, but will be far less relevant if it occurs in the Anamnesis or even Family History section. Hence, the categorization of single assertions according to clinically relevant context-defining episodes of the patient (anamnesis, diagnosis, therapy, medication, etc.) is of primary importance. This task is usually referred to in clinical NLP as section heading classification. Edinger et al.1 have already shown that retrieving patient cohorts based on searching sections rather than whole clinical documents increases precision and overall F-score, yet decreases recall.
One might argue that manually assigned section classifications already exist in clinical reports and thus should simply be reused. Yet even though medical practitioners already use subheadings to divide clinical documents into paragraphs, this division is not necessarily useful from a data quality point of view, since doctors often segment text portions in an arbitrary and even inconsistent manner1. It is also quite common for medical practitioners to not use any headings at all, i.e., their document divisions are incomplete. During our study we even found sentences and phrases which did not belong to the category they should according to the meaning of the subheading they were assigned to; such lost sentences are abundant and constitute a major source of false assertion assignments.
It thus becomes obvious that it will not suffice to resort to given headings, if one wants to properly contextualize and thus disambiguate the meaning of assertions in clinical documents. Neither will it be enough to merely annotate given paragraphs with certain headings since this level of granularity seems overly coarse. We therefore refrained from having our annotation team work on the paragraph level and instead instructed them to categorize each sentence of the corpus of medical documents independently.
Section classification is further hampered by the fact that, almost up to now (in Germany), each hospital and even each hospital department follows individual (though partially overlapping) site-specific section category schemes. Fortunately, in the meantime, an XML-based HL7 interoperability standard (CDA) has been defined but it is still not fully operational (in Germany, at least). So, in the near future, CDA-compliant document structures will be enforced by any clinical documentation activity. Accordingly, clinical NLP systems should already implement this de-facto standard and be prepared to process legacy documents which do not adhere to the CDA regulations.
The following paper documents the process of developing a list of useful categories for the annotation of CDA-compliant sections in discharge summaries and related documents—the result of which is an annotation guide for the annotation of sections in German discharge summaries and related documents. Based on these conventions, we further introduce a corpus of clinical documents in a section-annotated form as well as a baseline classifier to automatically label sections.
Related Work
Cho et al.2 worked with medical documents created in U.S. hospitals through dictate (by practitioner) and subsequent transcription by a specialist in clinical documentation, structuring the data into paragraphs and providing most of them with labels (headers). In a two-step process, their algorithm finds section boundaries and subsequently assigns labels to these sections, detecting both segmentations that already have a header and those that have not. In contrast to that, our data is less structured as the discharge summaries are directly written by practitioners (see the section ‘Data Extraction and Preparation’ for more information on the structure and idiosyncrasies of our corpus).
Apostolova et al.3 describe the construction of a segmentation classifier for 215,000 radiology reports created in U.S. hospitals. They define section labels such as Demographics containing basic data about the patient, History including the reason for treatment, Comparison, Technique with all procedures, Findings giving all observations, Impressions containing diagnosis and conclusion, Recommendations for future treatment and Sign off. Those section labels served as a first basis for our definitions of potential section labels.
In a data-driven approach, Denny et al.4 devise a new hierarchical terminology for the tagging of section headers (Clinical Document Sections Ontology (CDSO)), based on QMR (Quick Medical Reference),a LOINC and the help of clinicians and clinical textbooks which, amongst others, provided us with a basis for the development of the categories we used for our section annotation. Their corpus consists of template-based and thus highly structured data, dictation, hand-written notes and .doc files without any template. For our study, we follow their definition of a section as a “clinically meaningful grouping of synonyms, history, findings, results, or clinical reasoning that is not itself part of the unique narrative for a patient” and a section header as a phrase that “includes words that provide context for the encapsulated text but whose words themselves do not add specific clinical information”.4[p.157]
In a follow-up study, Denny et al.5 developed the SecTag algorithm which is used to identify both labeled and unlabeled sections. Their choice of section headers is based on “frequently used but nonstandardized terms” used as headers for paragraphs in clinical documents. However, it must be noted that, according to the authors, in the U.S. medical students are explicitly trained to write clinical documents such as discharge summaries. The authors thus assume some standard and logical sequence (i.e., from head to toe, from the beginning of treatment to its end) in the segmentation of these documents. This does not hold true for German medical education and thus further confuses our data. Nevertheless, we oriented the development of the categories for our annotation task on their findings as well.
Based on the findings of Denny et al.4, Tran et al.6 describe their Automated Section Annotator (OBSecAn) trained on semi-structured documents from Veterans Affairs hospitals, which can identify hierarchical relationships. For training purposes, they used a corpus of approximately one million clinical notes. To find unlabeled sections, they used Denny et al.’s CDSO4. OBSecAn operates in three steps. First, it reads and parses the documents to get sections with their hierarchy, then it detects and corrects errors and, finally, it generates the output with a parsed tree that contains nested sections. As an example, the authors evaluate the performance of the identification of Family History sections and show up with an accuracy of 99% for this use case.
Apart from the clues given within the textual data itself, Haug et al.7 also consider the formatting of a text to locate section boundaries. They are aware of the high variability of both headers and content of sections and thus asked their annotators to take into account both the header of a section and its content to devise topic identifiers.
Li et al.8 describe a sequence-based model with Hidden Markov Models to find sections and categorize them into 15 medical categories. They worked on a dataset of approximately 10,000 clinical notes from a U.S. hospital. Their corpus consists of different types of notes (e.g., consultation and follow-up notes) and clinical summaries. They also examined the structure of the documents: 33% of the sections do not contain any header at all.
Tepper et al.9 take into account that templates for discharge summaries are not binding and that “clinicians do not follow strict section naming conventions.” In contrast to earlier works, they do not work on the paragraph level but asked annotators to mark the line of a section header (i.e., define its location) within a clinical document and select one out of 33 section categories drawn from an ontology. With this data, the authors trained a line-based classifier to determine a section category label for each line of a document. As we found that there are many “lost sentences” in our data (see section ‘Data Extraction and Preparation’), we applied a similar approach and had our annotators work on the sentence-level.
A general supervised approach using an l1 -regularized multi-class Logistic Regression model to handle clinical text segmentation was published by Ganesan et al.10 The focus of the identified sections lies on Header, Footer and top-level sections, containing, for example, Allergies and Chief Complaint. The authors set contraints for the lines of a document, i.e., every line of the documents is either BeginHeader, ContHeader, BeginSections, ContSection or Footer. They further define rules that apply to these five elements, e.g., the first line always constitutes the beginning of a header (BeginHeader) or the beginning of a section (BeginSection).
Research on the annotation of sections is rare for clinical documents outside the U.S. and languages other than English. For French, Deleger et al.11 provide a basic study separating the content of clinical documents from administrative information at the beginning and the end of a text, yet they do not provide any further assistance for section classification proper. For Hungarian, Orosz et al.12 developed a hybrid text segmentation tool for clinical documents that combines a rule-based and an unsupervised statistical solution and performs above 90% F-score.
Data Extraction and Preparation
For the section annotation experiments, our annotation team worked on 1,106 discharge summaries and similar document types (short summaries and transfer letters), the Jena segment of a corpus of roughly 3,000 German clinical documents from three different German university hospital sites (3000PA13). Based on the approval by the local ethics committee (4639-12/15) and the data protection officer of Jena University Hospital documents were extracted from the HIS of the Jena University Hospital and further transformed.
The extraction workflow consisted of the following steps: We first sampled and selected documents contained in the HIS in a proprietary data format (using SAP BUSINESS WAREHOUSE) according to a defined set of inclusion criteria (all patients deceased, treated between 2010 and 2015, at least 5 days of hospital stay, treated internally or intensively, etc.). Next, we transformed those documents into .doc files with a predefined naming scheme using TALEND OPEN STUDIOb, and, finally, converted the .doc files into .txt files using APACHE POI.c As an initial preprocessing step, we automatically cut off the laboratory values from the documents because they are merely an (often inconsistent or incomplete) summary/copy of data from the structured data set from the HIS and do not contain the complete list of all laboratory values of the treatments. Table structures were retained with the character ‘|’ followed by four space characters as a delimiter for cell boundaries. The corpus altogether consists of 170,539 sentences, 760 of them are cell boundaries of tables.
Annotation of the Corpus
We will now describe in detail the procedure how we managed to arrive at a medically useful and reliable annotation of sections for German-language clinical reports. The steps we went through reflect a trial-and-error procedure likely to occur in other clinical environments as well. Especially, we tried to find empirical evidence for
the proper annotation unit (paragraph or sentence), and
the proper annotation language (set of section categories).
Such an experience report might, in the future, help and possibly speed up annotation campaigns at other clinical sites.
Manual Annotation of Sentences. In an annotation task preceding section annotation, we asked a team of five annotators to split up clinical documents into tokens and sentences—only the latter being of importance for the task at hand, since we used the sentence splits to provide proper annotation units for the sections. For a pre-segmentation of sentences, we used the UIMA-based tool suite JCORE,14d together with publicly available models trained on the confidential German clinical FRAMED corpus.15
Due to the special structure and writing style of discharge summaries, some specific instructions needed to be formulated for the sentence segmentation, including but not limited to cases for
the handling of colons (sometimes they terminated a sentence, sometimes they did not),
ongoing diagnosis sections running over multiple lines without punctuation marks,
medication lists,
listings, in general.
Annotation Language. For the major annotation study, the target annotation language consists of a (human-made) classification system for distinguishing different segments of German clinical texts (discharge summaries and closely related documents). The categories we used for our main annotation effort are based on the relevant literature and the Health Level 7 (HL7)e standard Clinical Document Architecture (CDA)16, especially relying on the implementation guide for German discharge letters, Arztbrief PLUS.f They were refined in the course of an iterative process with the support of experienced and highly qualified practitioners from the Jena University Hospital.
CDA is an XML-based healthcare interoperability standard for the structuring of clinical documents, as well as their exchange, and contains a definition system for segments, the CDA Section Level Templates. It is complemented by rules for the structured composition of medical documents—the section headings as well as the content of these sections (in the form of free text or structured and semantically annotated entries). These content-related rules are usually prepared by medical experts in the form of implementation guides defining the structure and content of specific document types (such as discharge letters). These conventions are not yet legally binding in Germany up until now, but on the rise as a future standard in the medical sector.g
Without the use of consented document templates based on interoperability standards such as CDA, medical practitioners themselves decide on the form and content of sections they want to include in their documents or are merely bound by those standards specific to their particular institution (not counting some codified or uncodified rules regarding the sequence and content of sections). Today, some hospitals already follow the CDA standard in the preparation of medical reports. However, legacy documents written before the propagation of CDA do not fit those standards but still contain information that might be of utmost importance for the treatment of patients, as well as for research purposes.
Annotation Tool. For our annotation campaign, we used the Web Annotation Tool for Segment Labeling (WAT-SL).17h It is similar to the well-known BRAT annotation tool18i, but—since we encountered systematic problems with BRAT when longer stretches of text incorporating several lines of a clinical document had to be annotated—was developed especially for the annotation of text segments and thus fits our task best. Together with the original development team of WAT-SL we added several features to the tool. As these extensions not only helped us with our work but might be of general interest for the scientific community, they will be presented in more detail in a forthcoming publication.
We configured WAT-SL according to our specific requirements, i.e., the annotation of discharge summaries and closely related documents. Each annotator was assigned a personal WAT-SL profile, enabling us to monitor each annotator’s work progress and specific decisions. Each annotator was provided with several tasks per round, each task consisted of one clinical document (discharge summary, transfer letter or short summary).
Annotation Iteration Rounds. In an iterative process, we developed annotation guidelines for the segmentation of clinical texts, incorporating the CDA implementation guide for discharge letters as much as possible while retaining a sufficiently high agreement between our annotators (necessary for subsequent NLP applications). In four preliminary rounds and one main annotation cycle, we ran our annotation campaign with eight annotators (all medical students after their first medical licensing exam).
As can be seen from Table 1, we assigned our annotation team overall 30 or 50 annotation tasks per round, respectively. Throughout the iterations, we first tested for the usefulness of several possible options for our annotation task. We especially tried to tackle the problem of lost sentences, i.e., sentences that content-wise do not belong to the section they actually appear in according to the meaning of the section’s header. As these sentences belong to another section as the header indicates, they thus are contained in the wrong section. Surprisingly, these sentences appear to be significantly frequent.
Table 1:
Experimental set-ups of the four pre-annotation rounds and the main annotation round (multiply annotated documents only) with count of documents (Doc.) and annotation decisions to be made (Items)
| Iteration 1 | Iteration 2 | Iteration 3 | Iteration 4 | Main Annotation | |
|---|---|---|---|---|---|
| Doc. Items | Doc. Items | Doc. Items | Doc. Items | Doc. Items | |
| Raw data | 10 2,071 | - | - | - | - |
| Paragraphs | 10 329 | 15 1,856 | - | - | - |
| JCoRe & Framed | 10 3,010 | 15 5,225 | - | - | - |
| Manual sentence annotation | - | 20 4,256 | 49 7,492 | 50 7,314 | 50 7,394 |
| £ per annotator | 30 5,410 | 50 11,337 | 49 7,492 | 50 7,314 | 50 7,394 |
| £ all annotators | 240 43,280 | 400 90,696 | 396 59,936 | 400 58,512 | 400 59,152 |
In the first two iterations, we thus had our annotators work on diverse versions of the data: 10/15 documents were pre-divided into paragraphs, i.e., our annotation team had to decide for each paragraph into which category it should belong and 10/15 documents were divided into sentences using the FRAMED model for sentence segmentation with JCORE components. In the first iteration, we also provided our annotators with “raw” data, i.e., they worked on data as they were exported from the original data using data split into sentences: Either, just as in iteration 1, with the help of JCORE components, or by manual annotation (see subsection ‘Manual Annotation of Sentences’, above).
We could confirm the stipulation that lost sentences as defined above were rather frequent and thus refrained from annotating on the paragraph level in the subsequent iterations. As the manually annotated sentences showed a higher quality, we decided to continue iteration 3 and 4 as well as the main annotation round with documents divided into sentences.j
Besides determining the right granularity level for units to be annotated, we used the four pre-annotation iterations to develop a list of content categories for the annotation of sections in discharge summaries and related documents (see Table 2). The first round of the four iterations was meant to be a more or less preliminary one as five members of the annotation team were not used to this specific task and three members were new to working as annotators altogether. Nevertheless, we incorporated findings about the quality of the annotations, as reflected in the time needed for each task per annotator and the inter-annotator agreement (IAA), into the design of the following iterations. As the measure for IAA we used Krippendorff’s alpha.19
Table 2:
Categories used in four pre-annotation rounds and the final main annotation round
| Iteration 1 | Iteration 2 | Iteration 3 | Iteration 4 | Main Annotation |
|---|---|---|---|---|
| 1. Preamble | 1. Preamble | 1. Salutation | 1. Salutation | 1. Salutation |
| 2. Anamnesis | 2. Anamnesis | 2. Reason For Referral | 2. Anamnesis | 2. Anamnesis |
| 3. Diagnostics | 3. Diagnostics | 3. History of present illness | (a) Patient history | (a) Patient history |
| 4. Therapy | 4. Therapy | 4. History of past illness | (b) Family history | (b) Family history |
| 5. Future | 5. Future | 5. Family history | 3. Diagnosis | 3. Diagnosis |
| 6. Appendix | 6. Appendix | 6. Hospital discharge studies sum. | (a) Admission diagnosis | (a) Admission diagnosis |
| 7. Mix | 7. Laboratory Result Observation | (b) Discharge diagnosis | (b) Discharge diagnosis | |
| 8. Admission diagnosis | 4. Hospital discharge studies sum. | 4. Hospital discharge studies sum. | ||
| 9. Discharge diagnosis | 5. Procedures | 5. Procedures | ||
| 10. Procedures | 6. Allergies intolerances risks | 6. Allergies intolerances risks | ||
| 11. Allergies intolerances risks | 7. Medication | 7. Medication | ||
| 12. Admission medication | (a) Admission medication | (a) Admission medication | ||
| 13. Medication during stay | (b) Medication during stay | (b) Medication during stay | ||
| 14. Discharge medication | (c) Discharge medication | (c) Discharge medication | ||
| 15. Remedies and Aids | 8. Hospital course | 8. Hospital course | ||
| 16. Immunizations | 9. Plan of care | 9. Plan of care | ||
| 17. Hospital course | 10. Final remarks | 10. Final remarks | ||
| 18. Plan of care | 11. Supplements | 11. Supplements | ||
| 19. Final remarks | 12. Mix | |||
| 20. Supplements | ||||
| 21. Mix |
In iteration 1, we started with 6 categories taken from the literature (see section ‘Related Work’): Preamble, Anamnesis, Diagnostics, Therapy, Future, Appendix. In iteration 2, we continued to use these categories but included a Mix category for those sentences that did not fully fit into one or the other category. The annotators were asked to give an explanation for these hard-to-decide categories in a “comments” field we provided. We were thus able to study which categories often co-occur in one sentence and, thereby, find out which categories might be overlapping or inconclusive in their definition. This proved even more useful for the following iteration where we introduced the categories from the CDA standard.
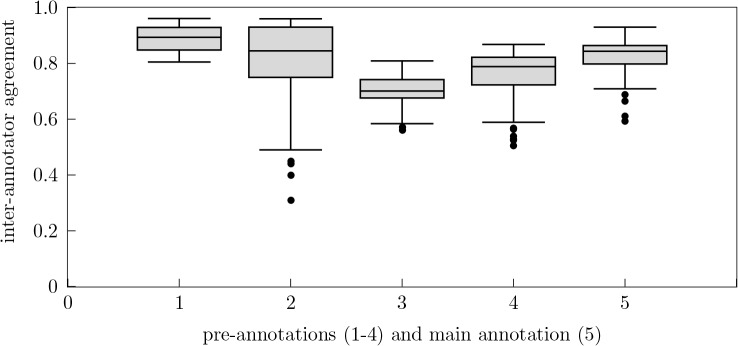
As the categories we had used so far to annotate the sections did not match the categories from the CDA specifications, in iteration 3, we chose to use the complete list of section headings of the German CDA implementation guide Arztbrief PLUS.k The categories given in the CDA implementation guide are meant to be as general as possible. Furthermore, it is not intended as a standard for reading clinical documents but rather for summarizing them. Thus, for the third iteration, we expected a loss in IAA and an increase in the time our annotators would need to annotate the 50 documents they were given. As can be seen from Table 3, this was indeed the case as Krippendorff’s alpha fell dramatically; the average value dropped from μα1 = 0.892 and μα2 = 0.795 to μα3 = 0.701. In addition, our annotators reported that, due to ambiguities and overlaps within the standard, it became extremely difficult to annotate the documents on the basis of these categories. This underlined the need to revise segment categories on the basis of the CDA and customize them to our dataset and our task of (automatically) reading clinical documents.
Table 3:
Inter-annotator agreement (measured by Krippendorff’s Alpha); we calculated the IAA for each document over all eight annotators
| Iteration 1 | Iteration 2 | Iteration 3 | Iteration 4 | Main Annotation | |
|---|---|---|---|---|---|
| μα | 0.892 | 0.795 | 0.701 | 0.752 | 0.821 |
| σα | 0.050 | 0.156 | 0.056 | 0.103 | 0.072 |
| minα | 0.805 | 0.313 | 0.562 | 0.506 | 0.594 |
| Lower Quartile | 0.848 | 0.755 | 0.675 | 0.730 | 0.799 |
| Median | 0.896 | 0.846 | 0.701 | 0.789 | 0.844 |
| Upper Quartile | 0.929 | 0.919 | 0.741 | 0.820 | 0.864 |
| maxα | 0.961 | 0.960 | 0.809 | 0.868 | 0.930 |
In iteration 4, we therefore took a selection from the CDA standard collaborating with expert staff from the Jena University Hospital. In contrast to the iterations before, here it was possible for our annotators to label sub-categories that were semantically more specific than their associated more general categories. We conceptually subsumed the categories Reason For Referral, History of Past Illness and Family History under the label Anamnesis, which could be specialized by the sub-categories Patient History and Family History. The categories Admission Diagnosis and Discharge Diagnosis were included as sub-categories of the category Diagnosis. The categories Admission Medication, Medication during Stay and Discharge Medication were defined as sub-categories of the Medication category. As they do not play a role for our work, we omitted the categories Remedies and Aids and Supplements altogether. The category Laboratory Result Observation was merged with Hospital Discharge Studies Summary and the section Immunizations was included in the sub-categories of Medications. The rearrangement of CDA section headings of iteration 4 yielded much better annotation performance as the IAA values increased up to μα4 = 0.752.
Main Annotation. With the input from all four iterations, we set up the final annotation guidelines and issued them to our annotation team for the main annotation cycle of the complete corpus. Just as in the fourth pre-annotation round, these guidelines describe labels and contents for eleven main categories and fine-tuned sub-categories for Anamnesis, Diagnosis and Medication. However, as the Mix category had barely been used in the final iteration (5 times, in total, in comparison to 224 in iteration 2 and 674 in iteration 3, respectively).l we considered our categories to be sufficiently well delineated overall and excluded it from further consideration.
For the main annotation round, we randomly chose 50 documents which had to be annotated by all eight annotators (to be compared later) and 132 or 133 additional documents, respectively, comprising one-eighth of the overall corpus. Thus, for the main annotation, each annotator was given either 182 or 183 documents, respectively. The average IAA of the 50 multiply annotated documents taken from the overall corpus (roughly 5%) amounts to μαmain = 0.821. This can be taken as an indication for the high quality of the whole annotation effort. From the 50 multiply annotated documents we randomly chose one document and added it to the overall corpus again. We thus come up with a corpus of 1,106 annotated documents, with 170,539 classified sentences and 17,848 related segments. Table 4 gives an overview on the frequency of categories and the amount of their sections within the corpus.
Table 4:
Annotated categories with their count of the main annotation, the count of segments and Precision, Recall and F-Scores of a baseline classifier
| Annotation Categories | Annotated Items | Count of Segments | Precision | Recall | F-score |
|---|---|---|---|---|---|
| Salutation | 12,913 (7.57%) | 977 (5.47%) | 0.84 | 0.86 | 0.85 |
| Anamnesis | 567 (0.33%) | 304 (1.70%) | 0.10 | 0.01 | 0.02 |
| Patient history | 6,033 (3.54%) | 948 (5.31%) | 0.66 | 0.63 | 0.65 |
| Family history | 28 (0.02%) | 14 (0.08%) | 0.00 | 0.00 | 0.00 |
| Diagnosis | 3,984 (2.34%) | 1,826 (10.23%) | 0.50 | 0.40 | 0.45 |
| Admission diagnosis | 9,235 (5.42%) | 1,568 (8.79%) | 0.62 | 0.60 | 0.61 |
| Discharge diagnosis | 4,788 (2.81%) | 1,432 (8.02%) | 0.46 | 0.33 | 0.38 |
| Hospital discharge studies summary | 87,116 (51.08%) | 1,920 (10.76%) | 0.89 | 0.96 | 0.93 |
| Procedures | 3,907 (2.29%) | 618 (3.46%) | 0.38 | 0.15 | 0.21 |
| Allergies intolerances risks | 188 (0.11%) | 133 (0.75%) | 0.53 | 0.36 | 0.43 |
| Medication | 409 (0.24%) | 135 (0.76%) | 0.13 | 0.02 | 0.04 |
| Admission medication | 53 (0.03%) | 21 (0.12%) | 1.00 | 0.12 | 0.22 |
| Medication during stay | 490 (0.29%) | 321 (1.80%) | 0.20 | 0.09 | 0.12 |
| Discharge medication | 11,636 (6.82%) | 1,069 (5.99%) | 0.93 | 0.94 | 0.94 |
| Hospital course | 19,770 (11.59%) | 2,914 (16.33%) | 0.82 | 0.84 | 0.83 |
| Plan of care | 3,610 (2.12%) | 2,119 (11.87%) | 0.69 | 0.68 | 0.68 |
| Final remarks | 4,810 (2.82%) | 1,135 (6.36%) | 0.98 | 0.98 | 0.98 |
| Supplements | 1002 (0.59%) | 394 (2.21%) | 0.80 | 0.25 | 0.38 |
| All Categories | 170,539 | 17,848 | 0.82 | 0.84 | 0.82 |
Baseline Classifier
After the main annotation round, we used the annotated data to train a baseline classifier. As features we extracted a bag-of-words statistics and used a logistic regression model with a 10-fold cross-validation for classification. This approach resulted in an average accuracy of 83.7% (+/- 1%). Our classifier reached an average F-score of 0.82 with a precision of 0.82 and a recall of 0.84. The F-score, precision and recall values of all individual categories are depicted in Table 4.
As can also be seen from Table 4, some categories barely appear in the given corpus (Family History, supercategory Medication). As expected, the classifier does not perform well for these categories (F-score ≈ 0). In the future, a more elaborate corpus might help solve this issue. As a baseline, however, the annotation occurrences of Salutation, Discharge Medication, Hospital Course with approximately 7%–12% of the whole corpus achieved reasonable F-score values in the range of 0.83–0.94; Final remarks with roughly 3% achieved an F-Score of 0.98. The section category Hospital Discharge Studies Summary which makes up more than half of the collection reached an F-score 0.93. These numbers compare nicely with results reported in the literature, e.g., by Denny et al.5 though they are not directly comparable (different languages, text genres, hospitals, country- and community-specific reporting habits).
Conclusion
We presented the evolution of an annotation scheme for German clinical discharge summaries, short summaries and transfer letters. Our focus was on narrowing down the appropriate granularity of the annotation unit (our choice: single sentences) and the set of relevant and feasible categories and their internal structure (our choice: see Table 2) compliant with HL7 CDA discharge letter section requirements.
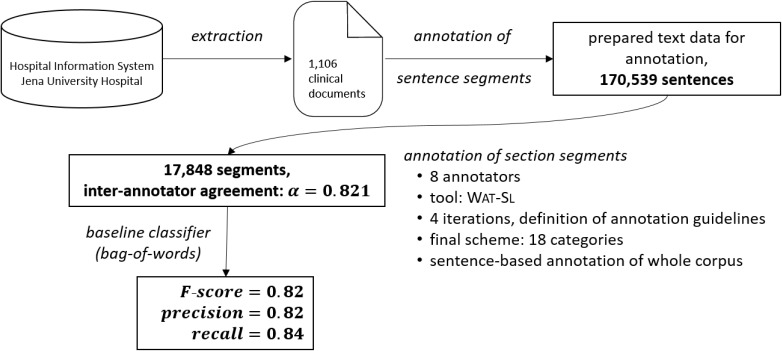
We then applied the final scheme in a manual annotation experiment to 1,106 German-language clinical documents from the years 2010 to 2015. The outcome constitutes the first section-annotated corpus of German discharge summaries and related documents. We further applied our categories in a machine learning application using a simple baseline classifier that already achieved promising results with an average F-score of 0.82. Figure 2 summarizes our approach in a nutshell.
Figure 2:
Overview of the work-flow from the set-up of the data, sentences segmentation, annotation rounds and development of a baseline classifier
In the future, we will try out our categories in more diverse settings for automated classification tasks in order to find the optimal set-up for German medical documents. Further, we will exploit our set-up for related types of clinical documents, documents from other departments and from other institutions. Of special interest will be the usage of the categories and the classifier on legacy and new/CDA-conformant data in a meshed up corpus. The results might serve as a basis for more advanced NLP applications, including named entity recognition and relation extraction.
Figure 1:
Overview of the aggregated inter-annotator agreement ranges of the four pre-annotations (1-4) and the main annotation (5)
Acknowledgements
This work was supported by Deutsche Forschungsgemeinschaft (DFG) under grants HA 2097/8-1 and KI 564/2-1 within the STAKI2B2 project (Semantic Text Analysis for Quality-controlled Extraction of Clinical Phenotype Information within the Framework of Healthcare Integrated Biobanking).
We thank all annotators and Karsten Krohn for their support to redefine the annotation guidelines and Johannes Kiesel for his support to adapt the WAT-SL annotation tool to our requirements.
Footnotes
ahttp://www.openclinical.org/aisp_qmr.html
In October 2017, a first agreement between representatives of the German Central Federal Association of Health Insurance Funds, the German Federal Association of Nursing Care Funds, the German National Association of SHI Physicians and the German Hospital Federation on discharge management of German hospitals was established http://www.kbv.de/media/sp/Rahmenvertrag_Entlassmanagement.pdf, p.10, Furthermore, the German “E-Health Act”, which came into force in 2016, implemented financial support for healthcare institutions sending structured digital discharge letters based on the CDA standard.
In iteration 3, there were some issues with the configuration of one task. Thus, we did not include the results of this one task into our evaluation.
This amounts to 0.247% in round 2, 0.011% in round 3 and less than 0.01% in round 4.
References
- 1.Edinger T, Demner-Fushman D, Cohen A. M, Bedrick S, Hersh W. R. in AMIA 2017 — Proceedings of the 2017 Annual Symposium of the American Medical Informatics Association. Precision Informatics for Health: The Right Informatics for the Right Person at the Right Time. Washington, D.C., USA, November 4-8, 2017. “Evaluation of clinical text segmentation to facilitate cohort retrieval,”; pp. 660–669. [PMC free article] [PubMed] [Google Scholar]
- 2.Cho P. S, Taira R. K, Kangarloo H. AMIA 2003 — Proceedings of the 2003 Annual Symposium of the American Medical Informatics Association. Biomedical and Health Informatics: From Foundations to Applications. Washington, D.C: USA; 2003. Nov 8-12, “Automatic section segmentation of medical reports,” in; p. 155159. [PMC free article] [PubMed] [Google Scholar]
- 3.Apostolova E, Channin D. S, Demner-Fushman D, Furst J, Lytinen S, Raicu D. “Automatic segmentation of clinical texts,” in. EMBC 2009 — Proceedings of the 31st Annual International Conference of the IEEE Engineering in Medicine and Biology Society; Minneapolis, Minnesota, USA. 2009. Sep 2-6, pp. 5905–5908. [DOI] [PubMed] [Google Scholar]
- 4.Denny J. C, Miller R. A, Johnson K. B, Spickard A., III . AMIA 2008 — Proceedings of the 2008 Annual Symposium of the American Medical Informatics Association. Washington, D.C., USA: 2008. Nov 8-12, “Development and evaluation of a clinical note section header terminology,” in; pp. 156–160. [PMC free article] [PubMed] [Google Scholar]
- 5.Denny J. C, Spickard A, III, Johnson K. B, Peterson N. B, Peterson J. F, Miller R. A. “Evaluation of a method to identify and categorize section headers in clinical documents,”. Journal of the American Medical Informatics Association. 2009;vol. 16(no. 6):806–815. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran L.-T. T, Divita G, Redd A. M, Carter M. E, Samore M. H, Gundlapalli A. V. AMIA 2015 — Proceedings of the 2015 Annual Symposium of the American Medical Informatics Association. San Francisco, California, USA: 2015. Nov 14-18, “Scaling out and evaluation of OBSECA N,an automated section annotator for semi-structured clinical documents, on a large VA clinical corpus,” in; pp. 1204–1213. [PMC free article] [PubMed] [Google Scholar]
- 7.Haug P. J, Wu X, Ferraro J. P, Savova G. K, Huff S. M, Chute C. G. AMIA 2014 — Proceedings of the 2014 Annual Symposium of the American Medical Informatics Association. Washington, D.C., USA: 2014. Nov 15-19, “Developing a section labeler for clinical documents,” in; pp. 636–644. [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Lipsky Gorman S, Elhadad N. IHI ‘10 — Proceedings of the 1st ACM International Health Informatics Symposium. Virginia, USA: Arlington; 2010. Nov 11-12, “Section classification in clinical notes using supervised Hidden Markov Model,” in; pp. 744–750. [Google Scholar]
- 9.Tepper M, Capurro D, Xia F, Vanderwende L, Yetisgen-Yildiz M. “Statistical section segmentation in free-text clinical records,” in. LREC 2012 — Proceedings of the 8th International Conference on Language Resources and Evaluation; Istanbul, Turkey. 2012. May 21-27, pp. 2001–2008. [Google Scholar]
- 10.Ganesan K, Subotin M. “A general supervised approach to segmentation of clinical texts,” in. BigData ‘14 — Proceedings of the 2014 IEEE Conference on Big Data; Washington, D.C., USA. 2014. Oct 27-30, pp. 33–40. [Google Scholar]
- 11.Deléger L, Névéol A. “Identification automatique de zones dans des documents pour la constitution d’un corpus médical en français,” in. TALN-RECITAL 2014 — 21ème Traitement Automatique des Langues Naturelles. Marseille, France, 1-4 July 2014. vol. 2:568–573. Short Papers. [Google Scholar]
- 12.Orosz G, Novák A, Prószéky G. “Hybrid text segmentation for Hungarian clinical records,” in. In: Castro F, Gelbukh A. F, González M, editors. Advances in Artificial Intelligence and Its Applications. MICAI 2013 — Proceedings of the 12th Mexican International Conference on Artificial Intelligence; Mexico: Mexico City Springer-Verlag; 2013, 2013. Nov 24-30, pp. 306–317. no. 8265 in Lecture Notes in Articificial Intelligence. [Google Scholar]
- 13.Hahn U, Matthies F, Lohr C, Löffler M. “3000PA: Towards a national reference corpus of German clinical language,” in. MIE 2018 — Proceedings of the 29th Conference on Medical Informatics in Europe: Building Continents of Knowledge in Oceans of Data. The Future of Co-Created eHealth; Gothenburg, Sweden. IOS Press; 2018, 2018. Apr 24-26, pp. 26–30. no. 247 in Studies in Health Technology and Informatics. [Google Scholar]
- 14.Hahn U, Matthies F, Faessler E, Hellrich J. “UIMA-based JCoRe 2.0 goes GitHub and Maven Central: State-of-the-art software resource engineering and distribution of NLP pipelines,” in. LREC 2016 — Proceedings of the 10th International Conference on Language Resources and Evaluation. Portorož; Slovenia. 2016. May 23-28, pp. 2502–2509. [Google Scholar]
- 15.Faessler E, Hellrich J, Hahn U. “Disclose models, hide the data: How to make use of confidential corpora without seeing sensitive raw data,” in. LREC 2014 — Proceedings of the 9th International Conference on Language Resources and Evaluation. Reykjavik; Iceland. 2014. May 26-31, pp. 4230–4237. [Google Scholar]
- 16.Boone K. W. The CDATM Book. Springer Science & Business Media; 2011. [Google Scholar]
- 17.Kiesel J, Wachsmuth H, al Khatib K, Stein B. “WAT-SL: A customizable Web annotation tool for segment labeling,” in. EACL 2017 — Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics: Software Demonstrations.; Valencia, Spain,. 2017. Apr 5-6,, pp. 13–16. [Google Scholar]
- 18.Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii J. “B RAT:A Web-based tool for NLP-assisted text annotation,” in. EACL 2012 — Proceedings of the 13th Conference of the European Chapter of the Association for Computational Linguistics: Demonstrations. Avignon,; France,. 2012. Apr 25-26,, pp. 102–107. [Google Scholar]
- 19.Krippendorff K. Content Analysis: An Introduction to its Methodology. Sage Publications, 2nd ed; 2004. [Google Scholar]