Abstract
Dosing errors due to erroneous body weight entry can be mitigated through algorithms designed to detect anomalies in weight patterns. To prepare for the development of a new algorithm for weight-entry error detection, we compared methods for detecting weight anomalies to human annotation, including a regression-based method employed in a real-time web service. Using a random sample of 4,000 growth charts, annotators identified clinically important anomalies with good inter-rater reliability. Performance of the three detection algorithms was variable, with the best performance from the algorithm that takes into account weights collected after the anomaly was recorded. All methods were highly specific, but positive predictive value ranged from < 5% to over 82%. There were 203 records of missed errors, but all of these were either due to no prior data points or errors too small to be clinically significant. This analysis illustrates the need for better weight-entry error detection algorithms.
Introduction
Ensuring the quality of electronic health record (EHR) data remains a significant challenge that prevents the full utilization of EHRs and puts patients at a higher risk of experiencing medication errors. Inaccurate body weight data is an important issue in pediatrics because of the high variability of pediatric growth, the frequent use of weight-based dosing, and the creation of latent weight errors propagated by EHRs. Evidence from previous studies indicates that 18-22% of pediatric medication errors result from “improper dose/quantity”1,2, a percentage that is significantly higher than in the adult setting3. In practice, body weight is usually measured by clinic staff via a weight scale that is not interfaced to EHR systems. Errors can occur when a scale is not operated properly or when weight data are inaccurately read and entered4. For example, a 30-pound child can be improperly dosed if the weight is recorded as “30 kg,” resulting in a 2.2-fold overdose.
Although weight errors can be detected through manual growth-chart review, this practice is inefficient and is not performed during busy clinic routines. Current normative alerting in EHRs can be rudimentary, insensitive, and nonspecific, which quickly leads users to ignore alerts about potentially anomalous weights. Therefore, there is a critical need to develop a computerized approach that detects pediatric weight errors with such a high sensitivity and specificity that a computer system can halt dangerous prescribing before it can do harm. This computerized algorithm could further be implemented as a clinical decision support (CDS) tool to assist clinicians in preventing weight entry errors and weight-based dosing errors in clinical routines.
With our goal to develop an advanced and novel machine learning algorithm to detect pediatric weight errors, it is critical to understand the characteristics of weight errors, or abnormal weight values in general, and train our machine learning model based on these findings. It is therefore important to collect a large amount of human-annotated weight data for the model to train on. In our previous work, we assessed the frequency of weight errors and developed a regression-based algorithm to detect such errors5,6. We also developed a visual annotation tool to facilitate the large collection of annotated weight errors7. Through our investigation, we found that identifying “true” weight errors through manual chart review and building error detection algorithms on them can limit the utility of such algorithms. Since our ultimate goal is to improve patient safety and prevent medication errors, it is imperative that the weight values that trigger review have high clinical importance, especially if the providers are going to review and act upon error notifications during busy workflows. As such, the success of our machine learning algorithm lies in its sensitivity and specificity to such “clinically important abnormal weight values.”
In the study, we aim to understand the characteristics of clinically important weight errors in order to provide a foundation for the development of our machine learning algorithm. Specifically, we are interested in learning: 1) what is the performance of existing weight error detection methods against these clinically important abnormal weight values, and 2) what are the characteristics of the patient growth charts and their abnormal weight values that cannot be captured by any of the existing methods?
Method
Data Collection
We compiled a set of weight values containing 3.8 million data points collected at Cincinnati Children’s Hospital Medical Center (CCHMC) since EHR implementation in 2010. This dataset was de-identified to remove any protected health information. The dataset contains the information regarding when the weight values were recorded (in terms of the age in days of the patient and the time of day to the minute, with no dates), which healthcare setting (login department of the user), who (provider type), and basic patient demographic information such as gender and age (in days of life). The use of the de-identified data to develop our annotation tool and machine learning algorithm was reviewed and approved by the University of Cincinnati Intuitional Review Board (UC IRB# 2017-2075).
Annotation
Since the abnormality and clinical importance of weight values are largely dependent on human judgement, it is necessary to collect the “reference standard” of such errors through human annotation. We enlisted domain experts with proper medical training and knowledge to annotate abnormal weight values that may cause dosing errors and significant patient harm and would warrant further review by providers in a clinical setting. To aid in this, we developed an annotation guideline for use on the dataset and invited three domain experts (including co-authors MN and SAS) to mark any abnormal weight values with a high likelihood of being inaccurate. The annotation process used for the dataset went through successive stages to minimize annotator variability. In the initial stage, the domain experts screened a convenience sample of 800 patient charts to develop the annotation guideline, which has two guiding principles: 1) weight values should be evaluated retrospectively and in aggregate, based on all available weights for a patient. 2) abnormal weight values were flagged as errors based on their clinical importance and risk for leading to patient harm.
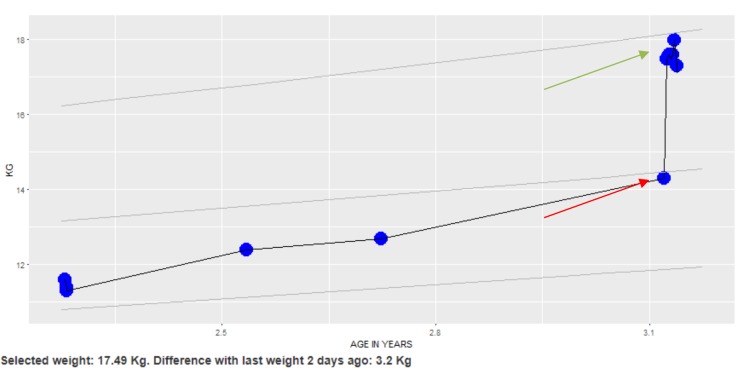
Specifically, abnormal weight values with high clinical importance were flagged as red (corresponding to the red arrow in Figure 1), while other abnormal weight values were flagged as orange. All weight values were considered normal and defaulted to blue. Figure 1 shows an example patient weight chart that was used for our annotation guideline implementation. In this example, the domain experts looked at the weight trend to determine that the upper weight value denoted by the green arrow was regarded as the correct weight and the weight value with the red pointer was likely an error (it is unlikely a patient would gain that much weight over a short time period).
Figure 1.
A weight chart display used to develop the annotation guideline. The trend allows one to distinguish that the upper cluster of points (green arrow) are likely correct, while the lower weight value (red arrow) is likely erroneous.
In the second stage, we randomly sampled 4,000 patients (age >= 24 months) with more than 50,000 weight values from the 3.8 million weight values. We excluded patients below 24 months to simplify the annotation tasks as newborns can have dramatic weight changes that are considered physiologically normal. This dataset was annotated by two of the three domain experts who are currently in their post-doc (medical school) training. These two annotators independently reviewed the 4,000 patient charts through our visual annotation tool and marked the color of a weight value to red or orange based on their perceived clinical importance. Weight values with annotator discrepancy were reviewed and reconciled by the third domain expert (SAS), who is an experienced clinical informatics physician- researcher. This annotated database then became the reference standard of the clinically important abnormal weight values for our subsequent experiments. The inter-rater reliability was calculated using Cohens Kappa8. Due to the fact that the majority of the recorded weight values are normal (more than 95%), we also reported the agreement based on the prevalence-adjusted and bias-adjusted kappa9,10. It is worth noting that the domain experts did not conduct manual chart reviews to identify the “true” weight errors in these 4,000 patient charts due to our focus on identifying clinically important weight errors.
Experiment
We adopted three weight error detection methods in our experiment to test the performance of each method against our reference standard of clinically important abnormal weight values. The first method is the modified weight-forage z-scores, for a child’s sex and age, based on the growth charts developed by the Centers for Decease Control and Prevention (CDC)11. This method has the default z-score range between -5 and 8, with the values outside of range being considered as abnormal weights. We used the SAS program released by the CDC to perform the calculation. The second method is an algorithm developed in our earlier study6, which uses regression to model change in weight based on age, gender, previous weight and time from previous weight and determines whether the current weight is considered to be an outlier. Our regression method (REG) is currently running in production as an EHR alert at Cincinnati Children’s hospital with additional rules to determine the alert timing.
The third method is developed by the researchers in the Children’s’ Hospital of Philadelphia (CHOP)12, which compares standard deviation scores of recorded values of weight against a weighted moving average for the child to determines “implausible” weight values based on the deviation between recorded and expected scores. The CHOP method aims to clean these implausible weight values for research purposes and has a parameter in their equation defaulted to 1.5. We downloaded the supplement code from the publication and extract the core calculation procedures to support our experiment. Of the various types of weight errors identified by the CHOP method, we allowed ‘duplicate’ (repeated) measurements on the same day and also allowed ‘carried forward’ (same value) weights within 90 days of prior measurement. To compare, the CDC method identifies weight errors only based on the difference between the current data point with its immediate previous one, while the CHOP and REG method consider all data points in a patient chart. We therefore expect that the CHOP and REG outperform the CDC method due to their considerations of overall weight trends. For another comparison, the CHOP method focuses on retrospective error detection and data cleaning for research, while our REG method has been implemented in clinical routines to perform real-time weight error detection although its performance is under evaluation.
Data Analysis
We scored the weights in the 4,000 patient charts using the three weight error detection methods, namely, the CDC, REG, and CHOP method. The performance of these methods was determined by the reference standard. We measured the performance in terms of sensitivity (true positive rate or recall), specificity (true negative rate), and positive predictive value (precision). We then extracted weight values that were considered by domain experts as errors but neglected by all three methods and perform a content analysis. We inspected these missed weight errors through our visual annotation tool and summarized their characteristics for future algorithm development.
Results
Table 1 shows the annotators’ agreement. All the marked weight values regardless of their clinical importance (red: high, orange: medium and low) are merged into the “Abnormal” category. This resulted in a total of 276 weight measurements (0.54%) classified as abnormal. Cohen’s Kappa shows that we achieved substantial agreement (Kappa=0.628). Since the majority (99.45%) of the weight values were considered “Normal” by both experts, we calculated the prevalence-adjusted and bias-adjusted kappa (PABAK=0.994) to justify the high annotator agreement.
Table 1.
Annotator agreement
| Expert 2 | ||||
|---|---|---|---|---|
| Normal | Abnormal | Total | ||
| Expert 1 | Normal | 50,680 (99.45%) | 31 (0.06%) | 50,711(99.51%) |
| Abnormal | 121 (0.24%) | 129 (0.25%) | 250 (0.49%) | |
| Total | 50,801 (99.69%) | 160 (0.31%) | 50,961 (100%) | |
As mentioned earlier, the disagreement among the two experts (a total of 152 records as shown in Table 1) was resolved by the third expert. This dataset then became the reference standard of our experiment. Table 2 shows the performance of the three selected methods against this reference standard. Here we have eight variations of the CDC method and five variations (models) of the CHOP method based on prescribed and tweaked values of the parameters in those methodologies. For example, CDC_n4p5 stands for the CDC method with the normal z-scores ranging from -4 to 5. Similarly, CHOP_20 stands for the CHOP method with the parameter P=2.0. The methods with the default parameter(s) are marked with an asterisk (*). From Table 2 we found that all these models had very high specificity, which is not surprising because of the large portion of the true negatives (>99%) in the dataset. However, all of their sensitivity scores are very low, with the lowest being 4.71% by the default CDC method and the highest being 18.84% by two CHOP models (parameter = 1.4 and 2.0). The positive predictive values (PPV, or precision) of the methods are at two extremes. CDC models had an overall PPV 8% or lower. The CHOP models achieved PPV slightly more than 80%. Our regression model (REG) performed much better than the CDC method but did not have the highest overall performance. CHOP_20 had the best performance although its precision did not exceed 90%. As expected, our regression method and the CHOP methods outperformed the CDC method in all measures. We believe this is because the REG and CHOP method consider the overall weight trend by incorporating multiple weight measurements gathered over time in their methodologies to identify abnormal values, rather than consider an individual weight measurement as in the CDC method. This observation is an important and essential consideration in developing an advanced machine learning algorithm-based approach.
Table 2.
Performance measures of the weight error detection methods
| Sensitivity (Recall) | Specificity | PPV (Precision) | |
| CDC_n4p5 | 11.59% | 98.35% | 3.68% |
| CDC_n4p6 | 9.78% | 98.97% | 4.90% |
| CDC_n4p7 | 9.06% | 99.24% | 6.10% |
| CDC_n4p8 | 7.97% | 99.40% | 6.79% |
| CDC_n5p5 | 8.33% | 98.66% | 3.27% |
| CDC_n5p6 | 6.52% | 99.28% | 4.69% |
| CDC_n5p7 | 5.80% | 99.55% | 6.58% |
| CDC_n5p8* | 4.71% | 99.72% | 8.28% |
| REG* | 13.77% | 99.90% | 43.18% |
| CHOP_10 | 18.48% | 99.98% | 80.95% |
| CHOP_14 | 18.84% | 99.98% | 81.25% |
| CHOP_15* | 18.48% | 99.98% | 80.95% |
| CHOP_16 | 18.48% | 99.98% | 82.26% |
| CHOP_20 | 18.84% | 99.98% | 82.54% |
Methods with default parameters.
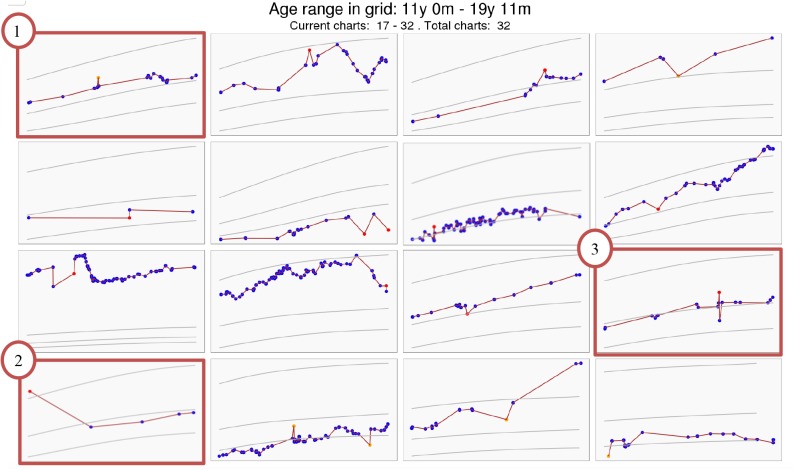
To inspect the weight errors not captured by all of the methods, we used the results of the models with default parameters (i.e. CDC_n5p8, REG, and CHOP_15). Other models were ignored in this process due to the insignificant difference of their performance. This selection led to a set of 203 missed weight errors in 163 patient charts that were not captured by any of the three models. These weight charts were extracted and displayed in our visual annotation tool as a set of 4 by 4 grids. Two experienced clinical informatics physician-researchers (ESK and SAS) examined patient charts and discussed the patterns. Through this discussion, we reassured the need to detect clinically important abnormal weight values and summarize three categories of abnormal weight values based on the content analysis. In the first category, the abnormal weight values had “minor change amidst dense data”. For example, looking at the orange point in the patient chart #1 at the top-left corner in Figure 2, it was clear to clinicians, due to the density of the data, that there was an anomaly in very temporally-close, but discrepant in magnitude weight values. We postulated that this type of abnormal weight value was not captured by the methods in our experiment simply because they did not pass a computational threshold. The second category is called “no prior data.” Chart #2 at the bottom left corner in Figure 2 shows that the likely error (first red point) is apparent because the annotators considered all weight values in a retrospective manner, but there is no preceding data to compare to and only temporally distant subsequent weight values. Lastly, the third category is termed “borderline cases,” where the abnormal weight values have unclear clinical importance and may or may not be clinically important depending on clinicians’ judgment. For example, the patient chart #3 on the far right of the third row in Figure 2 has a red dot clearly separated from its neighbors, but the weight value difference is so small that it is likely not to lead to clinically significant erroneous dosing in most contexts. However, the global trend of this chart suggests that it is well within the variability noted across the patient’s entire pattern.
Figure 2.
An example of clinically important abnormal weight values not captured by CDC, REG, and CHOP
Discussion
In this study, we focused on the clinical importance of weight errors and annotated 4,000 randomly sampled patient weight charts from a pediatric academic medical center. Our annotation achieved high inter-rater agreement. We selected three existing weight error detection methods and applied them to the annotated dataset. The results showed that none of the existing method achieved high sensitivity (or recall, 18% highest) and high positive predictive value (or precision, 82% highest). This highlights the need for a more sophisticated algorithm to capture this new type of weight errors. Our secondary content analysis summarizes three categories of errors that were not automatically captured by any of the three methods. From this content analysis we learned that the low sensitivity of the algorithms might result from the lower number of data points per chart or the longer time intervals between data points. We also learned that the overall trend of weight values would be a critical and decisive feature. However, determining the threshold of an error can be challenging because of the unclear relationship between clinical importance and mathematical significance. More research is needed to explore the features of these clinically important abnormal weight values.
Based on the current work, we plan to develop a more sophisticated system to detect weight errors, such as one based on machine learning methods. By summarizing weight dynamics with a variety of features (e.g., number of substantial increase/decrease, exceptions from average smoothing), we could apply traditional machine learning algorithms such as logistic regression and support vector machines to identify patient charts that have potential errors13,14. To identify errors for individual weight samples, we will leverage sequence labeling algorithms such as conditional random fields and recurrent neural networks15,16. We aim to design this algorithm based only on a patient’s gender, weight values, and age when the values are recorded to maximize its generalizability to other health organizations. If needed, we will extend the algorithms’ capacity in weight error detection by analyzing additional information such as the patient’s medical histories and medication usage.
This study has two limitations. First, the annotators did not consider the clinical importance of a weight value at the time when it was recorded. We have considered performing a simulation study on the temporality of these weight values. In this study, each patient charts will have multiple snapshots on different time points, and the annotators will mark the weight errors without seeing the subsequent values. However, doing so would significantly increase the number of charts to annotate and therefore is out of the scope of the current study. Second, we did not conduct manual chart reviews to determine whether an expert-annotated weight error is a true error or not. However, since our focus is on the clinical importance of weight errors, this would not change our findings and conclusion. It is for this reason that we called the annotation dataset as a reference standard rather than a gold standard throughout the manuscript.
Our future work involves collating a large dataset with expert-annotated weight errors with high clinical importance and exploring more patterns of such weight errors in addition to their overall trend. We will develop and evaluate multiple machine learning algorithms on this large annotation dataset and comparing their performance. We will also conduct evaluation studies to test our advanced algorithms on real-time and retrospect weight records. We are excited to continue this research topic and hope to develop a generalizable tool to help clinicians improve their care quality as well as patient safety.
Conclusion
We compared three existing methods to detect weight data errors. The CHOP method with a tuned parameter performed the best in our experiment. However, since the CHOP method is designed for retrospective dataset cleaning, its performance of real-time weight error detection is unknown. On the other hand, the performance of our REG method has room for improvement. Its precision needs to achieve at least 90% to be useful as an alert for clinical decision support. Currently, our REG method is only deployed and used at the pharmacist order verification level. Our future work aims to use the training sets created in the current study as the basis to develop an advanced machine- learning algorithm for weigh-entry error detection.
Acknowledgement
We thank Pieter-Jan van Camp, MD, a current PhD student in the Department of Biomedical Informatics at the University of Cincinnati, for creating the visual annotation tool and helping the data annotation. This study was supported by the first author’s startup fund at the University of Cincinnati and had no other funding support when submitting the manuscript.
References
- 1.Pham JC, Story JL, Hicks RW, et al. National study on the frequency types, causes, and consequences of voluntarily reported emergency department medication errors. J Emerg Med. 2011;40(5):485–492. doi: 10.1016/j.jemermed.2008.02.059. doi:10.1016/j.jemermed.2008.02.059. [DOI] [PubMed] [Google Scholar]
- 2.Rinke ML, Shore AD, Morlock L, Hicks RW, Miller MR. Characteristics of pediatric chemotherapy medication errors in a national error reporting database. Cancer. 2007;110(1):186–195. doi: 10.1002/cncr.22742. doi:10.1002/cncr.22742. [DOI] [PubMed] [Google Scholar]
- 3.Bates DW, Boyle DL, Vander Vliet MB, Schneider J, Leape L. Relationship between medication errors and adverse drug events. J Gen Intern Med. 1995;10(4):199–205. doi: 10.1007/BF02600255. [DOI] [PubMed] [Google Scholar]
- 4.Evans L, Best C. Accurate assessment patient weigh. Nurs Times. 2014;110(12):12–14. [PubMed] [Google Scholar]
- 5.Hagedorn PA, Kirkendall ES, Kouril M, et al. Assessing Frequency and Risk of Weight Entry Errors in Pediatrics. JAMA Pediatr. 2017;171(4):392–393. doi: 10.1001/jamapediatrics.2016.3865. doi:10.1001/jamapediatrics.2016.3865. [DOI] [PubMed] [Google Scholar]
- 6.Spooner S, Shields S, Dexheimer J, Mahdi C, Hagedorn P, Minich T. October 2016. Weight Entry Error Detection: A Web Service for Real-time Statistical Analysis. [Google Scholar]
- 7.Van Camp P, Newcomb M, Kirkendall E, Spooner SA, Wu DTY. May 2018. Developing a Visual Annotation Tool to Rapidly Collect Expert-Annotated Weight Errors in Pediatric Growth Charts. [DOI] [PubMed] [Google Scholar]
- 8.McHugh ML. Interrater reliability: the kappa statistic. Biochem Medica. 2012;22(3):276–282. [PMC free article] [PubMed] [Google Scholar]
- 9.Byrt T, Bishop J, Carlin JB. Bias, prevalence and kappa. J Clin Epidemiol. 1993;46(5):423–429. doi: 10.1016/0895-4356(93)90018-v. [DOI] [PubMed] [Google Scholar]
- 10.Chen G, Faris P, Hemmelgarn B, Walker RL, Quan H. Measuring agreement of administrative data with chart data using prevalence unadjusted and adjusted kappa. BMC Med Res Methodol. 2009;9:5. doi: 10.1186/1471-2288-9-5. doi:10.1186/1471-2288-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A SAS Program for the 2000 CDC Growth Charts (ages 0 to < 20 years) https://www.cdc.gov/nccdphp/dnpao/growthcharts/resources/sas.htm. Accessed March 6, 2018.
- 12.Daymont C, Ross ME, Russell Localio A, Fiks AG, Wasserman RC, Grundmeier RW. Automated identification of implausible values in growth data from pediatric electronic health records. J Am Med Inform Assoc JAMIA. 2017;24(6):1080–1087. doi: 10.1093/jamia/ocx037. doi:10.1093/jamia/ocx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shawe-Taylor J, Cristianini N. Kernel methods for pattern analysis. Cambridge, UK; New York: Cambridge University Press; 2004. [Google Scholar]
- 14.Bishop CM. Pattern recognition and machine learning. New York: Springer; 2006. [Google Scholar]
- 15.Jain LC. Recurrent neural networks: design and applications. Boca Raton FL: CRC Press; 2000. ed. [Google Scholar]
- 16.Lafferty J, McCallum A, Pereira F. 2001. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In Morgan Kaufmann. [Google Scholar]