Abstract
Digital rectal examination (DRE) is considered a quality metric for prostate cancer care. However, much of the DRE related rich information is documented as free-text in clinical narratives. Therefore, we aimed to develop a natural language processing (NLP) pipeline for automatic documentation of DRE in clinical notes using a domain-specific dictionary created by clinical experts and an extended version of the same dictionary learned by clinical notes using distributional semantics algorithms. The proposed pipeline was compared to a baseline NLP algorithm and the results of the proposed pipeline were found superior in terms of precision (0.95) and recall (0.90) for documentation of DRE. We believe the rule-based NLP pipeline enriched with terms learned from the whole corpus can provide accurate and efficient identification of this quality metric.
Introduction
Digital rectal exam (DRE) has been an important part of routine prostate cancer screening and clinical staging in the United States for decades.1-3 Most clinical guidelines include DRE as a pre-treatment assessment examination and a quality metric for prostate cancer treatment.4,5
However, DRE and its results are often not recorded systematically or included in billing or claims datasets and therefore DRE assessment is limited to labor-intensive approaches. Several studies have conducted manual chart reviews of prostate cancer patients to assess documentation of DRE.5,6 These reviews found that while DRE was well documented in the patient record, clinical details from the exam were often missing.
As we move into the era of digital healthcare, adoption and implementation of electronic health records (EHR) have consistently grown.7 New studies leverage information available in EHRs that not only enable clinicians to better record processes of care (e.g. DRE) in digital format, but also allow clinical research to move from time consuming manual chart reviews to automated and efficient text processing.8 However, the use of EHRs to assess DRE documentation and results is limited, making the assessment of this quality metric burdensome and inefficient. Therefore, natural language processing (NLP) techniques are needed to extract DRE information from the clinical narratives in EHRs to a format readable and analyzable for computers. NLP has emerged as a potential solution for bridging the gap between free-text and structured representations of clinical information. Since biomedical domains exhibit a high degree of terminological variation, NLP’s precision becomes even more valuable based on its ability to automatically recognize all variants of domain language.9 There are a few studies10-13 using NLP techniques to extract concepts related to prostate cancer from electronic health records for decision support, but the accuracy of DRE extraction was reported only in two studies12,13. The features related to DRE reporting was also missing in those studies.
There is a limited number of terms for DRE and its findings, but it is reported in a non-standardized format which makes it hard to export or capture the rich information found in the unstructured text. As key sources of knowledge, clinical ontologies and lexicons have supported many NLP applications. However, these semantic resources are limited in their scope and domain, and their manual construction is knowledge-intensive and time-consuming14. With the help of NLP, the similarity of words and phrases used in clinical notes can be compared using word embeddings, which are dense feature vectors that capture the semantics of words, and help build, combine, and expand the associated terminologies for applications15. Extraction of DRE reporting can be achieved using a straight through rule-based system. However, since its success is mostly dependent on the terminology used for the desired task, it is important to capture all representations of terms which can be learned from the corpus itself. Therefore, the primary aim of this study was to develop and evaluate an NLP framework for automatic identification of DRE from unstructured clinical notes using a rule-based approached enriched with terms learned from the whole corpus. This study addresses data preparation, the NLP development process and its strengths/weaknesses, evaluation method, and challenges.
Methods
Data Source
The Stanford prostate cancer research database was used for analysis, which is described in detail elsewhere.16 In brief, data were collected from a tertiary-care academic medical center Epic EHR system (Epic Systems, Verona, WI) and managed in an EHR-based relational database. Data were linked to the California Cancer Registry and include patients diagnosed with prostate cancer from 2005 to February 9, 2018. Prostate cancer patients were identified using ICD diagnostic codes, ICD-9-CM:185 and ICD-10-CM: C61. This study received the approval from the institute’s Institutional Review Board.
Training and test sets
A reference standard of 301 randomly selected patients’ clinical notes were manually reviewed and annotated as sentence level by two domain experts (a research nurse and one urology clinical fellow) as ‘examined, ‘historical’, ‘hypothetical, ‘deferred’ and ‘refused’. If DRE was performed and documented currently at the visit which the report was created it was tagged as “examined”, if in the same report physician also refers a previously performed DRE, it was tagged as “historical”, if DRE suggested for future visits it was tagged as “hypothetical” and if it was reported that DRE deferred or refused by patient it was tagged as “deferred/refused”. Reviewers had high inter-rater reliability in finding indications (Cohen’s K = .97). First, 101 randomly selected records from the chart reviews were used as the preliminary training set to determine what DRE information was recorded in the EHRs and what information could be captured via a rule-based NLP system. Next, we tested our rule-based pipeline, which explained in detail in the following section, on the remaining 200 records from the reference dataset. Error analysis was performed throughout the development process, and the pipeline was subsequently adjusted.
The proposed pipeline
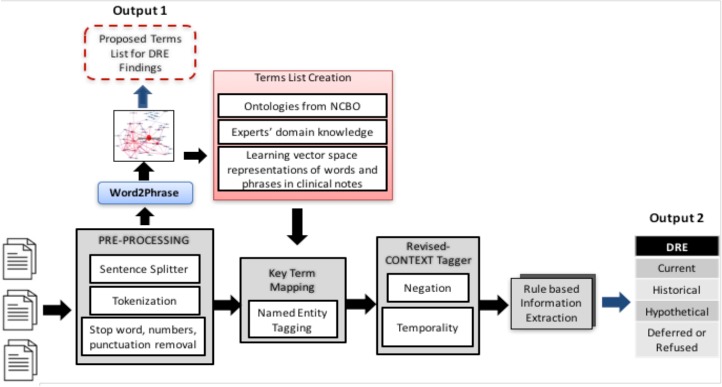
We decomposed our NLP problem into a set of subtasks, and we created a pipeline comprising a set of sequentially executed processing modules to automatically recognize the pertinent named entities in clinical narratives, e.g., “rectal exam” and “DRE”. Figure 1 presents the core processing blocks of the proposed DRE extraction pipeline.
Figure 1.
The Proposed Pipeline
Pre-Processing
A set of common pre-processing steps in the pipeline was implemented using the Natural Language Toolkit (NLTK, www.nltk.org) library, a suite of open source Python modules. The pre-processing steps included sentence boundary detection, tokenization, and removing all the stop words, punctuation characters, and 2 letter-words.
Creation of Terms List
We created a domain-specific dictionary for extracting DRE information from EHRs of prostate cancer patients in three phases (Figure 1: Terms List Creation). First, a list of terms was generated based on domain knowledge, prior experience in the field, and review of medical notes. For example, the following terms were used to identify when DRE was not performed: “deferred”, “not performed”, “not examined”, “declined” and “refused”. Next, terms were matched with existing ontologies from the National Center for Biomedical Ontology (NCBO)17. Finally, additional words were added to the dictionary using distributional information on words and phrases from the all notes in our database. Terms occurring in less than 50% of reviewed charts were excluded. The final list of terms was reviewed by the domain experts.
In order to learn how terms are used in a particular clinical text corpus in phase 3, we used distributional semantics which can be easily trained from a large corpus and derive representations for words in such a way that words occurring in similar contexts will have similar representations15,18,19. As the distributional semantics method, we used the popular word2vec package created by Mikolov et al. to build vector representations of all terms in our corpus.20 The skip-gram model with vector length 100 (the standard dimension for word2vec) and a context window width of 5 was used to recognize biomedical synonyms. We used a linear context window of width 5 since it is concluded that a prior study the optimal way.15,19,21 Because word2vec constructs a separate vector for each token in the text, there is no intuitive way to compose the vectors for “rectal” and “exam” into a combined vector for the term “rectal_exam”. We thus needed to create single entities for multiword expressions identified in the corpus and “merged” into single tokens (e.g. rectal exam” → rectal_exam) before running word2vec using bigrams and trigrams of words15,21.
In order to quantify closeness of the word vectors, we used the cosine similarity between two terms. Based on the calculated word similarity metric, additional synonyms and other lexical variants were identified in the clinical notes and the terms list was expanded after the expert confirmation of candidate terms. We used the NLTK and Gensim to preprocess text, build word vectors and calculate word similarities. In addition, as an exploratory data analysis to group and visualize a high-dimensional dataset, like word vectors produced by the word2vec model, we used t-SNE (t-Distributed Stochastic Neighbor Embedding), which is a well-suited technique for dimensionality reduction by giving each data point a location in a two or three-dimensional map. We use t-SNE scikit-learn implementation to represent visually the insight relation between the words in the text reports and explore possible sub-divisions of the terms list.
Determining the context of entities: negation, experiencer, and temporal status
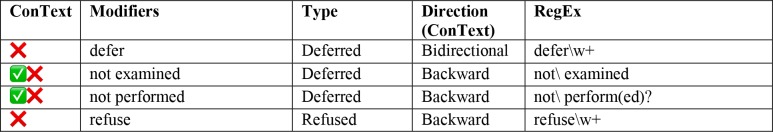
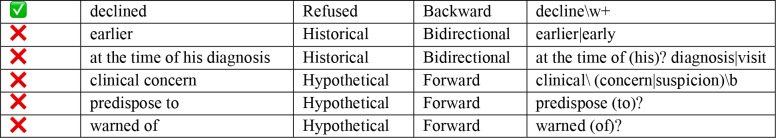
Clinical conditions can be modified by several contextual properties that are relevant for our information extraction task; ConText identifies three contextual values in addition to NegEx’s negation, including hypothetical, historical, and experiencer values.22 We implemented the ConText algorithm within our NLP system in two ways in order to tag DRE if it is reported as historical or negated (also includes deferred and refused). For instance, if in the clinical narrative it is reported as “earlier DRE showed that”, DRE documentation was extracted as “historical”. First, we used the default version of ConText to determine whether a DRE entity is negated and its temporal status. For example, the output of this input text: ‘his digital rectal exams were always normal. DRE: not examined’ returns to two aspects of DRE information: historical and currently negated. Next, we built our rule-based pipeline by extending the open-source ConText algorithm to include domain-specific terms. Although ConText comes with a set of triggering terms, some domain relevant terms, such as: ‘refused’ and ‘defer’, were not present in the dictionary. Therefore, we added seven additional modifiers learned from the corpus to ConText’s working dictionary (Table 1). The rule-based algorithm identified keywords from the document level output. Keywords were categorized as target (DRE related terms) and modifiers (historical, etc.). The general rules for extraction were: 1) In a sentence level if there is only one target word, check any modifier in the sentence and assign it to the target term. 2) If there is more than one target word, assign the closest modifier in terms of word offset. 3) In the absence of any target words, it was classified as not documented DRE.
Table 1.
Terms added to the ConText Modifier List.
|
|
Evaluation
We performed general descriptive analyses for documentation of DRE. We created a baseline pipeline which did not include any extended terminology and used default ConText modifiers. We compared results of the baseline and the proposed information extraction pipeline based on the manual chart review using our randomly sampled subset of records (n=200). We collected true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN), and the performance of the proposed pipeline was evaluated in terms of precision, recall and F-score.
Results
In our final cohort, we identified 3,766 patients older than 35 years old, who received initial prostate cancer treatment (prostatectomy, radiotherapy, hormone therapy, or chemotherapy) in our institute between 2005-2018. In our database, we found a total of 348,239 clinical notes with 12 different note types which belong to our final cohort from nine different providers. After excluding irrelevant notes (such as telephone encounter, radiology report, consult and etc.), our final corpus had a total of 185,356 notes. Characteristics of our cohort are summarized in Table 2.
Table 2.
Characteristics of the test set
| DRE (6 months period before treatment) | Examined | Historical | Deferred | Refused | Not reported |
|---|---|---|---|---|---|
| #Patients | 2434 | 1191 | 1676 | 41 | 1175 |
| #Notes | 9061 | 2216 | 4021 | 69 | 169989 |
| #Note types | |||||
| Clinical visits | 5884 | 1041 | 1323 | 40 | |
| H&P | 810 | 327 | 1384 | 7 | |
| Progress notes | 2287 | 838 | 1122 | 21 | |
| Assessment & Plan | 80 | 10 | 192 | 1 | |
In the evaluation phase, we compared the accuracy of our proposed pipeline with a baseline pipeline from a random sample of manual chart reviews. The results of the proposed pipeline were found superior in terms of precision (0.95) and recall (0.90) for documentation of DRE if it is examined at the current visit (Table 3). Both of the pipelines had lower accuracy in terms of detecting reported previous DREs than other sub-tasks, although pipeline 2 achieved a better performance in terms of precision (0.84) and recall (0.67). In the second pipeline, the impact of new concepts added to ConText modifiers was evaluated in order to detect DRE documentation in detail such as: historical, deferred and refused (Table 1-2). While there was a negation modifier in clinical terminologies, ‘defer’ and ‘refusal’ were not covered completely as separate modifiers. In addition, using negations created uncertainty in cases, such as ‘DRE no nodules or discrete areas of induration’, since it tagged DRE reporting as negated. We also calculated word similarities mathematically using the cosine similarities of terms and proposed a list of candidate terms divided into 5 different subcategories for future, more detailed assessments of DRE documentation.
Table 3.
Validation measures for automated DRE documentation extraction.
| PIPELINE 1 (Default ConText Vocabulary) | PIPELINE 2 (The proposed pipeline, Extended ConText Vocabulary) | p | ||
|---|---|---|---|---|
| EXAMINED | DRE + | 108 | 101 | <0.05 |
| DRE - | 19 | 83 | ||
| precision | 0.61 | 0.95 | ||
| recall | 0.96 | 0.90 | ||
| f1-score | 0.75 | 0.93 | ||
| HISTORICAL | DRE + | 13 | 41 | 0.062 |
| DRE - | 134 | 131 | ||
| precision | 0.72 | 0.84 | ||
| recall | 0.21 | 0.67 | ||
| f1-score | 0.33 | 0.75 | ||
| HYPOTHETICAL | DRE + | 0 | 62 | <0.05 |
| DRE - | 141 | 138 | ||
| precision | 0 | 1.00 | ||
| recall | 0 | 1.00 | ||
| f1-score | 0 | 1.00 | ||
| DEFERRED | DRE + | 0 | 59 | <0.05 |
| DRE - | 141 | 141 | ||
| precision | 0 | 1.00 | ||
| recall | 0 | 1.00 | ||
| f1-score | 0 | 1.00 | ||
| REFUSED | DRE + | 0 | 1 | <0.05 |
| DRE - | 199 | 199 | ||
| precision | 0 | 1.00 | ||
| recall | 0 | 1.00 | ||
| f1-score | 0 | 1.00 |
Discussion
In this study, we developed a rule-based NLP pipeline to aid in the identification and extraction of DRE features that are often buried in the unstructured text of clinical notes. The terms list was enriched with additional features learned automatically from the corpus and achieved superior results for pre-treatment DRE documentation of prostate cancer compared to standard NLP approaches. As current methods to measure and monitor quality metrics often embedded in unstructured text, such as DRE in prostate cancer patients, are frequently inefficient and burdensome, our approach is useful to multiple domains, such as prostate cancer, and it can accurately extract a high volume of features from clinical notes in the workflow. It has been shown in previous studies11,12,23,24 that NLP techniques have potential to extract prostate quality metrics hidden in unstructured clinical narratives; however, to our knowledge, there is no study which specifically focuses on DRE documentation.
We demonstrate that a rule-based approach can effectively capture the documentation and timing of exams from unstructured text in the EHR. In practice, NLP approaches can consist of rule-based and machine-learned frameworks25,26. In this study, since we were working in a very specific domain with small annotated corpus, we adapted a rule-based approach which has minimal set-up costs. Compared to a baseline NLP pipeline, we show that our proposed rule-based approach can enhance the efficiency and quality of data extraction. In addition, we created a terms-list based on usage patterns of words and phrases in our large corpus, which was confirmed as relevant by the domain experts. We believe automating this type of vocabulary creation and information extraction from clinical narratives may facilitate large scale text mining and data gathering tasks, and ultimately support efficient secondary use of routinely collected EHR data.
One of challenges in adapting an NLP system reported as the language diversity required extensive augmentation and revision to the NLP system’s dictionaries and rules.24 While ontologies, lexicons or domain-specific dictionaries might be used to deal with term variability, there is still a need for more comprehensive clinical ontologies.9 Recent studies showed that exploring word similarities using distributional semantics in order to create or expand terminologies might help to create broader learning ontologies.15 Therefore, in this study we calculated word similarities in order to create term lists. Since there is no previous collection of terms regarding categorization of DRE findings, we proposed a list of candidate terms for future detailed analysis.
Despite the apparent promising results from our system, our methods have several limitations. First, our algorithms have been developed and tested in a single academic center. However, the clinical terms used in our algorithms will be disseminated with a national repository (pheKB.org) and multiple clinicians have vetted the clinical terms used here. Future work will include testing our algorithms in another healthcare system to ensure their generalizability.
Conclusion
The overall goal of this study was to propose an approach to understand current documentation practices for DRE in the EHR for informing efforts to structure and standardize this important quality of care information for subsequent use. In this study, we demonstrate that with NLP techniques, it is feasible to accurately and efficiently identify and extract features associated with quality metrics (e.g. DRE) without increasing documentation burden for clinicians. As EHRs contain valuable information in unstructured format on this clinical assessment, it is important to capture and extract these data to produce precise, valid information to help assess medical practices that are considered important by clinicians in their daily routines.
References
- 1.Shoag J, Halpern JA, Lee DJ, et al. Decline in prostate cancer screening by primary care physicians: an analysis of trends in the use of digital rectal examination and prostate specific antigen testing. The Journal of urology. 2016;196(4):1047–1052. doi: 10.1016/j.juro.2016.03.171. [DOI] [PubMed] [Google Scholar]
- 2.Miller DC, Litwin MS, Sanda MG, et al. Use of quality indicators to evaluate the care of patients with localized prostate carcinoma. Cancer. 2003;97(6):1428–1435. doi: 10.1002/cncr.11216. [DOI] [PubMed] [Google Scholar]
- 3.Gori D DR, Blayney DW, Brooks JD, Fantini MP, McDonald KM, Hernandez-Boussard T. Jt Comm J Qual Patient Saf. 2018. Utilization of Prostate Cancer Quality Metrics for Research and Quality Improvement: a Systematic Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litwin MS, Steinberg M, Malin J, Naitoh J, McGuigan KA. 2000. Prostate cancer patient outcomes and choice of providers: development of an infrastructure for quality assessment RAND CORP SANTA MONICA CA. [Google Scholar]
- 5.Palmerola R, Smith P, Elliot V, et al. The digital rectal examination (DRE) remains important-outcomes from a contemporary cohort of men undergoing an initial 12-18 core prostate needle biopsy. The Canadian journal of urology. 2012;19(6):6542–6547. [PubMed] [Google Scholar]
- 6.Sayre EC, Bunting PS, Kopec JA. Reliability of self-report versus chart-based prostate cancer, PSA, DRE and urinary symptoms. The Canadian journal of urology. 2009;16(1):4463–4471. [PubMed] [Google Scholar]
- 7.Adler-Milstein J, DesRoches CM, Kralovec P, et al. Electronic health record adoption in US hospitals: progress continues, but challenges persist. Health Affairs. 2015;34(12):2174–2180. doi: 10.1377/hlthaff.2015.0992. [DOI] [PubMed] [Google Scholar]
- 8.Pan HY, Shaitelman SF, Perkins GH, Schlembach PJ, Woodward WA, Smith BD. Implementing a real-time electronic data capture system to improve clinical documentation in radiation oncology. Journal of the American College of Radiology. 2016;13(4):401–407. doi: 10.1016/j.jacr.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 9.Spasic I, Livsey J, Keane JA, Nenadic G. Text mining of cancer-related information: review of current status and future directions. International journal of medical informatics. 2014;83(9):605–623. doi: 10.1016/j.ijmedinf.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Thomas A, Zheng C, Jung H, et al. 83 Validity Of Natural Language Processing To Identify Patients With Prostate Cancer. The Journal of Urology. 2013;189(4):e34. [Google Scholar]
- 11.Gregg JR, Lang M, Wang LL, et al. Automating the Determination of Prostate Cancer Risk Strata From Electronic Medical Records. JCO Clinical Cancer Informatics. 2017;1:1–8. doi: 10.1200/CCI.16.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez-Boussard T, Kourdis P, Dulal R, et al. 2017. A natural language processing algorithm to measure quality prostate cancer care. In: American Society of Clinical Oncology. [Google Scholar]
- 13.Hong SN, Son HJ, Choi SK, et al. A prediction model for advanced colorectal neoplasia in an asymptomatic screening population. PloS one. 2017;12(8):e0181040. doi: 10.1371/journal.pone.0181040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu R, Guo J, Qin B, Che W, Wang H, Liu T. Learning semantic hierarchies via word embeddings. Paper presented at: Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers).2014. [Google Scholar]
- 15.Percha B, Zhang Y, Bozkurt S, Rubin D, Altman RB, Langlotz CP. Journal of the American Medical Informatics Association. 2018. Expanding a radiology lexicon using contextual patterns in radiology reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seneviratne M, Seto T, Hernandez-Boussard T. EGEMS (Wash DC) 2018. Architecture and implementation of a clinical research data warehouse for prostate cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musen MA, Noy NF, Shah NH, et al. The National Center for Biomedical Ontology. Journal of the American Medical Informatics Association : JAMIA. 2012;19(2):190–195. doi: 10.1136/amiajnl-2011-000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGregor S, Agres K, Purver M, Wiggins GA. From distributional semantics to conceptual spaces: A novel computational method for concept creation. Journal of Artificial General Intelligence. 2015;6(1):55–86. [Google Scholar]
- 19.Gupta A, Banerjee I, Rubin DL. Journal of biomedical informatics. 2018. Automatic Information Extraction from Unstructured Mammography Reports Using Distributed Semantics. [DOI] [PubMed] [Google Scholar]
- 20.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Paper presented at: Advances in neural information processing systems 2013. [Google Scholar]
- 21.Banerjee I, Chen MC, Lungren MP, Rubin DL. Radiology report annotation using intelligent word embeddings: Applied to multi-institutional chest CT cohort. Journal of biomedical informatics. 2018;77:11–20. doi: 10.1016/j.jbi.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. Journal of biomedical informatics. 2009;42(5):839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez-Boussard T, Kourdis PD, Seto T, et al. Mining Electronic Health Records to Extract Patient-Centered Outcomes Following Prostate Cancer Treatment. AMIA Annu Symp Proc. 2017;2017:876–882. [PMC free article] [PubMed] [Google Scholar]
- 24.Carrell DS, Schoen RE, Leffler DA, et al. Challenges in adapting existing clinical natural language processing systems to multiple, diverse health care settings. J Am Med Inform Assoc. 2017;24(5):986–991. doi: 10.1093/jamia/ocx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan WK, Hassanpour S, Heagerty PJ, et al. Acad Radiol. 2018. Comparison of Natural Language Processing Rules-based and Machine-learning Systems to Identify Lumbar Spine Imaging Findings Related to Low Back Pain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uzuner Ö, Stubbs A. Practical applications for natural language processing in clinical research: The 2014 i2b2/UTHealth shared tasks. J Biomed Inform. 2015;58(Suppl):S1–5. doi: 10.1016/j.jbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]