Abstract
The key to any computational drug repositioning is the availability of relevant data in machine-understandable format. While large amount of genetic, genomic and chemical data are publicly available, large-scale higher-level disease and drug phenotypic data are limited. We recently constructed a large-scale disease-comorbidity relationship knowledge base (dCombKB) and a comprehensive drug-treatment relationship knowledge base (TreatKB) from 21 million biomedical research articles and other resources. In this study, we demonstrated the potential of dCombKB and TreatKB in drug repositioning for schizophrenia, one of the top ten illnesses contributing to the global burden of disease. dCombKB contains 121,359 unique disease-disease comorbidity pairs for 23,041 diseases. TreatKB contains 208,330 unique drug-disease treatment pairs for 2,484 drugs and 24,511 diseases. We constructed a phenotypic comorbidity disease network (PDN) of 14,645 disease nodes and 101,275 edges based on dCombKB. We applied standard network-based ranking algorithm to find diseases that are phenotypically related to SCZ. We developed a drug prioritization system, PhenoPredict-CDN, to systematically reposition drugs for SCZ from diseases phenotypically related to SCZ. PhenoPredict-CDN found all 18 FDA-approved SCZ drugs and ranked them highly as tested in a de-novo validation setting (recall: 1.0, mean ranking: top 6.05%, median ranking: top 1.65%). When compared to PREDICT, a comprehensive drug repositioning system, for novel predictions, Pheno-Predict-CDN outperformed PREDICT in Precision-Recall (PR) curves across three different evaluation datasets. Compared to PREDICT, PhenoPredict-CDN showed a significant 110.0-230.0% improvements in mean average precision. In summary, large-scale higher-level disease-comorbidity relationships data extracted from biomedical literature has potential in drug discovery for SCZ, a complex disease with unknown pathophysiological mechanisms. All the data are publicly available: dCombKB at http://nlp.case.edu/public/data/dCombKB, TreatKB at http://nlp.case.edu/public/data/treatKB/, and predictions for SCZ at http://nlp.case.edu/public/data/SCZ_CDN/.
1. Introduction
Computational drug repositioning strategies can be categorized as drug-based, disease-based and profile-based [1-3]. Drug-based and disease-based approaches exploit drug-drug or disease-disease similarity and existing drug-treatment knowledge to infer new disease-drug associations [4-7]. Profile-based drug-repositioning approaches exploit profile similarities between drugs and diseases [8-12]. The key to all computation-based drug repositioning is the availability of relevant data in machine-understandable format. Existing drug repositioning systems mainly used genetic and genomic data of drugs and diseases [1-3], and in less-degree exploited phenotypic data of diseases and drugs [13-14]. While a large amount of genetic, genomic and chemical data are publicly available, large-scale higher-level disease and drug phenotypic data are limited. Human Phenotype Ontology (HPO), a standardized vocabulary of phenotypic abnormalities encountered in Mendelian diseases [15], is a commonly used disease phenotype data for drug repositioning [4, 13]. We have recently developed a drug repositioning strategy that used disease-manifestation associations from HPO [13]. The fact that HPO mainly contains Mendelian diseases, many of which have no drug treatments, greatly limited its potential in inferring candidate drugs from phenotypically related diseases.
We have recently constructed a large-scale disease-comorbidity relationship knowledge base (dCombKB) from 21 million biomedical research articles using natural language processing techniques[15]. dCombKB contains 121,359 unique disease-comorbidity pairs for 23,041 diseases. Different from HPO that is comprised of almost exclusively rare Mendelian disorders, dCombKB contains both common complex and Mendelian diseases. For example, dCombKB includes a total of 321 SCZ-comorbidity pairs, including both psychiatric comorbidities (e.g., epilepsy, anxiety, brain atrophy, psychosis) and non-psychiatric comorbidities (e.g., hyperprolactinemia, diabetes mellitus, obesity, dyslipi-demia, rheumatoid arthritis). We demonstrated in our previous study that diseases sharing comorbidities tend to share both underlying genetics and drug treatments [15]. In this study, we demonstrated that this unique large-scale disease-comorbidity relationship knowledge base dCombKB had great potential in drug repositioning using schizophrenia (SCZ) as a case study.
Schizophrenia is a psychiatric disorder involving chronic or recurrent psychosis. It is commonly associated with impairments in social and occupational functioning [16]. SCZ is among the most disabling and economically catastrophic medical disorders and is among top ten illnesses contributing to the global burden of disease [17]. Traditional drug discovery has produced many commercially successful antipsychotic drugs, but no new mechanisms of action have been discovered, nor have any gains in efficacy been made since the early 1960s [18]. Currently there exist no medications that can cure SCZ or treat its core symptoms. Despite the high prevalence and vast unmet medical need represented by the disease, many drug companies have moved away from the development of drugs for SCZ, not only because of the high costs, high failure rates, and lengthy development processes inherent to traditional drug development, but also due to a poor understanding of the molecular mechanisms underlying SCZ [19]. Under such circumstances, SCZ patients have little hope for new drug treatment. Therefore, alternative strategies for the discovery of truly innovative drug treatments for SCZ are needed [19-20].
We developed a phenome-driven drug repositioning system (PhenoPredict_CDN) and tested it in identifying repositioned candidate drugs for SCZ. PhenoPredict_CDN critically leveraged dCombKB to infer innovative drug treatments from diseases phenotypically related to SCZ. Our assumption is that if a drug treats many diseases that are phenotypi-cally related to SCZ , then this drug is likely a promising repositioned candidate to treat SCZ. Another critical component of PhenoPredict_CDN is TreatKB, a comprehensive drug-disease treatment relationship knowledge base that we recently constructed from multiple heterogeneous and complementary data resources using advanced computational techniques including natural language processing, text mining and data mining [15, 21-22]. All together, TreatKB contains 208,330 drug-disease treatment pairs for 2484 drugs and 24,511 diseases. We demonstrated in our recent studies the critical roles of TreatKB in computational drug repositioning [5-6, 13].
We compared PhenoPredict_CDN to PREDICT, a comprehensive drug repositioning system [4]. PREDICT used disease phenotypic similarities defined in HPO, and drug-drug similarities from other databases to construct a classifier to determine treatment associations between 593 drugs and 313 diseases, including SCZ. We compared our system to PREDICT in novel drug predictions using multiple evaluation datasets and demonstrated that PhenoPre-dict_CDN consistently achieved better performance than PREDICT. Compared to many existing mechanism-based drug repositioning systems that are based on known disease biology or drug mechanisms, PhenoPredict_CDN has the advantage of repositioning drug candidates to treat diseases with unknown pathophysiological mechanisms such as SCZ. To the best of our knowledge, our study represents the first drug repositioning system driven by large-scale disease-comorbidity relationships extracted from biomedical literature records. To clarify, the goal of this study is not to build a comprehensive drug repositioning systems for all disease, instead our goal is to demonstrate the potential of a literature-based large-scale disease-comorbidity relationship database in drug repositioning. We have made all the data publicly available, including dCombKB, TreatKB and predictions for SCZ, at http://nlp.case.edu/public/data/dCombKB, http://nlp.case.edu/public/data/treatKB/, and http://nlp.case.edu/public/data/SCZ_CDN/.
2. Data and Methods
2.1. Data
Disease-comorbidity relationship knowledge base (dCombKB) dCombKB was constructed from 21 million biomedical literature records using NLP techniques [15]. dCombKB contains 121,359 disease-comorbidity pairs for 23,041 unique diseases. For example, dCombKB contains a total of 321 SCZ-comorbidity pairs, including both psychiatric comorbidities (e.g., epilepsy, anxiety, brain atrophy, psychosis) and non-psychiatric comorbidities (e.g., hyper-prolactinemia, diabetes mellitus, obesity, dyslipidemia, rheumatoid arthritis). All disease terms were standardized based on UMLS terminologies. In addition, the disease-comorbidity pairs were weighted based on their occurrences with specific syntactic patterns (details were described in our published paper [15]). We showed that diseases sharing comorbidities also share genes and drug treatments [15]. dCombKB is publicly available at http://nlp.case.edu/public/data/dCombKB.
Drug-disease treatment relationship knowledge base (TreatKB) TreatKB includes 111,862 drug-disease pairs (1,336 drugs and 8,046 diseases) extracted from records of 4.8 million patients in FAERS, 9,216 drug-disease pairs (1,483 drugs and 1,381 diseases) extracted from 44,000 FDA drug labelings, 69,724 pairs (1,560 drugs and 7,970 diseases) extracted from 21 million MEDLINE abstracts, and 69,724 pairs (1,286 drugs and 11,848 diseases) from 180,000 clinical trial studies [15, 21-22]. We demonstrated that TreatKB was important for the computational drug repositioning [5-6,13]. treatKB is publicly available at http://nlp.case.edu/public/data/treatKB.
2.2. Methods
The experiment framework consists of three steps: (1) we constructed phenotypic comorbidity disease networks (PDNs) using disease-disease comorbidity relationships from dCombKB. We experimented with four different ways in constructing PDNs; We applied a network-based ranking algorithm to find diseases that are phenotypically related to SCZ; We tested the network construction and ranking algorithms by examining the distribution of mental disorders among ranked SCZ-related diseases; (2) We developed a drug prioritization algorithm to systematically reposition drugs from SCZ-related diseases to treat SCZ; We evaluated PhenoPredict_CDN using18 FDA-approved SCZ drugs in a de-novo setting. We compared PhenoPredict_CDN to PREDICT in novel predictions using three evaluation datasets; and (3) We evaluated top-ranked drug candidates by manually reviewing published literature and clinical trial reports.
2.2.1. Construct phenotypic comorbidity disease network (PDNs) and find SCZ-related diseases from PDNs
Construct phenotypic comorbidity disease network (PDNs) We explored four different ways in building PDNs: (1) PDN1, wherein two nodes were directly linked if they are disease-comorbidity (D-C) pairs in dCombKB. The edge weights were determined by the weights of D-C pairs (as defined in dCombkB); (2) PDN2, wherein two diseases (D1 and D2) were linked if they shared any comorbidities. The edge weights were determined by the number of shared comorbidities; (3) PDN3 was the same as PDN2 except that the edge weights were determined the Jaccard similarity [23] of disease-associated comorbidities; and (4) PDN4, which was similar to PDN2 and PDN3 except that the weights were determined by the cosine similarity [23] of disease-associated comorbidities. In this study, we did not discount the weights of common diseases or comorbidities since for drug repositioning purpose, common disease may be as important as rare disorders. We also generated random PDNs for each real PDN by randomly shuffling its edges. These random PDNs were used in the subsequent drug repositioning algorithms.
Find phenotypically related diseases for a given input (e.g., SCZ We applied the standard network-based ranking algorithm to find phenotypically related diseases for a given input (SCZ in this study). We have recently applied this algorithm to prioritize genes for a given disease [24-25] and to prioritize diseases for a given microbial metabolite [26-27]. The iterative network-based ranking algorithm is defined as: , wherein M is the column-normalized adjacency matrix of PDN, γ is a preset probability of restarting from the initial seed node (γ=0.1 in this study), and pt is a vector in which the ith element holds the normalized ranking score of disease i at tth iteration. The initial probability vector p0 contains normalized probability values for input. In our study, p0 contained SCZ, with a probability of 1.0. Diseases were ranked according to values in the steady-state probability vector, which was obtained by iterating the algorithm until the change between pt+1 and pt was less than 10–6.
Evaluate and analyze SCZ-related diseases We tested both the network construction and ranking algorithm by examining rankings of mental disorders among retrieved SCZ-related diseases. SCZ is a mental disorder and is known to share both phenotypes and genetics with other mental disorders. We expect that top-ranked SCZ-related disease will contain more mental disorders than bottom-ranked diseases. Using SCZ as the seed, we retrieved four ranked lists of diseases from four PDNs. We classified these diseases into mental diseases (“Mental, behavioural disorders”) using the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD10), a disease classification scheme designated by the World Health Organization (WHO) [28]. Since the term usage in dCombKB is often different from that in ICD10, we mapped disease terms to their corresponding unified medical language system (UMLS) unique concept identifiers [29] and classified diseases based on the unique concept identifiers. At ten ranking cutoffs (10%, 20%,. . . 100%), we calculated percentages of mental diseases among retrieved diseases.
2.2.2. Reposition drugs
Drug repositioning algorithm We developed a drug prioritization approach to systematically reposition drugs from SCZ-related diseases to treat SCZ. We first ranked drugs based on the number of SCZ-related diseases that they could treat as well as the ranking scores of these diseases. The drug prioritization algorithm is defined as: , wherein n is the number of SCZ-related diseases that a drug can treat and R_disease_J is the disease ranking score (output from the network-based disease ranking algorithm). During our study, we found that certain drugs were consistently ranked highly when both actual and random PDNs were used. For example, the drug “chlordiazepoxide” was ranked at top 0.32% based on the actual PDNs and at top 0.36% based on random PDNs. We designed a reprioritization strategy by taking into accounts of drug ranking scores derived from random networks. A drug was ranked highly if and only if it was ranked highly for actual PDNs and low for the random PDNs. The drug reprioritization algorithm is defined as: RRdrug = Rdrug/R′drug, where Rdrug is the ranking score of a drug based on the actual PDN and R′drug is the ranking score of the same drug based on random PDNs.
2.2.3. Evaluation and comparison
De-novo validation using 18 known SCZ drugs as evaluation dataset We evaluated PhenoPredict_CDN using 18 FDA-approved SCZ drugs. Since SCZ and its associated drug treatment pairs were removed from the inputs to the repositioning algorithm, the evaluation was in fact a de-novo validation. We calculated rankings of 18 FDA-approved SCZ drugs among all retrieved drugs. The recall, mean and median rankings of these drugs were calculated. We compared validation performances across four TreatKBs separately and in combination.
Compare PhenoPredict-CDN to PREDICT in novel predictions Since the ultimate goal of any drug repositioning algorithms is to find novel drugs for a given disease, we compared PhenoPredict_CDN to PREDICT in novel predictions instead of validation of FDA-approved SCZ drugs. We evaluated the performance using the following three evaluation datasets individually and combined: (1) 195 SCZ drugs that were extracted from 172,888 clinical trials; (2) 50 SCZ drugs that were extracted from 43,811 ongoing clinical trials initiated in 2012 and after; (3) 114 SCZ drugs that were extracted from over 21 million MEDLINE abstracts; and (4) 263 SCZ drugs extracted from all clinical trials and MEDLINE abstracts. The 18 FDA-approved drugs were removed from these evaluation datasets.
The output from PhenoPredict_CDN was a ranked list of 2,484 drugs. Note that these predictions were made with the prior knowledge of SCZ was removed from the inputs: SCZ was removed from SCZ-related diseases and SCZ-drug treatment pairs were removed from TreatKB. The novel predictions from PREDICT was a list of drugs that were classified as positive (classification probability greater than 0.50). A total of 593 drugs were included in PREDICT, among which 79 drugs were classified as positives for SCZ. The 79 drugs along with their corresponding probabilities (ranging from 0.543-0.994) are publicly available. We assumed that the rest 524 drugs were predicted as negatives for treating SCZ. We assigned each negative a value that was randomly picked from 0.0 to 0.5. We repeated this process for ten times and generated ten ranked list of drugs for PREDICT.
We used Precision-Recall (PR) curves instead of Receiver Operator Characteristic (ROC) curves to evaluate and compare PhenoPredict_CDN to PREDICT. Studies have shown that in domains where the number of negatives greatly exceeds the number of positives such as in drug repositioning and many other biomedical classification domains, ROC curves, not PR curves, can present an overly optimistic view of an algorithm’s performance [30]. Using each of the three evaluation datasets as gold standard, we calculated precisions at 10 different recall cutoffs (0.1, 0.2, ... 1.0) for both PhenoPredict_CDN and PREDICT and plotted the PR curves. The PR curves for PREDICT were then averaged across ten datasets ( the PR curves for these ten datasets were very similar, therefore we did not generate more datasets). Mean Average Precision (MAP), which approximates the area under the precision-recall curve [31], was used to compare the performance of PhenoPredic_CDN and PREDICT.
2.2.4. Analyze top-ranked repositioned drug candidates
We manually examined top 20 repositioned drug candidates by searching for supporting evidence from FDA approved drugs, clinical trials, and published biomedical literature.
3. Results
3.1. Compare four disease networks in ranking mental disorders among SCZ-related diseases
SCZ is a mental disorder and is known to share both phenotypes and genetics with other mental disorders including bipolar disorders and depression. We used the rankings of mental disorders among SCZ-related diseases at different ranking cutoffs to evaluate network construction and disease-ranking algorithms. As shown in Fig.1, SCZ-related diseases retrieved from all four PDNs have similar ranking curves, with mental diseases enriched among top-ranked diseases. Since PDN1 contains 14,645 diseases, which is 41% more diseases than other three PDNs, we used PDN1 in subsequent experiments.
Figure 1:
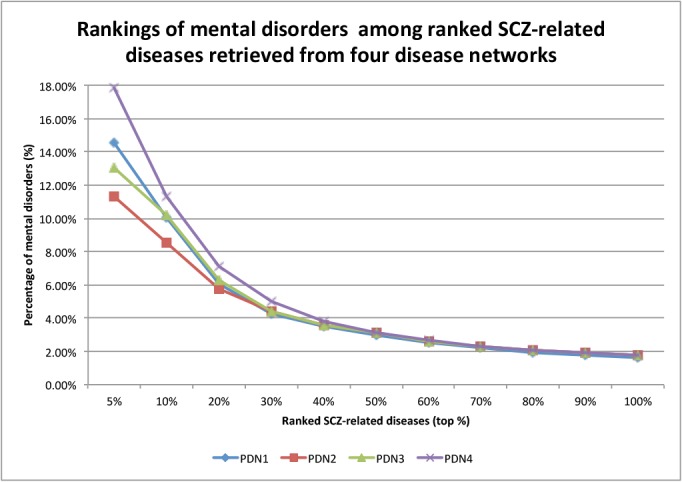
Comparison of the rankings of mental disorders among ranked SCZ-related diseases retrieved from four disease networks (PDN1, PDN2, PDN3, PDN4).
3.2. PhenoPredict_CDN found all 18 FDA-approved SCZ drugs and ranked them highly
We validated the drug repositioning algorithm using 18 FDA-approved SCZ drugs. Since the TreatKB is an essential component of PhenoPredict_CDN, we compared the performance of the validation across four TreatKBs. As shown in Table 1, when all four TreatKBs were combined, PhenoPredict_CDN achieved a recall of 1.00, an average ranking of 6.05%, and a median ranking of 1.65%. The performances for individual TreatKBs were lower. These results demonstrated that a comprehensive drug-disease treatment knowledge base was a critical component of PhenoPredict_CDN.
Table 1:
Comparing recalls, mean, and median rankings of 18 FDA-approved SCZ drugs across four TreatKBs.
| TreatKB | Recall | Mean | Median |
|---|---|---|---|
| FDA drug label | 1.00 | 14.16% | 8.63% |
| Post-market | 1.00 | 5.76% | 2.58% |
| Clinicaltrials | 0.83 | 20.69% | 8.53% |
| Literature | 1.00 | 13.71% | 1.98% |
| Combined | 1.00 | 6.05% | 1.65% |
There was a big difference between the median ranking of 1.65% and the mean ranking of 6.05%, demonstrating a skewed ranking distribution of these FDA-approved SCZ drugs. As shown in Fig.2, 17 of the 18 SCZ drugs were ranked within top 10% except for prochlorperazine. Prochlorperazine is used to treat severe nausea and vomiting, which are not SCZ comordities according to dCombKB, which explains why prochlorperazine was ranked low.
Figure 2:
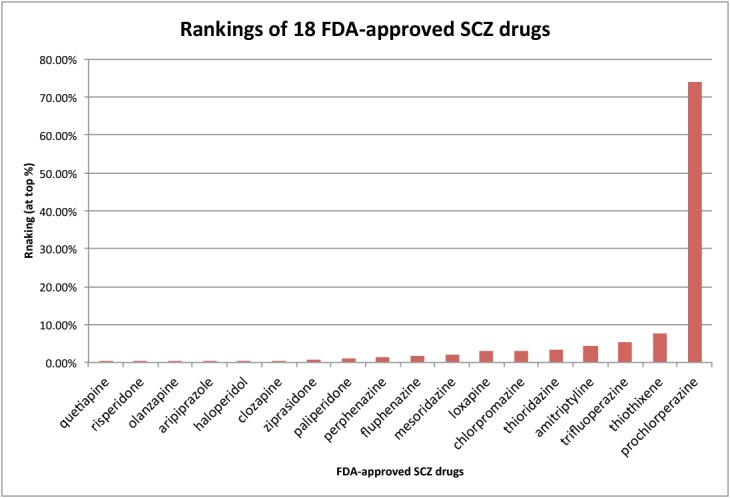
Percent rankings of 18 FDA-approved drugs among all 2484 drugs. The combined TreatKB was used.
3.3. Compare PhenoPredict CDN to PREDICT in novel predictions
We plotted PR curves for PhenoPredict_CDN and for PREDICT using 263 novel SCZ drugs that were extracted from 172,888 clinical trial reports and from 21 million MEDLINE records (Fig.3). As shown in the figure, PhenoPredict-CDN performed better than PREDICT. The mean average precision (MAP), which approximates the area under the PR curve as 0.413 for PhenoPredict_CDN and 0.197 for PREDICT, representing a 110.0% improvement.
Figure 3:
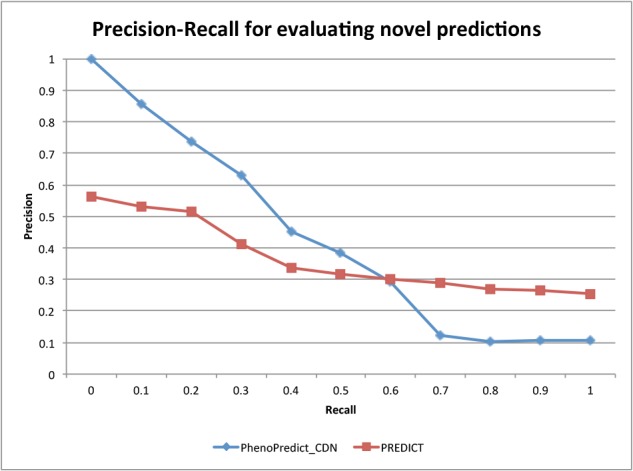
The Precision-Recall curves evaluated with 263 novel SCZ drugs from 172,888 clinical trial reports and 21 million MEDLINE records.
Fig. 4 shows the PR curves using 195 novel SCZ drugs extracted from 172,888 clinical trials. PhenoPredict_CDN performed better than PREDICT as shown in the PR curves. The MAP for PhenoPredict_CDN was 0.332, representing a 137.1% improvement as compared to the MAP of 0.140 for PREDICT. Fig. 5 shown the PR curves when 50 novel SCZ drugs extracted from ongoing clinical trials were used. These 50 drugs may represent a newer generations of SCZ drugs. The MAPs for both algorithms were lower than previous ones. PhenoPredict_CDN performed better than PREDICT as measured by PR curves. The MAP is 0.093 for PhenoPredict_CDN and 0.030 for PREDICT, representing a 210.0% improvement. Fig. 6 shows the PR curves when 114 novel SCZ drugs extracted from biomedical literature were used. PhenoPredict_CDN performed better than PREDICT as shown by PR curves. The MAP for PhenoPredicLCDN was 0.343, representing a 230.0% improvement as compared to MAP of 0.104 for PREDICT.
Figure 4:
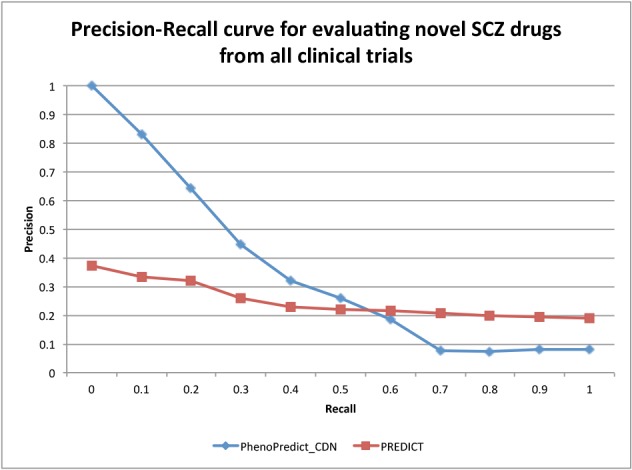
The Precision-Recall curves evaluated with 195 novel SCZ drugs from 172,888 clinical trials.
Figure 5:
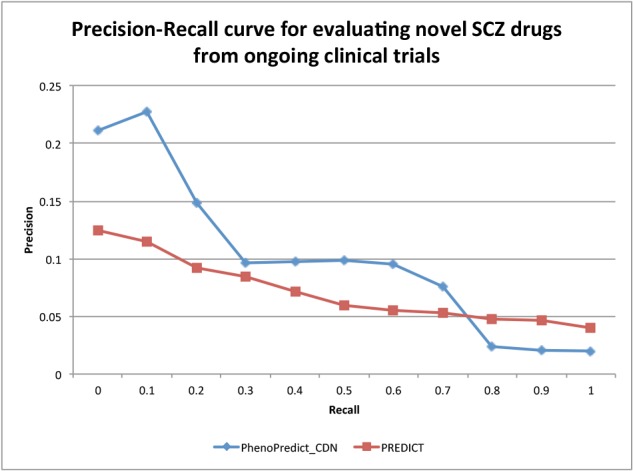
The Precision-Recall curves evaluated with 50 novel SCZ drugs from 43,811 ongoing clinical trials.
Figure 6:
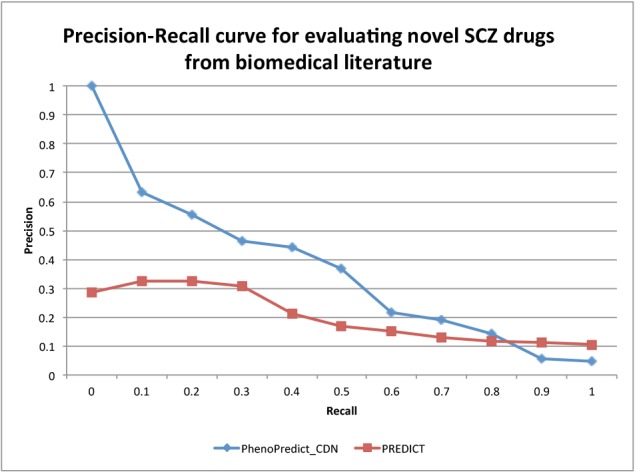
The Precision-Recall curves evaluated with 114 novel SCZ drugs from 21 million MEDLINE abstracts.
In summary, PhenoPredict_CDN performed better than PREDICT across all evaluation datasets. Two facts may account for this significant improvement. First, PREDICT used disease-disease similarity matrix from HPO that mainly contains phenotypic description of rare Mendelian disorders. One of the major limitations in using interrelationships among rare Mendelian disorders for drug repositioning is that many of these diseases have no drug treatments themselves, which could greatly limit the potential in transferring drug treatments among phenotypically related diseases. PhenoPredict_CDN instead used disease interrelationships from dCombKB, which contains 23,041 diseases including both common complex diseases and rare Mendelian disorders. Second, we used a comprehensive treatKB that consists of drug-disease treatment pairs extracted from multiple complementary resources and PREDICT used drug-treatment pairs derived from FDA drug labels only. We have shown that drug repositioning using this comprehensive TreatKB performed better than using TreatKB derived from FDA drug labels alone (Table 1).
3.4. Top-ranked drug candidates
Table 2 shows top 20 repositioned drug candidates, all of which have supporting evidences from FDA drug labels, clinical trials, or biomedical literature for their potential treatment benefits in SCZ patients. Among these 20 drugs, seven are FDA-approved drugs. These specific examples demonstrated the potential of our disease-comorbidity guided drug repositioning strategy. The complete list is at http://nlp.case.edu/public/data/SCZ_CDN.
Table 2.
Top 20-ranked repositioned drug candidates. NCT**: SCZ drugs from clinical trials. PMID**: SCZ drugs from biomedical literature. FDA-approved SCZ drugs are highlighted.
| Rank | Drug | Evidence | Rank | Drug | Evidence |
|---|---|---|---|---|---|
| 1 | quetiapine | FDA-approved | 11 | clozapine | FDA-approved |
| 2 | sertraline | NCT00169988, NCT00531518 | 12 | trazodone | NCT00659919 |
| 3 | risperidone | FDA-approved | 13 | valproic acid |
NCT00194025, NCT01094249, NCT02011750 |
| 4 | alprazolam | PMID3289523, PMID1348161, PMID12516314 |
14 | lithium |
NCT00202306 NCT00183443 NCT00202293 |
| 5 | olanzapine | FDA-approved | 15 | donepezil |
NCT01490567, NCT00465283, NCT00206947 |
| 6 | fluoxetine |
NCT00531518, NCT02022709 |
16 | memantine |
NCT02001103 NCT00757978 NCT00097942 |
| 7 | aripiprazole | FDA-approved | 17 | sulpiride |
NCT00654576, NCT02307396 |
| 8 | citalopram |
NCT00893256, NCT00047450, NCT01032083, NCT01032083 |
18 | bupropion |
NCT01111149, NCT00307203 |
| 9 | haloperidol | FDA-approved | 19 | lorazepam |
NCT00797277, NCT00431184, NCT00159133 |
| 10 | levetiracetam | PMID12609283, PMID19265183 |
20 | ziprasidone | FDA-approved |
4. Discussion
The key to any computational drug repositioning systems is the availability of relevant data in machine-understandable format. While large amount of genetic, genomic and chemical data are publicly available, large-scale higher-level disease and drug phenotypic data are limited. In this study, we demonstrate that the disease-comorbidity relationship data that we extracted from biomedical literature has great potential in drug repositioning for complex diseases with unknown pathophysiological mechanisms such as SCZ. Intuitively, disease-comorbidity data does not necessarily work well for all diseases. In the future, we will systematically test PhenoPredict_CDN on other diseases and examine its performances across disease classes. Even though we demonstrate that PhenoPredict_CDN had better performance than PREDICT in predicting drug candidates for SCZ across different evaluation datasets, our goal for this study is not to build a comprehensive drug repositioning system. Instead, our goal is to demonstrate the usefulness of dCombKB in drug repositioning. To build a more encompassing prediction system, other levels of disease-and drug-related data such as genetics and genomics may be necessary. In addition, other disease and drug phenotypic data, for example drug side effects and disease manifestations, may further improve prediction performances.
Author’s contributions
Xu and Wang have jointly conceived the idea, designed and implemented the algorithms and prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgements
RX is funded by the NIH Director’s New Innovator Award under the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health (DP2HD084068, Xu), NIH National Institute of Aging (1 R01 AG057557-01, Xu), American Cancer Society Research Scholar Grant (RSG-16-049-01 - MPC, Xu), the Landon Foundation-AACR INNOVATOR Award for Cancer Prevention Research (15-20-27-XU), Mary Kay Foundation Grant (057-15, Xu), and Pfizer 2015 ASPIRE Rheumatology and Dermatology Research Award (WI206753, Xu).
References
- 1.Dudley J. T, Deshpande T, Butte A. J. Exploiting drug?disease relationships for computational drug repositioning. Briefings in bioinformatics. (2011);12(4):303–311. doi: 10.1093/bib/bbr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Zheng S, Chen B, Butte A. J, Swamidass S. J, Lu Z. A survey of current trends in computational drug repositioning. Briefings in bioinformatics. (2015);17(1):2–12. doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodos R. A, Kidd B. A, Shameer K, Readhead B. P, Dudley J. T. In silico methods for drug repurposing and pharmacology. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. (2016);8(3):186–210. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb A, Stein G. Y, Ruppin E, Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Molecular systems biology. (2011);7(1) doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Xu R. The 2017 Annual American Medical Informatics Association Symposium. Wahshington DC: (2017). Nov 14-18, Drug repositioning for prostate cancer: using a data-driven approach to gain new insights. [PMC free article] [PubMed] [Google Scholar]
- 6.Xu R, Wang Q. A genomics-based systems approach towards drug repositioning for rheumatoid arthritis. BMC genomics. (2016);17(7):518. doi: 10.1186/s12864-016-2910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai X, Chen Y, Gao Z, Xu R. AMIA Summits on Translational Science Proceedings. (2016), 2016. Explore Small Molecule-induced Genome-wide Transcriptional Profiles for Novel Inflammatory Bowel Disease Drug; p. 22. [PMC free article] [PubMed] [Google Scholar]
- 8.Nagaraj A. B, Wang Q. Q, Joseph P, Zheng C, Chen Y, Kovalenko O, Xu R*, Difeo* A. Using a novel computational drug-repositioning approach (DrugPredict) to rapidly identify potent drug candidates for cancer treatment. Nature Oncogene. (2018);37(3):403. doi: 10.1038/onc.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sirota M, Dudley J. T, Kim J, Chiang A. P, Morgan A. A, Sweet-Cordero A, Butte A. J. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science translational medicine. (2011);3(96):96ra77–96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudley J. T, Sirota M, Shenoy M, Pai R. K, Roedder S, Chiang A. P, Butte A. J. Com-putational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Science translational medicine. (2011);3(96):96ra76–96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Cai X, Xu R. (2015). Combining Human Disease Genetics and Mouse Model Phenotypes towards Drug Repositioning for Parkinson’s disease. In AMIA Annual Symposium Proceedings (Vol. 2015, p. 1851). American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Gao Z, Wang B, Xu R. Towards precision medicine-based therapies for glioblastoma: interrogating human disease genomics and mouse phenotypes. BMC genomics. (2016);17(7):516. doi: 10.1186/s12864-016-2908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R, Wang Q. PhenoPredict: a disease phenome-wide drug repositioning approach towards schizophrenia drug discovery. Journal of biomedical informatics. (2015);56:348–355. doi: 10.1016/j.jbi.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duran-Frigola M, Aloy P. Recycling side-effects into clinical markers for drug repositioning. Genome Med. (2012);4(3) doi: 10.1186/gm302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Li L, Wang Q. Towards building a disease-phenotype knowledge base: extracting disease-manifestation relationship from literature. Bioinformatics. (2013);29(17):2186–2194. doi: 10.1093/bioinformatics/btt359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arlington V. A. American Psychiatric Association. American Psychiatric Association. (2013). Diagnostic and statistical manual of mental disorders. [Google Scholar]
- 17.Murray C, Lopez A. D. Cambridge, Massachusetts: Harvard University Press; (1996). The global burden of disease. [Google Scholar]
- 18.Hyman S. E. Time for new schizophrenia Rx. Science. (2014);343(6176):1177–1177. doi: 10.1126/science.1252603. [DOI] [PubMed] [Google Scholar]
- 19.Insel T. R. Next-generation treatments for mental disorders. Science translational medicine. (2012);4(155):155ps19–155ps19. doi: 10.1126/scitranslmed.3004873. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan P. F. Puzzling over schizophrenia: schizophrenia as a pathway disease. Nature medicine. (2012);18(2):210–211. doi: 10.1038/nm.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, Wang Q. Large-scale extraction of accurate drug-disease treatment pairs from biomedical literature for drug repurposing. BMC bioinformatics. (2013);14(1):1. doi: 10.1186/1471-2105-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu R, Wang Q. Automatic signal extraction, prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA Adverse Event Reporting System (FAERS) Journal of biomedical informatics. (2014);47:171–177. doi: 10.1016/j.jbi.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Kamber M, Pei J. Elsevier; (2011). Data mining: concepts and techniques: concepts and techniques. [Google Scholar]
- 24.Chen Y, Xu R. Bioinformatics. (2017), 2017. Context-sensitive network based disease genetics prediction and its implications in drug discovery; p. btw737. DOI:10.1093/bioinformatics/btw737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Li L, Zhang G. Q, Xu R. Phenome-driven disease genetics prediction toward drug discovery. Bioinformatics. (2015);31(12):i276–i283. doi: 10.1093/bioinformatics/btv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu R, Wang Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC genomics. (2015);16(7):S4. doi: 10.1186/1471-2164-16-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R, Wang Q. Towards understanding brain-gut-microbiome connections in Alzheimer?s disease. BMC systems biology. (2016);10(3):63. doi: 10.1186/s12918-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ICD-10: International Statistical Classification of Diseases and Related Health Problems: 10th revision. World Health Organization; 1992. [Google Scholar]
- 29.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. (2004);32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis J, Goadrich M. The relationship between Precision-Recall and ROC curves. Proceedings of the 23rd international conference on Machine learning; 2006. Jun, pp. 233–240. [Google Scholar]
- 31.Manning C. D, Raghavan P, Schutze H. Cambridge: Cambridge university press; (2008). Introduction to information retrieval (Vol. 1, p. 6) [Google Scholar]


